Introduction
Esophageal carcinoma is a common type of cancer of
the digestive system, with an estimated 456,000 new cases and
400,000 cases of mortality worldwide during 2012, and with 50% of
all cases occurring in China alone (1,2). In
patients receiving early comprehensive treatment, the 5-year
relative survival rate of esophageal carcinoma approaches 90%.
However, the overall 5-year survival rate of patients with
esophageal carcinoma remains poor as it is most commonly diagnosed
at an advanced stage (with lymph node metastases) and only few
symptoms appear at an early stage (3,4).
Therefore, there is an urgent requirement for novel and reliable
biomarkers that facilitate the detection of preneoplastic
esophageal carcinoma lesions.
Typically, microRNA (miRNA) genes are transcribed
under the regulation of a promoter and an operon. Certain miRNAs
are positioned proximal to each other on the chromosome and form a
cluster (5–7). These clusters are considered to be
transcribed simultaneously, but may mediate synergistic or
antagonistic regulatory functions (8,9). An
increasing number of studies have demonstrated that miRNA clusters
serve important roles in certain biological processes and during
oncogenesis (10–13). A few well-studied miRNA clusters
include the miR-17/92, miR-221/222 and miR-1/133a clusters
(14–16). Additional studies have demonstrated
that miRNA clusters function more efficiently compared with
individual miRNAs alone. For instance, Li et al (17) demonstrated that miR-424 and
miR-503, forming the miR-424-503 cluster, cooperate to inhibit the
expression of Smad7 and Smurf2, thus increasing the transforming
growth factor-β signaling and the metastatic potential of breast
cancer cells. In addition, Wystub et al (18) observed that miR-1 and miR-133a,
forming the miR-1/133a cluster, cooperate to modulate the
cardiomyogenic lineage by suppressing the expression levels of
MYOCD and KCNMB1 genes, respectively. Previous miRNA profiling data
demonstrated that two members of the miR-144/451 cluster, namely
miR-144-3p and miR-451a, were significantly downregulated in tumor
tissues (P=0.039 and 0.005, respectively) compared with adjacent
non-tumor tissues (19).
Furthermore, studies have provided evidence to suggest that
miR-144-3p and miR-451a may serve tumor suppressive or oncogenic
roles in different tissues (16,20,21).
The majority of studies investigating the
miR-144/451 cluster have focused on elucidating the function of the
individual miRNAs, but not the cluster as a whole. In addition,
there is currently no detailed information about the miR-144/451
cluster in esophageal carcinoma (22–24).
It is possible that individual miRNAs within the miR-144/451
cluster may mediate opposing or similar functions as part of
complex regulatory networks. Clustered miRNAs appear to be more
stable and reliable than individual miRNAs as diagnostic biomarkers
(18,19,25).
In addition, detailed information on the expression levels and
functional roles of the miR-144/451 cluster may facilitate an
improved understanding of the mechanisms involved in tumor
development and maintenance, which may also be beneficial for
therapy. Promoting or suppressing the expression of miRNAs in the
miR-144/451 cluster may be a promising therapeutic strategy, with
high efficiency and low treatment resistance, for patients with
esophageal carcinoma.
In the present study, the expression levels of
miR-144/451 cluster members in esophageal carcinoma tissues were
examined, and the correlation between the expression levels of
individual miRNAs were determined. To the best of our knowledge,
this is the first study that has reported the expression levels of
hsa-miR-144-3p, hsa-miR-144-5p, hsa-miR-451a, hsa-miR-4732-3p and
hsa-miR-4732-5p, which comprise the miR-144/451 cluster, in
esophageal carcinoma. Pearson correlation analyses were performed
to evaluate the association between the expression levels of all
five miRNAs. The association between abnormal expression of the
miR-144-451 cluster and the risk of esophageal carcinoma was
further analyzed using principal component regression analysis. The
possible targets and functions of the miR-144-451 cluster were
analyzed by bioinformatics. The results of the present study may
advance our understanding of the expression pattern and functional
role of the miR-144/451 cluster in esophageal carcinoma.
Materials and methods
Study subjects
Samples from 102 patients with esophageal carcinoma,
diagnosed by pathological analyses (26) and recruited from the First People's
Hospital of Huaian (Huaian, China), were used in the present study.
Among these patients, 73 (71.6%) cases were male and 29 (28.4%)
were female, with an age range of 44–78 years and a mean age of
63.17±7.25 years. Esophageal carcinoma tissues and their
corresponding normal non-tumor tissues were collected surgically
between 2011 and 2012, and stored in tubes at −80°C. Written
informed consent was obtained from all subjects prior to
recruitment to the study. Ethical approval was provided by the
Institutional Review Board of the Southeast University-Affiliated
Zhongda Hospital (Nanjing, China).
RNA isolation and analysis
Total RNA was extracted from the tissue samples
using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's
instructions. The quality and concentration of the extracted RNA
was assessed with the 260/280 absorbance ratio using the NanoDrop
ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Bulge-Loop™ miRNA RT-qPCR Primer kits (catalog nos.
MQP-0101 and MQP-0201) were obtained from Guangzhou RiboBio Co.,
Ltd. (Guangzhou, China). A total of 0.5 µg RNA template, 1
µl miRNA-specific stem-loop RT-primers and RNase-free water
(Tiangen Biotech, Co., Ltd., Beijing, China) were combined. Next,
5.5 µl mixtures were incubated at 70°C for 10 min, and then
immediately placed on ice for 2 min. Reverse transcription of RNA
into cDNA was subsequently performed by addition of 2.5 µl
5X RT buffer (Promega Corp., Madison, WI, USA), 1 µl dNTPs
(2.5 mM; Tiangen Biotech, Co., Ltd.), 0.25 µl M-MLV reverse
transcriptase (200 U/µl; Promega Corp.), 0.25 µl
ribonuclease inhibitor (40 U/µl; Fermentas; Thermo Fisher
Scientific, Inc.) and 3 µl RNase-free water. The sample was
subsequently incubated for 1 h at 42°C, followed by 70°C for 10 min
and then held at 4°C. qPCR was performed using the StepOnePlus
system (Applied Biosystems; Thermo Fisher Scientific, Inc.), and U6
expression (Guangzhou RiboBio Co., Ltd.) was selected to normalize
the RNA input. cDNA (1 µl) was added to a mixture containing
4.5 µl SYBR Green PCR Master Mix (Toyobo Co., Ltd., Osaka,
Japan), 0.8 µl forward primer, 0.8 µl reverse primer
and 3.7 µl RNase-free water to a total volume of 10
µl. The thermal cycling conditions consisted of an initial
denaturation step at 95°C for 5 min, followed by 40 cycles of 95°C
for 15 sec, 60°C for 30 sec and 72°C for 30 sec. Fluorescence data
were collected at 60°C during each cycle, while melting curve
analysis was conducted from 60 to 95°C. The ΔΔCq formula
was used to quantify the relative target miRNA expression levels
(the fold change in target gene expression was equal to
2−ΔΔCq) (27).
