Introduction
Prostate cancer (PCa) is the most common malignancy
in males in western countries and the second leading cause of
cancer-associated mortality (1).
Long-range epigenetic activation (2) and long-range epigenetic silencing
(3) have been discovered in the
genome of PCa cells, which indicated there were structural
re-arrangements in PCa. Post-translational histone modifications,
which represent a classic epigenetic aberration, have been shown to
participate in PCa metastasis and androgen-independent transition.
A study on castration-resistant PCa patients revealed that the
blockade of histone deacetylases re-sensitized PCa cells to
androgen ablation therapy (4).
Histone methylation, controlled by histone methyltransferases
(HMTs) and histone demethylases (HDMs), was also proved to
participate in PCa metastasis and androgen-independent transition.
HMTs, such as CAM1 and EZH2, were found to be over-expressed in
PCa, with EZH2 being regarded as a co-activator of the androgen
receptor (AR), showing oncogenic activities in PCa (5). Following the discovery of
lysine-specific histone demethylase 1A in 2004 (6), histone methylation was initially
thought to be irreversible, while its interaction with the AR in
vitro and in vivo and its promotion of AR-dependent
transcription was demonstrated in 2005 (7). It has been indicated that histone
modifications are involved in the regulation of early cell fate
decisions, cell differentiation and tissue development, and are
also linked with numerous of diseases, particularly cancer
(4). HMTs and HDMs participate in
a variety of processes which are aberrant in cancers, including DNA
damage repair, cell replication and apoptosis (4,5).
Altered expression of HMTs and HDMs has been reported to be
associated with clinical and pathological outcomes of cancer and to
be implicated in the formation of androgen-independent PCa,
indicating that these enzymes may represent promising therapeutic
targets for PCa.
However, the functions of HMTs and HDTs in cancers
as well as the underlying molecular mechanisms have largely
remained elusive. The present study performed an RNA interference
screening using a lentivirus-directed small hairpin (sh) RNA
library targeting all human HMTs and HDTs in order to
systematically elucidate the function of HMTs and HDTs in PCa cell
growth and viability. PRDM16 was identified to be associated with
the evasion of PCa cells from apoptosis, and its spliced form,
sPRDM16, was found to be aberrantly expressed in PCa cells.
Materials and methods
Cell culture, antibodies and shRNA
library
The RWPE-1 prostate epithelial (PR) cell line, the
BPH-1 benign prostate hyperplasia (BPH) cell line and the DU145,
PC-3, PC-3M and LNCaP PCa cell lines were purchased from the
American Type Culture Collection (Manassas, VA, USA) and maintained
at 37°C in a humidified atmosphere containing 5% CO2.
The cell culture media of DU145, PC-3, PC-3M, LNCaP, RWPE1, and BPH
cell lines was RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The fetal bovine serum was purchased by Gibco
(Thermo Fisher Scientific, Inc). Antibodies against PRDM16 (cat.
no. ab106410; 1:500), B-cell lymphoma 2 (Bcl-2; cat. no. ab182858;
1:1,000), Bcl-2 homologous antagonist killer (Bak; cat. no.
ab32371; 1:1,000), cleaved caspase-3 (cat. no. ab13847; 1:500) and
glyceralde-hyde-3-phosphate dehydrogenase (GAPDH; cat. no.
ab181602; 1:5,000) were purchased from Abcam (Cambridge, MA, USA).
All the antibodies were polyclonal raised in rabbit. The customized
lentiviral-mediated shRNA library targeting 88 HMTs and HDMs was
purchased from 3D-HTS (Shanghai, China). Each gene was targeted by
two pools of four distinct shRNA species with specificity for
different sequences of the target transcript.
RNA interference (RNAi) screening
DU145 cells (were plated in 96-well plates at 3,000
per well and transfected using a virus concentration of MOI=30,
with the shRNA library for 24 h. An
3-(4,5-dimethylthiazol-2-yl)-5-(3-carbo
xymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay was
then performed as described below. Cells were screened in
independent triplicates. Four individual shRNA species which
targeted each gene were then assessed in order to validate and
narrow down the results of the primary screening. RNAi-mediated
gene silencing was then confirmed by reverse-transcription
quantitative polymerase chain reaction (RT-qPCR) and western blot
analysis.
