Introduction
Osteosarcoma, the most common primary malignant
tumor of bone, has an annual worldwide incidence of between one and
three cases per 1,000,000, and arises primarily in children and
adolescents, with a second peak in incidence in those >50 years
old (1–6). Although osteosarcoma has been treated
with surgery and chemotherapy for>30 years, patients with
recurrent or metastatic osteosarcoma have a poor prognosis
(7–11). Further elucidating the molecular
mechanism of the disease not only enables further understanding of
the pathogenesis and progression of the disease, but also assists
in identifying novel targets for effective therapies.
It has become increasingly recognized that mRNA 3′
end formation is subject to dynamic regulation under diverse
physiological conditions (12–15)
and the processing is crucial for mRNA maturation in eukaryotic
cells, promoting mRNA stability, efficient nuclear transport and
translation (16–18). This process involves six primary
protein complexes in mammals: The cleavage and polyadenylation
specificity factor, cleavage stimulatory factor, cleavage factor I
and II (CFIm and CFIIm), poly(A) polymerase and poly(A) binding
protein, together with RNA polymerase II (19,20)
and additional proteins (21). The
downregulation of CFIm25 in glioblastoma cells enhances their
tumorigenic properties and increases tumor size, whereas the
overexpression of CFIm25 reduces these properties and inhibits
tumor growth (22). In addition,
the global shortening of mRNAs through alternative polyadenylation
(APA), which occurs during enhanced cellular proliferation,
represents an important mechanism of regulated gene expression, and
a subset of CFIm25-regulated APA genes with shortened
3′untranslated regions (UTRs) have been identified in glioblastoma
tumors with reduced expression of CFIm25 (12,22,23).
These findings identified a pivotal role of CFIm25 in governing
APA, however, the role of CFIm25 in osteosarcoma remains to be
elucidated.
MicroRNAs (miRNAs/miRs) are a class of non-coding
RNAs, which are able to regulate gene expression at the
post-transcriptional level by binding to the 3′UTR of target mRNAs
through partial sequence homology, and inhibiting translation
and/or mRNA degradation (24–26).
Aberrant miRNA expression has been linked to osteosarcoma (27–29),
and it has been found to be involved in several cellular processes,
including proliferation, differentiation, migration and apoptosis
(30–35). miRNA-181a is upregulated in
osteosarcoma. It facilitates the proliferation and invasion, and
suppresses the apoptosis of osteosarcoma cells. However, the
mechanism of miR-181a as an oncogene in osteosarcoma remains to be
fully elucidated.
In the present study, it was first confirmed that
the expression of miR-181a was upregulated in osteosarcoma,
compared with adjacent normal tissues. Silencing miR-181a inhibited
the proliferation and promoted the apoptosis of osteosarcoma cells.
Target genes of miR-181a were screened using miRanda, which is a
commonly used prediction algorithm. It was found that miR-181a
targeted the 3′UTR of CFIm25 mRNA. Subsequent experiments confirmed
that miR-181 downregulated the expression of CFIm25 in osteosarcoma
cells. Finally, the present study demonstrated that CFIm25 protein
was downregulated in osteosarcoma tissues, in addition to
inhibiting proliferation and promoting apoptosis in the cells.
Materials and methods
Cell culture and expression
plasmids
MG63 human osteosarcoma cells (American Type Culture
Collection, Rockville, MD USA) were maintained in minimum essential
medium (Life Technologies; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) containing 10% fetal bovine serum (Biowest, Riverside, MO,
USA), 1% non-essential amino acids, and 1% penicillin/streptomycin
in a 5% CO2 incubator at 37°C. The expression plasmid CFIm25 and
empty vector (pcDNA.3.1) were purchased from Tiangene (Tianjin,
China). In the transfection experiments, the cells were cultured at
a density of 2×105 in serum-free medium without antibiotics at 60%
confluence for 24 h in a 5% CO2 incubator at 37°C, and then
transfected with transfection reagent (Lipofectamine 2000;
Invitrogen; Thermo Fisher Scientific, Inc.), according to
manufacturer's protocol. Following incubation for 6 h, the medium
was removed and replaced with normal culture medium for 48 h in a
5% CO2 incubator at 37°C.