Bioinformatics analysis
TargetScan (www.targetscan.org/), miRDB (www.mirdb.org/), PicTar (pictar.mdc-berlin.de/) and miRanda (www.microrna.org/) software were used to predict the
potential targets of hsa-miR-144-3p, hsa-miR-144-5p and
hsa-miR-451a. The Database for Annotation, Visualization and
Integrated Discovery (DAVID) Bioinformatics Resources 6.7
(david.abcc.ncifcrf.gov/) was used for
functional enrichment analysis. In addition, the Kyoto Encyclopedia
of Genes and Genomes (KEGG) database (www.kegg.jp/kegg/pathway.html) was used for pathway
analysis.
Statistical analysis
Statistical analysis of the data was performed using
SAS version 9.2 software (SAS Institute, Cary, NC, USA). Any data
records with missing values were excluded from the analysis. The
following statistical tests were used to analyze the data:
Conditional logistic regression, Pearson correlation, principal
component analysis, multiple logistic regression and paired
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression levels of miR-144/451 cluster
members in esophageal carcinoma
The paired sample t-test was used to compare
the mean miRNA expression levels between esophageal tumor and
adjacent non-tumor tissues (Table
I). The results indicated that hsa-miR-144-3p, hsa-miR-144-5p
and hsa-miR-451a were significantly downregulated in tumor tissues
compared with the adjacent non-tumor tissues (P<0.001 for all
three miRNAs) and the fold-change (2−ΔΔCq) was found to
be 0.376, 0.463 and 0.443, respectively. By contrast, no
statistically significant differences in hsa-miR-4732-5p and
hsa-miR-4732-3p expression levels were observed between esophageal
carcinoma and adjacent non-tumor tissues.
 | Table IExpression levels of miRNA members of
the miR-144/451 cluster in esophageal carcinoma and adjacent
non-tumor tissue samples. |
Table I
Expression levels of miRNA members of
the miR-144/451 cluster in esophageal carcinoma and adjacent
non-tumor tissue samples.
| miRNA | Group | Sample no. | ΔCq |
ΔΔCq |
2−ΔΔCq | t-value | P-value |
|---|
| hsa-miR-144-3p | T | 101 | 19.090±2.459 | 1.412±1.760 | 0.376 | 8.062 | <0.001 |
| N | 101 | 17.678±2.788 | | | | |
| hsa-miR-144-5p | T | 99 | 17.522±2.239 | 1.111±1.764 | 0.463 | 6.267 | <0.001 |
| N | 99 | 16.411±2.301 | | | | |
| hsa-miR-451a | T | 99 | 9.300±2.666 | 1.176±1.843 | 0.443 | 6.348 | <0.001 |
| N | 99 | 8.124±3.057 | | | | |
|
hsa-miR-4732-5p | T | 100 | 17.716±3.191 | −0.161±1.845 | 1.118 | −0.871 | 0.386 |
| N | 100 | 17.876±2.858 | | | | |
|
hsa-miR-4732-3p | T | 93 | 19.780±2.281 | −0.041±2.311 | 1.029 | −0.171 | 0.865 |
| N | 93 | 19.821±2.376 | | | | |
Correlation analysis of the expression
levels of miR-144/451 cluster members
Pearson correlation analysis was performed to
estimate the degree of association between the members of the
miR-144/451 cluster. A significant correlation was observed among
the relative expression levels (ΔCq) of the majority of
miRNA members. Notably, strong correlations were observed among the
expression levels of hsa-miR-144-5p, hsa-miR-4732-5p and
hsa-miR-451a, with the correlation coefficients of two canonical
pairs (miR-144-5p and miR-451a; miR-4732-5p and miR-451a) being
0.729 (P<0.001) and 0.608 (P<0.001), respectively (Fig. 1). However, no significant
correlation was detected between the expression levels of
hsa-miR-144-3p and hsa-miR-4732-3p.