RT-qPCR
DU145 cells were seeded into six-well plates and
transfected with shRNA for 96 h. TRIzol was used to extract the
total RNA and complementary DNA was generated using a ReverTra Ace
qPCR RT kit (Toyobo Co., Ltd., Osaka, Japan). A SYBR Green PCR kit
(Takara Bio Inc., Otsu, Japan) was then used to amplify the cDNA by
PCR. The thermocycling conditions used were, pre-denaturation at
94°C for 5 min, 30 cycles of denaturation at 94°C for 30 sec,
annealing at 56°C for 30 sec and extension at 72°C for 30 sec,
final step at 72°C for 20 min. The primers were purchased from
Dingguo Changsheng Biotechnology, Co., Ltd. (Beijing, China) and
the following sequences were used: PRDM16, forward (F)
5′-TTCTCTGGACGCTTGGTTGA-3′ and reverse (R)
5′-GAGGCCCTAGAGGTGGTTGAT-3′; PRDM16-L, F 5′-CAAGGAGGAGGAGAGAGATT-3′
and R 5′-CGGTTGGGCTCATACATA-3′; and sPRDM16 F
5′-TGCACACCCAGCAACACC-3′and R 5′-GCTGCGCTAGAGAAAAGCGT-3′. The
expression of GAPDH was also evaluated to calculate the relative
target gene expression. The 2−ΔΔCq method was used for
quantification (8).
Western blot analysis
Cells cultured in six-well plates were lysed with
300 µl sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) lysis buffer. The bicinchoninic acid
protein assay (OriGene Technologies, Inc., Beijing, China) was used
to evaluate the protein concentration. A total of 30 µg
protein of each sample was subjected to 8% SDS-PAGE (OriGene
Technologies, Inc.) and then transferred onto polyvinylidene
difluoride membranes. Membranes were then blocked in 5% non-fat
milk in Tris-buffered saline/Tween 20 for 1 h at 37°C, followed by
incubation with the indicated antibodies for 1 h at 37°C.
Subsequently, the membranes were incubated with horseradish
peroxidase-conjugated secondary antibody (cat. no. ab6721; 1:300),
and blots were visual-ized using enhanced chemiluminescence reagent
(OriGene Technologies, Inc.).
Cell proliferation assay
Following transfection of DU145 cells with the shRNA
library as described above, the transfection medium in each well
was replaced with 100 µl cell culture medium. 20 µl
One Solution Reagent (Promega Corp., Madison, WI, USA) containing
19 µl MTS and 1 µl phenazine methosulfate was added
into each well, followed by incubation at 37°C for 1 h. The optical
density value of the supernatant was then measured at 490 nm using
a spectrophotometer.
Flow cytometry
DU145 cells were collected and washed with
phosphate-buffered saline (PBS) following 120 h of transfection.
They were centrifuged at 400 × g and washed twice. The cells were
seeded at a density of 1.0×106 cells/ml and 1X
Annexin-binding buffer was added. Alexa Fluor 488 Annexin V
(OriGene Technologies, Inc., Beijing, China) and 100 µg/ml
propidium iodide working solution were added to each 100 µl
of cell suspension. The cells were incubated at room temperature
for 15 min. Next, 400 µl 1X Annexin-binding buffer was added
and mixed gently. The fluorescence emission was measured at 530 nm
using a flow cytometer (FC 500 MCL/MPL, Beckman Coulter, Inc.,
Brea, CA, USA).
Statistical analysis
SPSS version 13.0 (SPSS, Inc., Chicago, IL, USA) and
GraphPad Prism 5 (GraphPad Software Inc., California, USA) were
used for the statistical analyses. Z-score and a Student's t-test
were performed. P<0.05 was considered to indicate a
statistically significant difference between values.