miRNA precursors and anti-miRNA
oligonucleotides
The locked nucleic acid-modified oligonucleotide
inhibitor (anti-miR-181a) used for miRNA knockdown and the scramble
control were purchased from Exiqon, Inc. (Woburn, MA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis of miRNA
Total RNA, including miRNA from tissue samples and
cells was isolated using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The concentration of RNA was measured using a
NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Thermo
Fisher Scientific, Inc.). The mRNA was reverse transcribed using a
reverse transcription kit (Takara Biotechnology Co., Ltd., Dalian,
China), according to the manufacturer's protocol. The relative
levels of miR-181a were examined using altered stem-loop RT-PCR
with specific RT and PCR primers. U6 served as a control. The
reverse transcription primers for miR-181a and U6 are as follows:
RT primer
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACTCACCGA-3′,
specific primer 5′-GAACATTCAACGCTGTCGGTG-3′ and U6
5′-ATCCAGTGCAGGGTCCGAGGTA-3′. Relative quantification of mRNA
expression levels was performed according to the manufacturer's
instructions (Bio Rad, Hercules, CA, USA). Detection of the mature
form of the miRNAs was performed using an mirVana qRT-PCR miRNA
detection kit and primer sets, according to the manufacturer's
protocol (Ambion; Thermo Fisher Scientific, Inc.). U6 small nuclear
RNA was used as an internal control.
Colony formation
Colony formation analysis was performed as described
previously (36). Briefly, when
the transfected cells were growing in the logarithmic phase they
were trypsinized with 0.05% Trypsin-EDTA (Gibco; Thermo Fisher
Scientific, Inc.) and seeded into 6-well plates at a density of
2,000 cells/well. The cells were maintained in an incubator at 37°C
for 7 days. On day 8, the colonies were washed with
phosphate-buffered saline (PBS), fixed with formalin (10%), and
stained with methyl violet, all were purchased from Beyotime
Institute of Biotechnology (Haimen, China). The methyl violet dye
was then washed with PBS and the number of colonies were counted
under a microscope (IX53; Olympus Corporation, Tokyo, Japan). The
following calculations were then performed: Colony inhibition rate
= [(1 - number of colonies in experimental groups) / control group]
× 100%; and colony forming efficiency = 1 colony inhibition
rate.
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
(MTT) assay
The analysis of proliferation was performed using an
MTT assay as described previously (37). Cell viability was determined by MTT
assay. Briefly, transfected cells were seeded into 96-well plates
at a density of 1.0×105 cells/well and 10 µl MTT (concentration, 5
mg/ml; Sigma Aldrich, St Louis, MO, USA) was added into 100 µl
medium at indicated time points. The cells were incubated with MTT
for ~4 h at 37°C, followed by removal of MTT and addition of 150 µl
DMSO. Following incubation with DMSO for 10 min in the dark,
absorbance was measured at 570 nm (A570nm) with a microplate reader
(Biorad-168-1000XC; Bio-Rad Laboratories, Hercules, CA, USA).
Western blot analysis
The cells were seeded into 6-well plate at a density
of 3.0×105 cells/well and the cells were transfected when the cell
density reached ~80% confluency in the second day. At 48 h
following transfection, the cells were lysed using RIPA buffer for
30 min at 4°C. The protein concentration was measured by the BCA
method. Western blot analysis was performed as described previously
(38). Briefly, following
incubation with primary antibody anti-BCL2 (cat. no. ab32124;
1:500; Abcam, Cambridge, MA, USA), anti-myeloid cell leukemia 1
(MCL1) (cat. no. ab32087; 1:500; Abcam, Cambridge, MA, USA),
anti-c-myc (cat. no. ab32072; 1:500; Abcam), anti-p53 (cat. no.
ab1431; 1:500; Abcam), anti-Ki-67 (cat. no. ab16667; 1:500; Abcam),
anti-proliferating cell nuclear antigen (PCNA; cat. no. ab1431;
1:500; Abcam), anti-cyclin D1 (cat. no. EPR2241; 1:500; Abcam) and
anti-β-actin (cat. no. ab5694; 1:500; Abcam) overnight at 4°C,
incubation with IRDyeTM-800 conjugated anti-rabbit secondary
antibodies (caat. no. 925-32211; Li-COR Biosciences, Lincoln, NE,
USA) was performed for 30 min at room temperature. The specific
proteins were visualized using an Odyssey™ infrared
imaging system (Gene Company, Ltd., Lincoln, NE, USA).