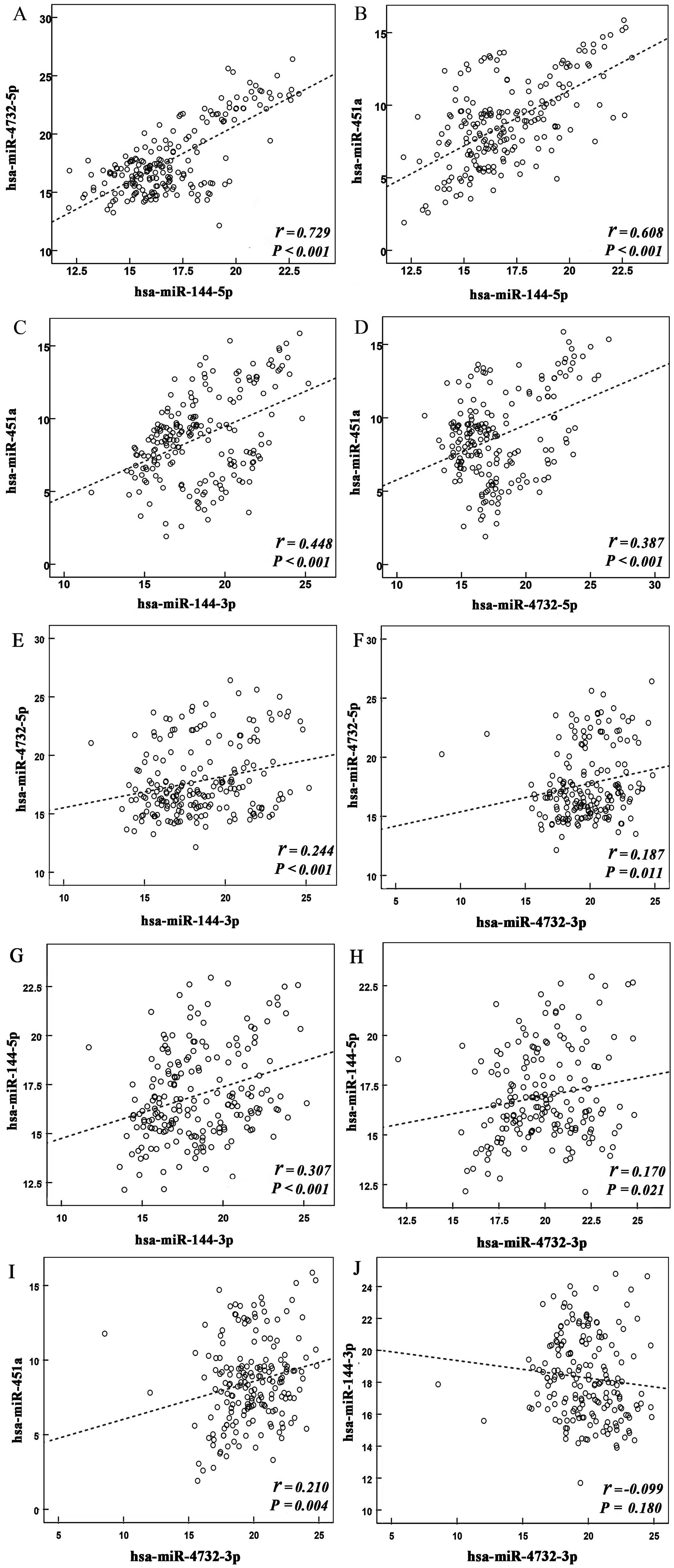 | Figure 1Correlation between the expression
levels of individual miRNAs in the miR-144/451 cluster. Scatter
diagrams demonstrate the association between the expression levels
of (A) hsa-miR-4732-5p and hsa-miR-144-5p, (B) hsa-miR-451a and
hsa-miR-144-5p, (C) hsa-miR-451a and hsa-miR-144-3p, (D)
hsa-miR-451a and hsa-miR-4732-5p, (E) hsa-miR-144-3p and
hsa-miR-4732-5p, (F) hsa-miR-4732-3p and hsa-miR-4732-5p, (G)
hsa-miR-144-3p and hsa-miR-144-5p, (H) hsa-miR-4732-3p and
hsa-miR-144-5p, (I) hsa-miR-4732-3p and hsa-miR-451a, and (J)
hsa-miR-4732-3p and hsa-miR-144-3p. The axes represent the ΔCq of
the relative expression levels of miRNAs. R2 values for each
scatter diagram are shown. miR, microRNA. |
Association between the expression levels
of miR-144/451 cluster members and the risk of esophageal
carcinoma
Conditional logistic regression models that coped
with 1:1 case-control matching were used to analyze the association
between the abnormal expression levels of miR-144/451 cluster
members and the risk for esophageal carcinoma (Table II). The results demonstrated that
reduced hsa-miR-144-5p, hsa-miR-144-3p and hsa-miR-451a expression
levels were significantly associated with an increased risk for
esophageal cancer [odds ratio (OR), 0.494, 0.370 and 0.474,
respectively]; however, no association was observed for
hsa-miR-4732-5p and hsa-miR-4732-3p. Therefore, hsa-miR-144-5p,
hsa-miR-144-3p and hsa-miR-451a may serve important roles in the
progression of esophageal carcinoma, which requires further
validation in future studies.
 | Table IIConditional logistic regression
analysis to determine the association between the expression levels
of miR-144/451 cluster members and esophageal carcinoma risk. |
Table II
Conditional logistic regression
analysis to determine the association between the expression levels
of miR-144/451 cluster members and esophageal carcinoma risk.
| miRNA | Group | β | SE | Wald | P-value | OR | 95% CI |
|---|
|
hsa-miR-4732-5p | T | 0.096 | 0.110 | 0.753 | 0.386 | 1.100 | 0.887–1.365 |
| N | | | | | 1 | |
| hsa-miR-144-5p | T | −0.706 | 0.152 | 21.658 | <0.001 | 0.494 | 0.367–0.665 |
| N | | | | | 1 | |
| hsa-miR-144-3p | T | −0.995 | 0.191 | 27.055 | <0.001 | 0.370 | 0.254–0.538 |
| N | | | | | 1 | |
| hsa-miR-451a | T | −0.748 | 0.159 | 22.137 | <0.001 | 0.474 | 0.347–0.647 |
| N | | | | | 1 | |
|
hsa-miR-4732-3p | T | 0.016 | 0.090 | 0.030 | 0.864 | 1.015 | 0.851–1.212 |
| N | | | | | 1 | |
Principal component analysis
In order to eliminate multi-collinearity of
individual miRNAs in the miR-144/451 cluster, principal component
analysis was performed to select the principal components of
clustered miRNAs (Table III).
Two principal components were extracted, and their cumulative
contribution of variance accounted for 70%, which is sufficient to
reflect the original factor's information of the cluster. The
results demonstrated that hsa-miR-4732-5p, hsa-miR-451a and
hsa-miR-144-5p primarily contributed to the F1 value, with scoring
coefficients of 0.788, 0.801 and 0.891, respectively (Table IV).
 | Table IIIPrincipal component analysis of
miR-144/451 cluster members. |
Table III
Principal component analysis of
miR-144/451 cluster members.
| Factor | Initial eigenvalue
| Extracted
eigenvalue
|
|---|
| Eigenvalue | Difference | Cumulative | Eigenvalue | Difference | Cumulative |
|---|
| F1 | 2.443 | 0.489 | 0.489 | 2.443 | 0.489 | 0.489 |
| F2 | 1.103 | 0.221 | 0.710 | 1.103 | 0.221 | 0.710 |
| F3 | 0.739 | 0.148 | 0.858 | | | |
| F4 | 0.515 | 0.103 | 0.961 | | | |
| F5 | 0.201 | 0.040 | 100.0 | | | |
 | Table IVComponent matrix for the principal
component analysis. |
Table IV
Component matrix for the principal
component analysis.