Results
RNAi screening for HMTs and HDMs
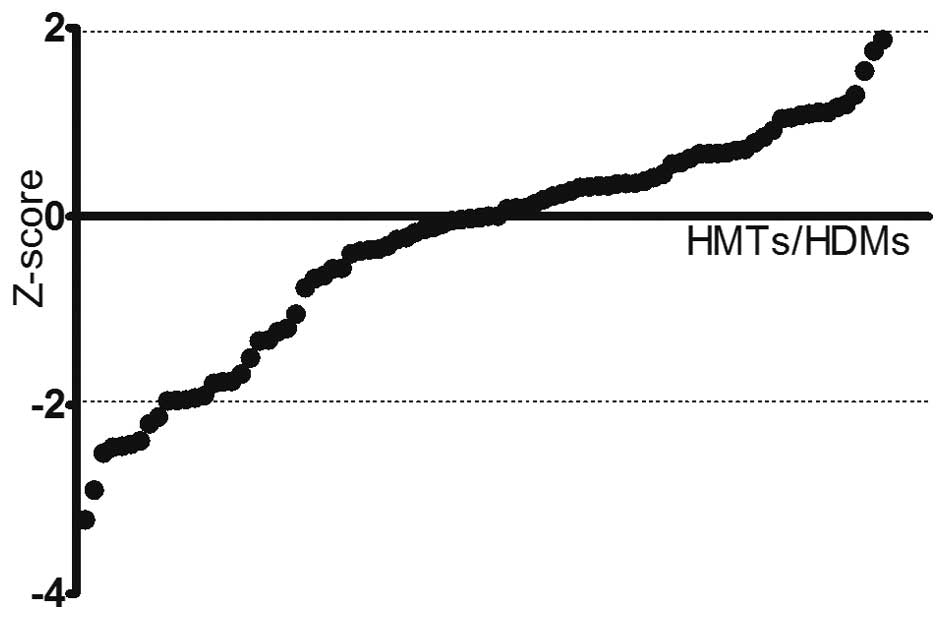
associated with PCa cell viability
To identify HMTs and HDMs implicated in PCa cell
viability, a systematic RNAi-mediated screening assay was
performed. DU145 cells were transfected with an array of 176 shRNAs
targeting a total of 88 HMTs and HDMs. After 24 h of transfection,
the cell viability in each well was evaluated to assess the effects
of each pool of shRNAs on the cell growth. Pools were classified as
hits if the absolute value of the Z-score was >1.96 compared to
the negative control (Fig. 1). A
total of nine genes were identified by this screen with a bottom
Z-score of −2.217 (PRDM16) and a top Z-score of −3.211 (FBXO11)
(Table I).
 | Table IGenes identified to be associated with
DU145 cell viability in the RNA interference screening. |
Table I
Genes identified to be associated with
DU145 cell viability in the RNA interference screening.
| Gene | FBXO11 | PRDM10 | JMJD8 | MLL | SETD4 | JMJD7 | PRMT2 | MEN1 | PRDM16 |
|---|
| Z-score | −3.221 | −2.903 | −2.509 | −2.446 | −2.435 | −2.415 | −2.380 | −2.202 | −2.127 |
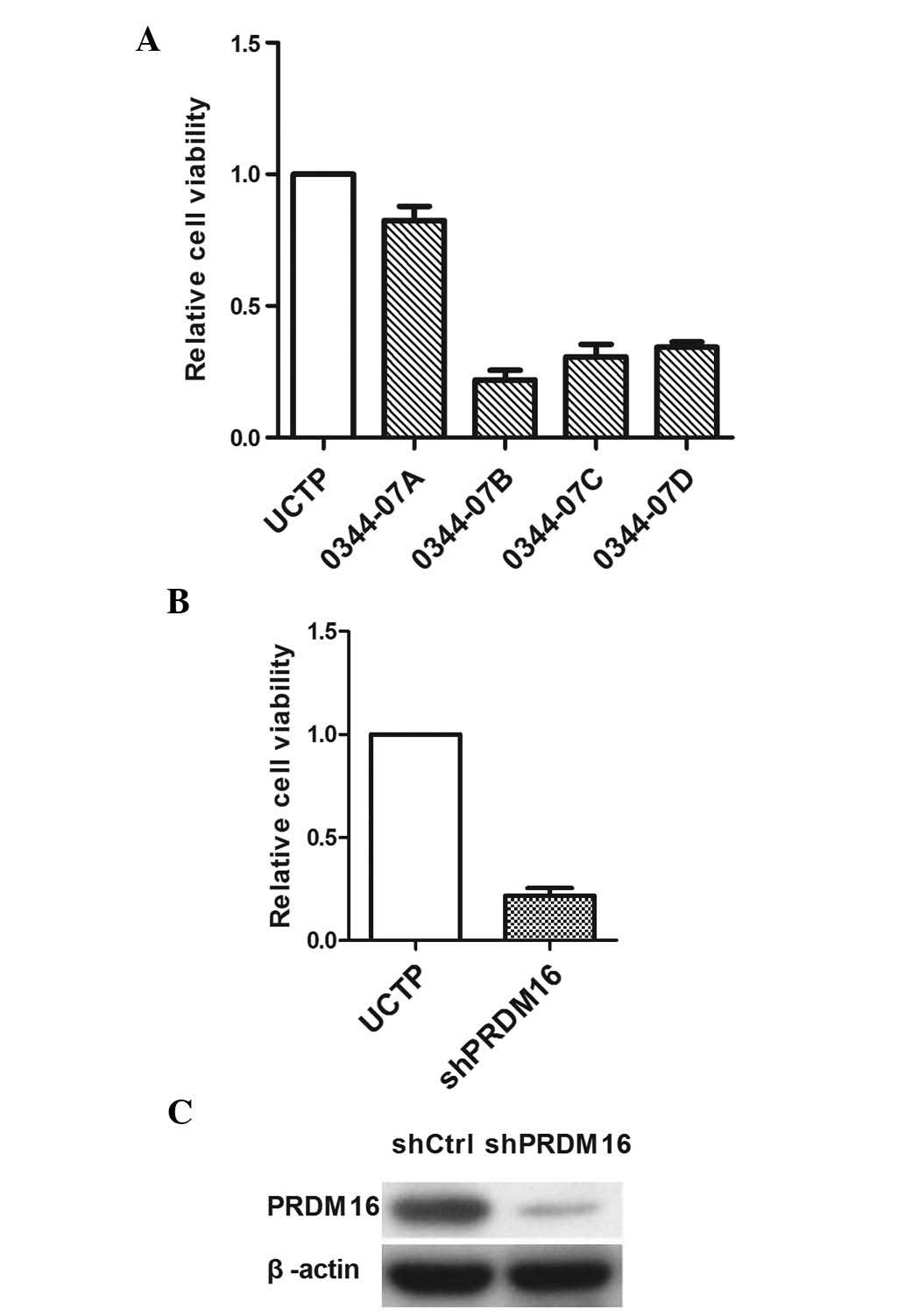
PRDM16 inhibition decreases DU145 cell
viability
The present study next evaluated the effects of
PRDM16 inhibition on the viability of DU145 cells and explored its
implication in apoptosis. To exclude the possibility that the
inhibition of DU145 viability was induced by the knockdown
experiments due to off-target effects, each well of DU145 cells was
transfected with one of four different shRNA lentiviral vectors. As
three of the four lentiviral vectors reduced the cell viability by
>70% (Fig. 2), the presence of
an off-target effect could be excluded. The results revealed that
0034-07D exerted the highest inhibitory effect on DU145 cells;
therefore, this shRNA was used in all subsequent experiments. PCR
and western blot analyses confirmed a reduction of PRDM16
expression by 0034-07D at the mRNA and protein level.