| Load factor | F1 | F2 |
|---|
| miR-4732-5p | 0.788 | 0.122 |
| miR-451a | 0.801 | −0.012 |
| miR-144-5p | 0.891 | −0.012 |
| miR-144-3p | 0.537 | −0.624 |
| miR-4732-3p | 0.311 | 0.835 |
Multiple logistic regression
analysis
Considering the strong correlation among the
expression levels of hsa-miR-4732-5p, hsa-miR-451a and
hsa-miR-144-5p, these miRNAs were grouped together and represented
as F-miRNAs. Multiple logistic regression analysis was conducted on
F-miRNAs, as well as hsa-miR-144-3p, in order to verify the
previous findings (Table V). The
OR values for hsa-miR-144-3p and F-miRNAs were 0.85 and 0.84,
respectively, while the downregulation of hsa-miR-144-3p and
F-miRNA expression levels was found to be associated with an
increased risk of esophageal carcinoma development. These results
suggest that the miR-144/451 cluster may serve as a biomarker for
the early detection of esophageal carcinoma.
 | Table VMultiple logistic regression analysis
for the miR-144/451 cluster. |
Table V
Multiple logistic regression analysis
for the miR-144/451 cluster.
| miRNA | β | SE | Wald | P-value | OR | 95% CI |
|---|
| hsa-miR-144-3p | −0.161 | 0.059 | 7.566 | 0.006 | 0.850 | 0.757–0.954 |
| F-miRNAsa | −0.175 | 0.070 | 6.300 | 0.012 | 0.840 | 0.733–0.962 |
| Constant | 5.946 | 1.431 | 17.272 | <0.001 | 382.204 | |
Target prediction for selected
miRNAs
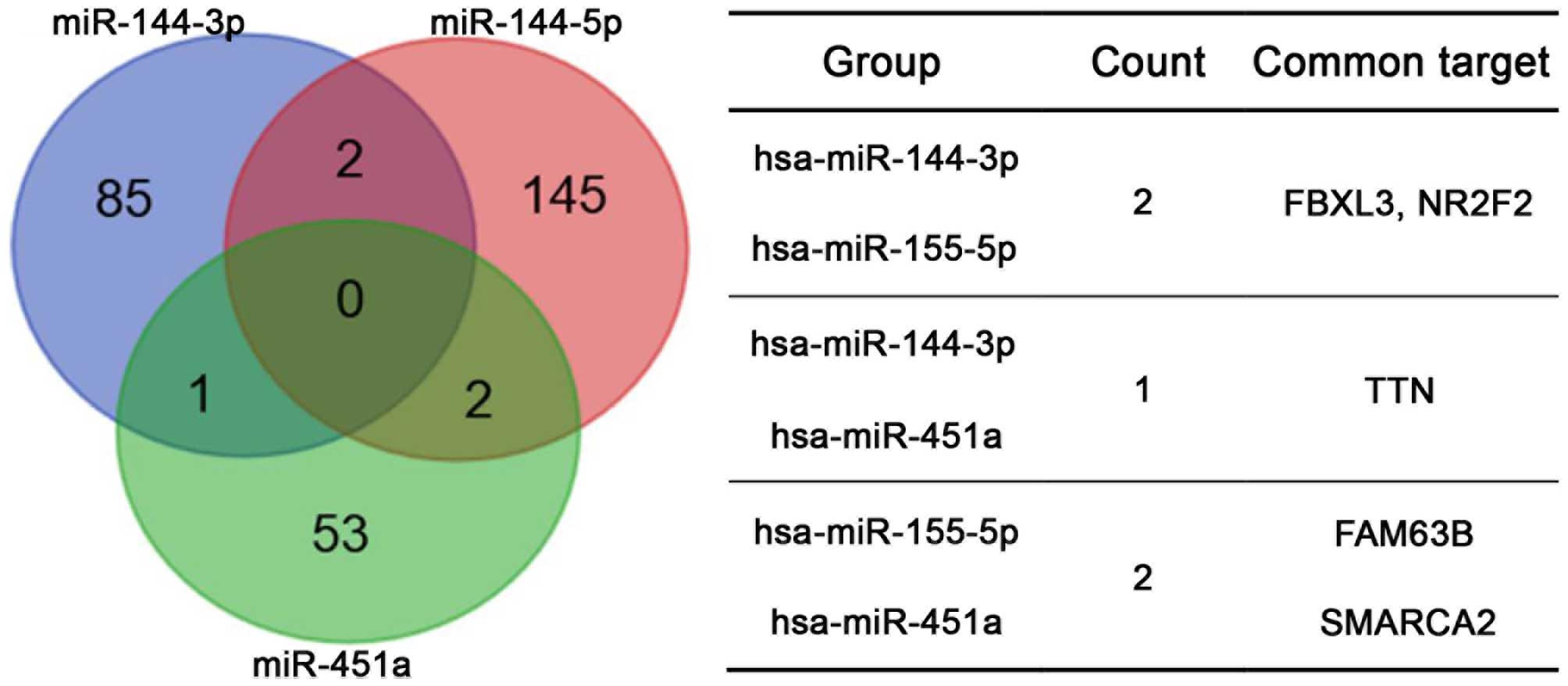
The TargetScan, miRDB, PicTar and miRanda algorithms
were used to predict the potential targets of hsa-miR-144-3p,
hsa-miR-144-5p and hsa-miR-451a, which were abnormally expressed in
esophageal carcinoma tissues. Targets identified by more than two
prediction tools were selected for further analysis. For
hsa-miR-144-3p, hsa-miR-144-5p and hsa-miR-451a, a total of 88, 149
and 56 potential targets were selected, respectively. The analysis
did not identify any common potential targets of all three miRNAs
or more than two common potential targets between each pair of
miRNAs (Fig. 2).