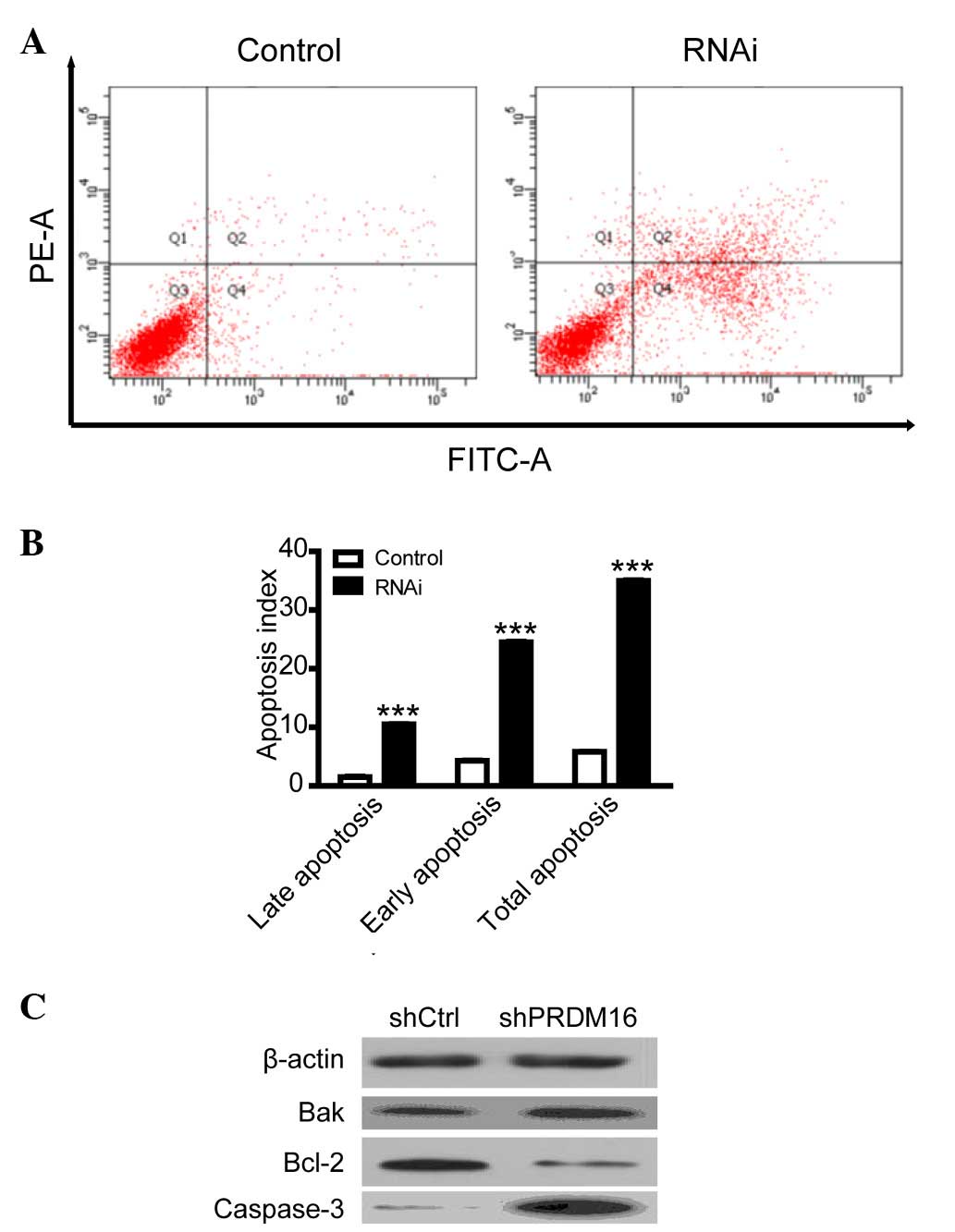
PRDM16 inhibition upregulates DU145 cell
apoptosis
To explore the mechanism by which PRDM16 regulates
cell viability, flow cytometric analysis was used to evaluate the
apoptotic rate of DU145 cells. The results indicated that
inhibition of PRDM16 induced DU145 cell apoptosis (Fig. 3); suggesting that PRDM16 is
involved in the evasion of apoptosis of PCa cells, while the study
of its effects on cell proliferation exceeded the margin of the
present study. To explore the mechanism by which PRDM16 regulates
apoptosis, the expression of several apoptotic proteins was
evaluated using western blot analysis. The results indicated that
the increase of apoptosis after PRDM16 inhibition was parallelled
with downregulation of the expression of the anti-apoptotic Bcl-2
and upregulation of the pro-apoptotic Bak and cleaved
caspase-3.