Enrichment analysis for selected
miRNAs
Enrichment analysis of gene ontology terms for
potential targets of hsa-miR-144-3p, hsa-miR-144-5p and
hsa-miR-451a was performed using the DAVID Bioinformatics Resources
6.7 database, and the Benjamini-Hochberg test indicated a false
discovery rate value of 0.01. The enriched functions were ranked
according to the number of predicted targets of the selected
miRNAs. The most enriched function associated with hsa-miR-144-3p
and hsa-miR-144-5p was the regulation of transcription, whereas
cell proliferation was the most enriched function for hsa-miR-451a
(Table VI).
 | Table VIResults of enrichment analysis for
predicted targets of selected miRNAs. |
Table VI
Results of enrichment analysis for
predicted targets of selected miRNAs.
| miRNA | GO term | Description | P-value |
|---|
| hsa-miR-144-3p | 0045449 | Regulation of
transcription | 0.0057 |
| 0032989 | Cellular component
morphogenesis | 0.0022 |
| 0000902 | Cell
morphogenesis | 0.0047 |
| 0014706 | Striated muscle
tissue development | 0.0001 |
| 0060537 | Muscle tissue
development | 0.0001 |
| hsa-miR-144-5p | 0045449 | Regulation of
transcription | 0.0016 |
| 0006350 | Transcription | 0.0001 |
| 0051252 | Regulation of RNA
metabolic process | 0.0043 |
| 0006355 | Regulation of
transcription, DNA-dependent | 0.0071 |
| 0010604 | Positive regulation
of macromolecule metabolic processes | 0.0001 |
| hsa-miR-451a | 0042127 | Regulation of cell
proliferation | 0.0034 |
Pathway of potential targets of selected
miRNAs
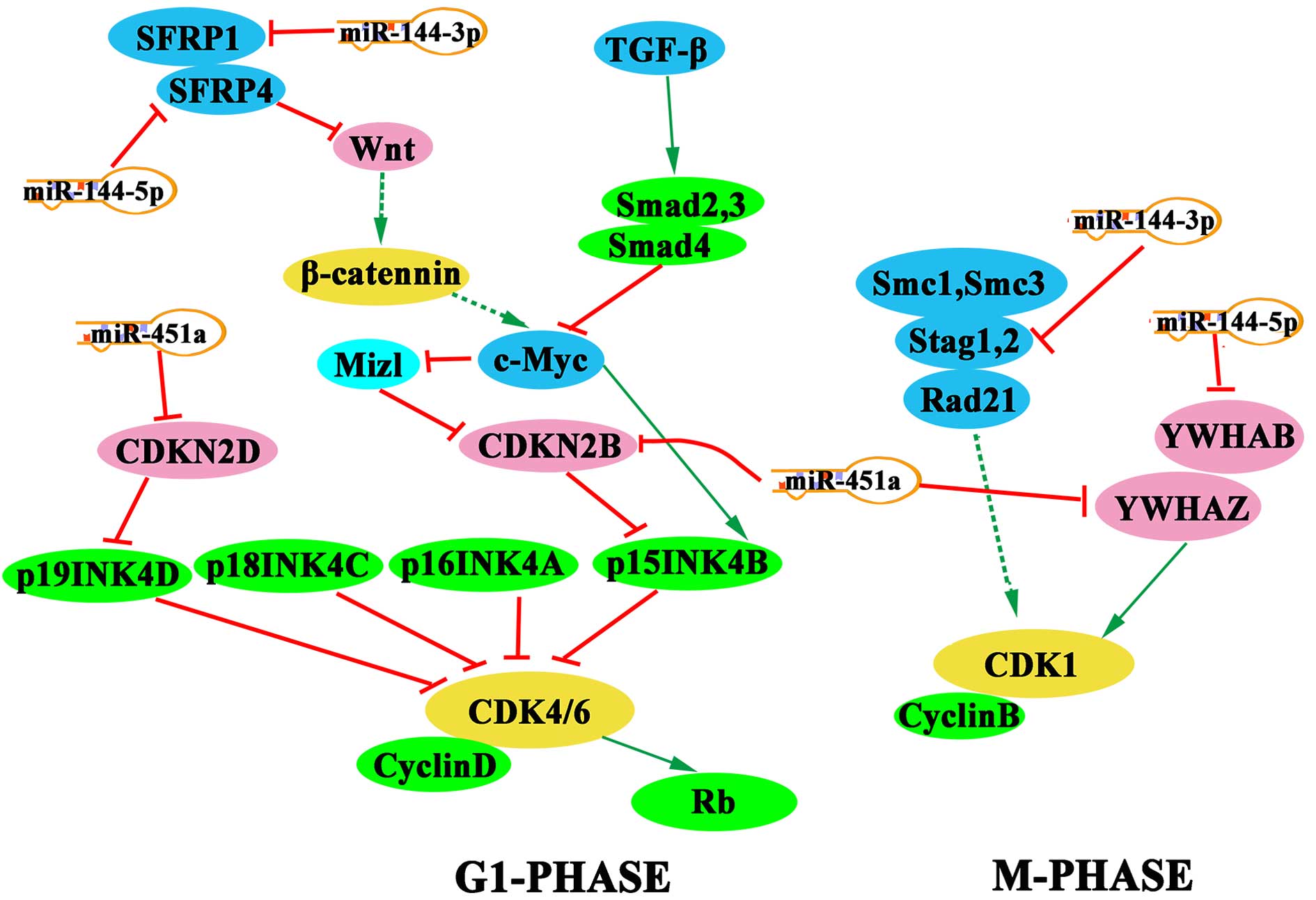
Pathway analysis was performed using the KEGG
pathway database, and the Benjamini-Hochberg test indicated a false
discovery rate value of 0.2. Notably, a number of cell cycle
regulators, including SFRP1, SFRP4, YWHAZ, YWHAB, CDKN2B, CDKN2D
and STAG1, were found to be targeted by miR-451a, miR-144-5p and
miR-144-3p (Fig. 3).
Discussion
The vast majority of identified miRNA clusters are
intragenic and commonly derived from polycistronic mRNA sequences,
which are considered to be transcribed as independent units
(28–30). The miR-144/451 cluster gene is
located on chromosome 17q11.2 and encodes hsa-miR-144-3p,
hsa-miR-144-5p, hsa-miR-451a, hsa-miR-4732-3p and hsa-miR-4732-5p,
as identified in the miRBase database (www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000460).