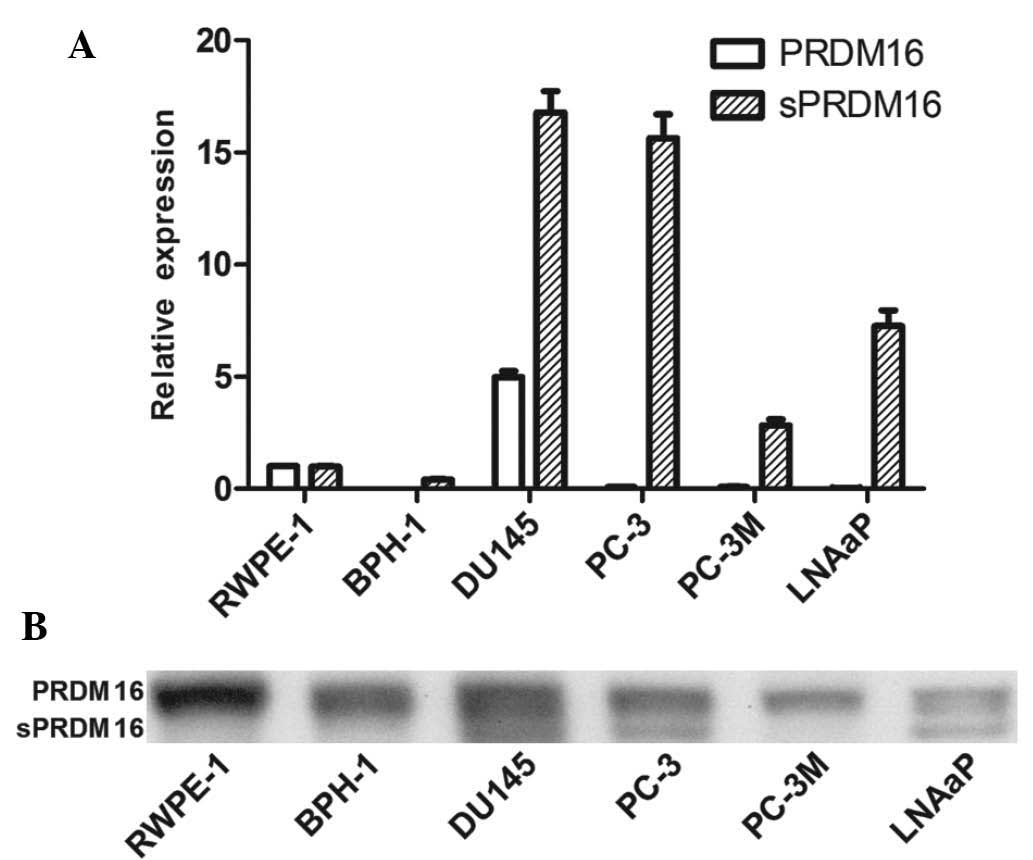
sPRDM16/MEL1S has an oncogenic role in
PCa
Next, the present study evaluated the expression of
PRDM16/MEL1 and its spliced version, sPRDM16/MEL1S, in the RWPE-1
PE cell line, the BPH-1 BPH cell line and four prostate cancer cell
lines, DU145, PC-3, PC-3M and LNCaP (Fig. 4). RT-qPCR showed that PRDM16/MEL1
levels were decreased in most PCa cell lines, while sPRDM16/MEL1S
was enhanced compared with that in the PE cell line. Furthermore,
western blot analysis showed that PRDM16/MEL1 was present in each
cell line, while sPRDM16/MEL1S was only detected in three out of
four PCa cell lines (DU145, PC-3 and LNCaP), which indicated an
oncogenic role of sPRDM16/MEL1S in PCa.
Discussion
To the best of our knowledge, the present study was
the first to use a RNAi screening-based approach to systematically
assess the effects of all human HMTs and HDTs on PCa cell
viability. By using this approach, nine genes associated with PCa
cell viability were identified, of which FBOX11, PRMT2, MEN1, MLL,
PRDM16 and SETD4 have already been reported to be abnormally
expressed in various cancer types (9–14).