This miRNA cluster is highly conserved across different types of
species (http://genome.ucsc.edu/). In addition,
members of the miR-144/451 cluster are located in the same intronic
region between FLOT2 and FAM222B, which is 361 base pairs in
length; therefore, these miRNA sequences may share common
promoters. A study by Jiang et al (25) reported that the overexpression of
miR-144-3p, miR-144-5p and miR-451a promoted pancreatic cell
proliferation by targeting the PTEN/AKT signaling pathway. In
addition, Wang et al (21)
demonstrated that absence of the miR-144/451 cluster activated
Rac-1-mediated oxidative stress signaling in cardiovascular cells,
which impaired ischemic preconditioning-mediated cardioprotection.
Zhang et al (31)
demonstrated that miR-144 and miR-451 individually augmented
cardiomyocyte survival and mediated cooperative functions by
promoting the expression of GATA-4 protein. Furthermore, reduced
levels of miR-144 and miR-451 have been shown to be inversely
correlated with increased expression and phosphorylation of protein
kinase AMPK (32). Considering the
aforementioned studies, members of the miR-144/451 cluster may
serve important roles in numerous biological pathways and may
potentially be used as biomarkers for diagnosis or therapy of
cancer. However, to date, the majority of studies involving the
miR-144/451 cluster have focused on investigating the functional
roles of individual miRNA members (33–45).
Aberrant expression of miR-451a has been observed in
lung, stomach and breast cancer, in addition to glioma and leukemia
(33–36). Fukumoto et al (33) reported that miR-451a inhibits the
invasion and migration of hypopharyngeal squamous cell carcinoma
cells by activating ESDN and DCBLD2. In addition, circulating
miR-451a was demonstrated to be a potential biomarker for the
diagnosis of papillary thyroid carcinoma (37). Through MIF signaling pathway,
miR-451a enhanced tamoxifen sensitivity and inhibited the
proliferation of breast cancer cells (38). Furthermore, miR-144-3p and
miR-144-5p, encoded by the miR-144 gene, were down-regulated in a
variety of tumor tissues and cells, and their overexpression was
correlated with poor outcome in several human cancer types
(39–41). miR-144-3p has been shown to inhibit
the expression of ZEB1 and ZEB2, thereby promoting the expression
of cadherin and inhibiting cell invasion (42). Guo et al (43) demonstrated that miR-144 inhibits
the expression of E2H2, thus affecting the Wnt/β-catenin signaling
pathway and cell proliferation. A study by Matsushita et al
(44) demonstrated that miR-144-5p
targets CCNE1/2 directly to suppress the proliferation of bladder
cancer cells. The miR-4732-3p and miR-4732-5p members of the
miR-144/451 cluster were identified after the other members, and
therefore, few studies currently exist about these miRNAs (45,46).
Omura et al (45) suggested
that miR-4732-5p may facilitate the prediction of disease
recurrence following S-1 chemotherapy treatment through mutational
analysis and sequence similarity. Furthermore, Pouladi et al
(46) demonstrated that, by
binding with the 5′-untranslated region of WRAP53, hsa-miR-4732-5p
promoted breast cancer progression. Ultimately, these studies
provide evidence demonstrating that members of the miR-144/451
cluster appear to serve important roles in multiple biological
pathways, and that individual miRNAs of the miR-144/451 cluster may
function as part of different biological pathways in different
types of tissues.
A notable finding of the present study was that the
expression levels of all five members of the miR-144/451 cluster
were associated with each other, particularly miR-144-5p,
miR-4732-5p and miR451a. Thus, it is possible that members of the
miR-144/451 cluster are expressed from the same primary transcript.
By conducting a search of the NCBI database (http://www.ncbi.nlm.nih.gov/), sequences encoding
miR-144-5p, miR-4732-5p and miR-451a were found to be located at
the 5′-end of precursors, indicating that they may share the same
transcription and regulatory processes, which may enable an
improved understanding of the expression patterns of the
miR-144/451 cluster. In addition, low expression levels of
hsa-miR-144-3p and hsa-miR-144-5p were found to be associated with
an increased risk for esophageal carcinoma. Thus, hsa-miR-144-3p
and hsa-miR-144-5p may function together and be a more stable and
reliable biomarker for the early screening of high-risk populations
for early diagnosis.
Previous studies have demonstrated that clustered
miRNAs may mediate cooperative functions by targeting the same or
multiple genes simultaneously (15,16,47).
Using bioinformatics analyses, the potential common or similar
molecular functions of miR-144-3p, miR-144-5p and miR-451a, which
are aberrantly expressed in tumor tissues, were explored in the
current study. KEGG pathway analysis also demonstrated that
miR-451a suppresses CDK4/6 by targeting CDKN2D and CDKN2B during
G1-phase, which may induce G1 arrest. Notably, Zang et al
(22) validated these findings in
the EC9706 esophageal carcinoma cell line, whereby miR-451 arrested
cells in G1 phase by targeting CDKN2D. The results of the present
study suggest that miR-144-3p, miR-144-5p and miR-451a may disrupt
the expression of CDK1 by inhibiting the activity of STAG1, YWHAB
and YWHAZ, respectively. In addition, miR-144-3p and miR-144-5p
participate in the Wnt signaling pathway by inhibiting SFRP1 and
SFRP4, respectively, and the G1-phase may be affected through
interference with the activity of c-Myc. The members of the
miR-144/451 cluster mediate synergistic regulatory effects on the
cell cycle during different phases, and therefore may present
potential biomarkers for the early diagnosis of esophageal
carcinoma. The miR-144/451 cluster appears to be more sensitive
than a single miRNA, as it provides more comprehensive information.
By promoting the expression of miR-144/451 cluster members, it is
hypothesized that relative targets will be regulated together,
which will help to eliminate resistance generated by targeting a
single miRNA of miR-144/451 cluster. Considering the limitations of
bioinformatics prediction and the complexity of biological
regulatory networks, further experimental studies should be
conducted to validate the regulatory mechanisms of the miR-144/451
cluster in esophageal carcinoma.