The PRDM16 gene is rearranged in AML and MDS (13). Approximately 25% of primary
prostate tumors that progress to metastasis have gained this at the
MEN1 locus (11). SETD4 was
important for breast carcinogenesis and may be a novel molecular
target for the diagnosis and treatment of breast carcinoma
(14). While JMJD7 and -8 were
identified by the screening of the present study, JMJD2 family
proteins are well known to be associated with a variety of cancer
types, including PCa (15). Among
the nine genes identified by the present study, MEN1 has been
previously reported to be involved in the tumorigenesis of PCa
(11), which indicated high
accuracy of the screening performed and suggested that RNAi
screening is a powerful tool to identify genes involved in
cancer.
Furthermore, the present study revealed that PRDM16
inhibition decreased DU145 cell viability by increasing the
apoptotic rate. To explore the underlying mechanisms by which
PRDM16 regulates apoptosis, the expression of several apoptotic
genes was evaluated using western blot analysis. The results
revealed that RNAi of PRDM16 induced apoptosis by reducing the
expression of Bcl-2 and by upregulating the expression of Bak.
Furthermore, the induction of apoptosis was associated with
activation of caspase-3 by its cleavage.
PRDM16 is a member of the family of PR
domain-containing proteins, which consists of 17 members and is
characterized by the PR domain with a variable number of Zn-fingers
(16). They function as regulators
of chromatin function and transcription factors which are involved
in the determination of cell fate. PRDM16 serves important
functions in adipose tissue differentiation, and has also been
described as an oncoprotein in myelodysplastic syndrome, acute
myelocytic leukemia, adult T-cell leukemia and gastric carcinoma
(13,17,18).
sPRDM16, an alternatively spliced form of PRDM16 lacking the PR
domain, was described to be aberrantly expressed in AML and gastric
carcinoma. sPRDM16, but not PRDM16, was able to prevent
TGF-β-induced growth inhibition in mouse T cells (17,18).
The present study provided the first evidence that sPRDM16 is
aberrantly expressed in PCa cell lines (DU145, PC-3 and LNCaP),
while its expression could not be detected in the RWPE-1 PE cell
line and the BPH-1 BPH cell line. These results suggested that
sPRDM16 may have an oncogenic function in PCa and may represent a
novel therapeutic target.
The screening approach of the present study provided
nine genes involved in the regulation of PCa cell viability. The
detailed results on PRDM16, and previous results on MEN1 (11), which were among these nine genes,
indicate that the other seven genes may also have oncogenic roles
in PCa, which requires further study. As BOX11, PRMT2, MLL and
SETD4 have been previously indicated to be linked with other cancer
types, their association with PCa is likely (8,9,11,13).
Furthermore, JMJD7 and -8 and PRDM10 are members of the JMJD and
PRDM family, respectively indicating similar functions to those of
JMJD2 (15) and PRDM16.
It has been previously suggested that histone
methylation, controlled by HMTs and HDMs, occurs during PCa
metastasis and androgen-independent transition. The present study
indicated that nine HMT/HDM genes are involved in the regulation of
PCa cell viability, and that the inhibition of PRDM16 decreased
DU145-cell viability via enhancement of the apoptotic rate.
sPRDM16, an alternatively spliced form of PRDM16 lacking the PR
domain, was identified to be overexpressed in three out of four PCa
cell lines. These results suggested that sPRDM16 has an oncogenic
role in PCa. While MEN1 has been previously reported to be
associated with PCa, the likely oncogenic roles of the other seven
genes, which have been partly indicated in other types of cancer,
remain to be examined by further studies.
Acknowledgments
The present study was funded by a project supported
by the Province-Ministry Incubation Program (no. 2014PYA004),
Appropriate Technical Transformation of Zhejiang province (nos.
2013ZHB001 and 2014ZHB001) and the Natural Science Funds Of
Zhejiang Province (no. LY16H160034).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bert SA, Robinson MD, Strbenac D, Statham
AL, Song JZ, Hulf T, Sutherland RL, Coolen MW, Stirzaker C and
Clark SJ: Regional activation of the cancer genome by long-range
epigenetic remodeling. Cancer Cell. 23:9–22. 2013. View Article : Google Scholar
|
|
3
|
Coolen MW, Stirzaker C, Song JZ, Statham
AL, Kassir Z, Moreno CS, Young AN, Varma V, Speed TP, Cowley M, et
al: Consolidation of the cancer genome into domains of repressive
chromatin by long-range epigenetic silencing (LRES) reduces
transcriptional plasticity. Nat Cell Biol. 12:235–246.