In conclusion, the present study provided evidence
demonstrating that clustered miRNAs encoded by miR-144/451 gene can
interact with a number of tumor-associated factors. The expression
levels of miR-144/451 cluster members were downregulated in
esophageal carcinoma tissues compared with adjacent non-tumor
tissues. Notably, the expression levels of all five miR-144/451
cluster members were found to be associated with each other. In
addition, downregulation of hsa-miR-144-3p and hsa-miR-144-5p was a
potential risk factor for esophageal carcinoma development.
hsa-miR-144-3p and hsa-miR-144-5p, representing the miR-144/451
cluster, may serve as potential biomarkers for the early detection
of esophageal carcinoma. Further bioinformatics analyses indicated
that members of the miR-144/451 cluster may function together in
the progression of esophageal carcinoma. However, these molecular
mechanisms remain to be verified by further experimental studies.
More detailed understanding of the miR-144/451 cluster may provide
new diagnostic and therapeutic approaches for patients with
esophageal carcinoma.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81172747, 81573108
and 81573191), and the New Century Excellent Talents in University
from the Ministry of Education (grant no. NCET-13-0124).
References
|
1
|
Stewart W and Wild P: World Cancer Report
2014. IARC Press; Lyon, France: 2015
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shadfan A, Hellebust A, Richards-Kortum R
and Tkaczyk T: Confocal foveated endomicroscope for the detection
of esophageal carcinoma. Biomed Opt Express. 6:2311–2324. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang S, Du Z, Luo J, Wang X, Li H, Liu Y,
Zhang Y, Ma J, Xiao W, Wang Y and Zhong X: Inhibition of heat shock
protein 90 suppresses squamous carcinogenic progression in a mouse
model of esophageal cancer. J Cancer Res Clin Oncol. 141:1405–1416.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yue J and Tigyi G: Conservation of
miR-15a/16-1 and miR-15b/16-2 clusters. Mamm Genome. 21:88–94.
2010. View Article : Google Scholar :
|
|
8
|
Chan WC, Ho MR, Li SC, Tsai KW, Lai CH,
Hsu CN and Lin WC: MetaMirClust: Discovery of miRNA cluster
patterns using a data-mining approach. Genomics. 100:141–148. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Olive V, Li Q and He L: miR-17-92: A
polycistronic oncomir with pleiotropic functions. Immunol Rev.
253:158–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mohan S, Wergedal JE, Das S and Kesavan C:
Conditional disruption of miR17-92 cluster in collagen type
I-producing osteoblasts results in reduced periosteal bone
formation and bone anabolic response to exercise. Physiol Genomics.
47:33–43. 2015. View Article : Google Scholar :
|
|
11
|
Luo T, Cui S, Bian C and Yu X: Crosstalk
between TGF-β/Smad3 and BMP/BMPR2 signaling pathways via miR-17-92
cluster in carotid artery restenosis. Mol Cell Biochem.
389:169–176. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brockway S and Zeleznik-Le NJ: WEE1 is a
validated target of the microRNA miR-17-92 cluster in leukemia.
Cancer Genet. 208:279–287. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bazot Q, Paschos K, Skalska L,
Kalchschmidt JS, Parker GA and Allday MJ: Epsteinbarr virus
proteins EBNA3A and EBNA3C together induce expression of the
oncogenic MicroRNA cluster miR-221/miR-222 and ablate expression of
its target p57KIP2. PLoS Pathog. 11:e10050312015. View Article : Google Scholar
|
|
14
|
Zhu H, Han C, Lu D and Wu T: miR-17-92
cluster promotes cholangiocarcinoma growth: Evidence for PTEN as
downstream target and IL-6/Stat3 as upstream activator. Am J
Pathol. 184:2828–2839. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Besser J, Malan D, Wystub K, Bachmann A,
Wietelmann A, Sasse P, Fleischmann BK, Braun T and Boettger T:
MiRNA-1/133a clusters regulate adrenergic control of cardiac
repolarization. PloS One. 9:e1134492014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gits CM, van Kuijk PF, Jonkers MB, Boersma
AW, Smid M, van Ijcken WF, Coindre JM, Chibon F, Verhoef C,
Mathijssen RH, et al: MicroRNA expression profiles distinguish
liposarcoma subtypes and implicate miR-145 and miR-451 as tumor
suppressors. Int J Cancer. 135:348–361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Li W, Ying Z, Tian H, Zhu X, Li J
and Li M: Metastatic heterogeneity of breast cancer cells is
associated with expression of a heterogeneous TGFβ-activating
miR424-503 gene cluster. Cancer Res. 74:6107–6118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wystub K, Besser J, Bachmann A, Boettger T
and Braun T: miR-1/133a clusters cooperatively specify the
cardiomyogenic lineage by adjustment of myocardin levels during
embryonic heart development. PLoS Genet. 9:e10037932013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang M, Liu R, Sheng J, Liao J, Wang Y,
Pan E, Guo W, Pu Y and Yin L: Differential expression profiles of
microRNAs as potential biomarkers for the early diagnosis of
esophageal squamous cell carcinoma. Oncol Rep. 29:169–176.
2013.
|
|
20
|
Liu L, Wang S, Chen R, Wu Y, Zhang B,
Huang S, Zhang J, Xiao F, Wang M and Liang Y: Myc induced
miR-144/451 contributes to the acquired imatinib resistance in
chronic myelogenous leukemia cell K562. Biochem Biophys Res Commun.
425:368–373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Zhu H, Zhang X, Liu Y, Chen J,
Medvedovic M, Li H, Weiss MJ, Ren X and Fan GC: Loss of the
miR-144/451 cluster impairs ischaemic preconditioning-mediated
cardioprotection by targeting Rac-1. Cardiovasc Res. 94:379–390.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zang WQ, Yang X, Wang T, Wang YY, Du YW,
Chen XN, Li M and Zhao GQ: MiR-451 inhibits proliferation of
esophageal carcinoma cell line EC9706 by targeting CDKN2D and
MAP3K1. World J Gastroenterol. 21:5867–5876. 2015.PubMed/NCBI
|
|
23
|
Wang T, Zang WQ, Li M, Wang N, Zheng YL
and Zhao GQ: Effect of miR-451 on the biological behavior of the
esophageal carcinoma cell line EC9706. Dig Dis Sci. 58:706–714.