2010.PubMed/NCBI
|
|
4
|
Crea F, Sun L, Mai A, Chiang YT, Farrar
WL, Danesi R and Helgason CD: The emerging role of histone lysine
demethylases in prostate cancer. Mol Cancer. 11:522012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Albert M and Helin K: Histone
methyltransferases in cancer. Semin Cell Dev Biol. 21:209–220.
2010. View Article : Google Scholar
|
|
6
|
Shi Y, Lan F, Matson C, et al: Histone
Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1.
Cell. 119:941–953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Metzger E, Wissmann M, Yin N, Müller JM,
Schneider R, Peters AH, Günther T, Buettner R and Schüle R: LSD1
demethylates repressive histone marks to promote
androgen-receptor-dependent transcription. Nature. 437:436–439.
2005.PubMed/NCBI
|
|
8
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
9
|
Duan S, Cermak L, Pagan JK, Rossi M,
Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R and Pagano
M: FBXO11 targets BCL6 for degradation and is inactivated in
diffuse large B-cell lymphomas. Nature. 481:90–93. 2012. View Article : Google Scholar :
|
|
10
|
Baldwin RM, Morettin A, Paris G, Goulet I
and Côté J: Alternatively spliced protein arginine
methyltransferase 1 isoform PRMT1v2 promotes the survival and
invasiveness of breast cancer cells. Cell Cycle. 11:4597–4612.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Paris PL, Sridharan S, Hittelman AB,
Kobayashi Y, Perner S, Huang G, Simko J, Carroll P, Rubin MA and
Collins C: An oncogenic role for the multiple endocrine neoplasia
type 1 gene in prostate cancer. Prostate Cancer Prostatic Dis.
12:184–191. 2009. View Article : Google Scholar
|
|
12
|
Angelova S, Jordanova M, Spassov B,
Shivarov V, Simeonova M, Christov I, Angelova P, Alexandrova K,
Stoimenov A, Nikolova V, et al: Amplification of c-MYC and MLL
genes as a marker of clonal cell progression in patients with
myeloid malignancy and trisomy of chromosomes 8 or 11. Balkan J Med
Genet. 14:17–24. 2011.PubMed/NCBI
|
|
13
|
Duhoux FP, Ameye G, Montano-Almendras CP,
et al: PRDM16 (1p36) translocations define a distinct entity of
myeloid malignancies with poor prognosis but may also occur in
lymphoid malignancies. Br J Haematol. 156:76–88. 2012. View Article : Google Scholar
|
|
14
|
Faria JA, Corrêa NC, de Andrade C, et al:
SET domain-containing protein 4 (SETD4) is a newly identified
cytosolic and nuclear lysine methyltransferase involved in breast
cancer cell proliferation. J Cancer Sci Ther. 5:58–65.
2013.PubMed/NCBI
|
|
15
|
Berry WL and Janknecht R: KDM4/JMJD2
histone demethylases: Epigenetic regulators in cancer cells. Cancer
Res. 73:2936–2942. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pinheiro I, Margueron R, Shukeir N, Eisold
M, Fritzsch C, Richter FM, Mittler G, Genoud C, Goyama S, Kurokawa
M, et al: Prdm3 and Prdm16 are H3K9me1 methyltransferases required
for mammalian heterochromatin integrity. Cell. 150:948–960. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoshida M, Nosaka K, Yasunaga J, Nishikata
I, Morishita K and Matsuoka M: Aberrant expression of the MEL1S
gene identified in association with hypomethylation in adult T-cell
leukemia cells. Blood. 103:2753–2760. 2004. View Article : Google Scholar
|
|
18
|
Takahata M, Inoue Y, Tsuda H, Imoto I,
Koinuma D, Hayashi M, Ichikura T, Yamori T, Nagasaki K, Yoshida M,
et al: SKI and MEL1 cooperate to inhibit transforming growth
factor-beta signal in gastric cancer cells. J Biol Chem.
284:3334–3344. 2009. View Article : Google Scholar
|