2013. View Article : Google Scholar
|
|
24
|
Xie Z, Chen G, Zhang X, Li D, Huang J,
Yang C, Zhang P, Qin Y, Duan Y, Gong B and Li Z: Salivary microRNAs
as promising biomarkers for detection of esophageal cancer. PloS
One. 8:e575022013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang X, Shan A, Su Y, Cheng Y, Gu W, Wang
W, Ning G and Cao Y: miR-144/451 promote cell proliferation via
targeting PTEN/AKT pathway in insulinomas. Endocrinology.
156:2429–2439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rice TW, Blackstone EH and Rusch VW: 7th
Edition of the AJCC cancer staging manual: Esophagus and
esophagogastric junction. Ann Surg Oncol. 17:1721–1724. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)). Method. 25:402–408. 2001. View Article : Google Scholar
|
|
28
|
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek
SH and Kim VN: MicroRNA genes are transcribed by RNA polymerase II.
EMBO J. 23:4051–4060. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du
ZW and Zhang JW: Human microRNA clusters: Genomic organization and
expression profile in leukemia cell lines. Biochem Biophys Res
Commun. 349:59–68. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tian Y, Pan Q, Shang Y, Zhu R, Ye J, Liu
Y, Zhong X, Li S, He Y, Chen L, et al: MicroRNA-200 (miR-200)
cluster regulation by achaete scute-like 2 (Ascl2): Impact on the
epithelial-mesenchymal transition in colon cancer cells. J Biol
Chem. 289:36101–36115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang X, Wang X, Zhu H, Zhu C, Wang Y, Pu
WT, Jegga AG and Fan GC: Synergistic effects of the GATA-4-mediated
miR-144/451 cluster in protection against simulated
ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell
Cardiol. 49:841–850. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Turczynska KM, Bhattachariya A, Säll J,
Göransson O, Swärd K, Hellstrand P and Albinsson S:
Stretch-sensitive down-regulation of the miR-144/451 cluster in
vascular smooth muscle and its role in AMP-activated protein kinase
signaling. PloS One. 8:e651352013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fukumoto I, Kinoshita T, Hanazawa T,
Kikkawa N, Chiyomaru T, Enokida H, Yamamoto N, Goto Y, Nishikawa R,
Nakagawa M, et al: Identification of tumour suppressive
microRNA-451a in hypopharyngeal squamous cell carcinoma based on
microRNA expression signature. Br J Cancer. 111:386–394. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Moreira FC, Assumpção M, Hamoy IG, Darnet
S, Burbano R, Khayat A, Gonçalves AN, Alencar DO, Cruz A, Magalhães
L, et al: MiRNA expression profile for the human gastric antrum
region using ultra-deep sequencing. PloS One. 9:e923002014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W,
Chang G, Li X, Li Q, Wang S and Wang W: MicroRNA profiling implies
new markers of chemoresistance of triple-negative breast cancer.
PloS One. 9:e962282014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Babapoor S, Fleming E, Wu R and Dadras SS:
A novel miR-451a isomiR, associated with amelanotypic phenotype,
acts as a tumor suppressor in melanoma by retarding cell migration
and invasion. PloS One. 9:e1075022014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li M, Song Q, Li H, Lou Y and Wang L:
Circulating miR-25-3p and miR-451a may be potential biomarkers for
the diagnosis of papillary thyroid carcinoma. PloS One.
10:e01324032015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu Z, Miao T, Feng T, Jiang Z, Li M, Zhou
L and Li H: miR-451a inhibited cell proliferation and enhanced
tamoxifen sensitive in breast cancer via macrophage migration
inhibitory factor. Biomed Res Int. 2015:2076842015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Katsuura S, Kuwano Y, Yamagishi N,
Kurokawa K, Kajita K, Akaike Y, Nishida K, Masuda K, Tanahashi T
and Rokutan K: MicroRNAs miR-144/144* and miR-16 in
peripheral blood are potential biomarkers for naturalistic stress
in healthy Japanese medical students. Neurosci Lett. 516:79–84.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Keller A, Leidinger P, Vogel B, Backes C,
ElSharawy A, Galata V, Mueller SC, Marquart S, Schrauder MG, Strick
R, et al: miRNAs can be generally associated with human pathologies
as exemplified for miR-144. BMC Med. 12:2242014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen S, Li P, Li J, Wang Y, Du Y, Chen X,
Zang W, Wang H, Chu H, Zhao G and Zhang G: MiR-144 inhibits
proliferation and induces apoptosis and autophagy in lung cancer
cells by targeting TIGAR. Cell Physiol Biochem. 35:997–1007. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guan H, Liang W, Xie Z, Li H, Liu J, Liu
L, Xiu L and Li Y: Down-regulation of miR-144 promotes thyroid
cancer cell invasion by targeting ZEB1 and ZEB2. Endocrine.
48:566–574. 2015. View Article : Google Scholar
|
|
43
|
Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang
Z, Qiu F and Lin J: miR-144 downregulation increases bladder cancer
cell proliferation by targeting EZH2 and regulating Wnt signaling.
FEBS J. 280:4531–4538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Matsushita R, Seki N, Chiyomaru T,
Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S,
Itesako T, et al: Tumour-suppressive microRNA-144-5p directly
targets CCNE1/2 as potential prognostic markers in bladder cancer.
Br J Cancer. 113:282–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Omura T, Shimada Y, Nagata T, Okumura T,
Fukuoka J, Yamagishi F, Tajika S, Nakajima S, Kawabe A and Tsukada
K: Relapse-associated microRNA in gastric cancer patients after S-1
adjuvant chemotherapy. Oncol Rep. 31:613–618. 2014.
|
|
46
|
Pouladi N, Kouhsari SM, Feizi MH, Gavgani
RR and Azarfam P: Overlapping region of p53/Wrap53 transcripts:
Mutational analysis and sequence similarity with microRNA-4732-5p.
Asian Pac J Cancer Prev. 14:3503–3507. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng
H, Mastick GS, Xu C and Yan W: Two miRNA clusters, miR-34b/c and
miR-449, are essential for normal brain development, motile
ciliogenesis, and spermatogenesis. Proc Natl Acad Sci USA.
111:E2851–E2857. 2014. View Article : Google Scholar : PubMed/NCBI
|