Methods of bioinformatics
analysis
The analysis of potential miR target sites was
performed using the commonly used prediction algorithm, miRanda
(http://www.microrna.org/microrna/home.do).
Immunofluorescence analysis
For the immunofluorescence analysis of MG63 human
osteosarcoma cells. The cells were plated on glass coverslips in
6-well plates at density of 3.0×105 cells/well and transfected with
30 nM anti-miR-181a or the scramble control. At 36 h
post-transfection, the coverslips were stained with anti-CFIm25
antibodies. Alexa Fluor 488 goat anti-mouse IgG antibody or goat
anti-rabbit IgG antibody were used as secondary antibodies
(Invitrogen; Thermo Fisher Scientific, Inc.). The coverslips were
counterstained with DAPI (Invitrogen; Thermo Fisher Scientific,
Inc.) for the visualization of nuclei. Microscopic analysis was
performed with a confocal laser-scanning microscope (Leica
Microsystems, Bensheim, Germany). Fluorescence intensities were
measured in a number of areas with 200–300 cells per coverslip, and
analyzed using ImageJ 1.37v software (http://rsb.info.nih.gov/ij/index.html).
RT-qPCR analysis of CFIm25
Total RNA was isolated from the cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). First-strand
cDNA was synthesized from the total RNA using M-MLV reverse
transcriptase (Promega, Madison, WI, USA) and random hexamer
primers (Sangon Biotech, Shanghai, China). The thermal cycle
profile was as follows: Denaturation for 30 sec at 95°C, annealing
for 45 sec at 52–58°C depending on the primers used, and extension
for 45 sec at 72°C. The PCR products were visualized on 2% agarose
gels stained with ethidium bromide under UV transillumination.
RT-qPCR was performed using Power SYBR Green PCR Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The primer sequences for CFIm25 were:
Forward (F) 5′-AGTGTTTACAGGCACAAGAGG-3′ and reverse (R)
5′-TTCAGGAACAGGCAAGGA-3. GAPDH primers were: F
5′-GAAGGTCGGAGTCAACGGATTTG-3′ and R 5′-ATGGCATGGACTGTGGTCATGAG-3′.
The products were then separated by electrophoresis using 1.5%
agarose gels. The bands were visualized using the BioSpectrum 410
multispectral imaging system with a Chemi HR camera 410 (UVP,
Upland, CA, USA) and quantified as previosly described (39).
TUNEL staining
For the analysis of apoptosis, TUNEL staining was
performed as described previously (40). The TUNEL assays were performed with
the TMR red In Situ Cell Death Detection kit (supplemented with
0.1% sodium citrate; Hoffmann La Roche Inc., Nutley, NJ, USA),
according to the manufacturer's instructions.
Statistical analysis
Data are presented as the mean ± standard error of
the mean. Student's t-test (two-tailed) was used to compare between
the two groups. Statistical analysis was performed using SPSS
version 17 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of miR-181a is upregulated
in osteosarcoma tissues, promoting the proliferation and inhibiting
the apoptosis of osteosarcoma cells
In order to identify and compare the expression
levels of miR-181a between osteosarcoma tissues and adjacent normal
tissues, RT-qPCR was performed on the cancer tissues and normal
tissues. The RT-qPCR analysis revealed that the expression of
miR-181a in six osteosarcoma tissue samples were significantly
higher, compared with those of the adjacent normal tissues
(Fig. 1A). These data suggested
that miR-181a is an oncogene in osteosarcoma.
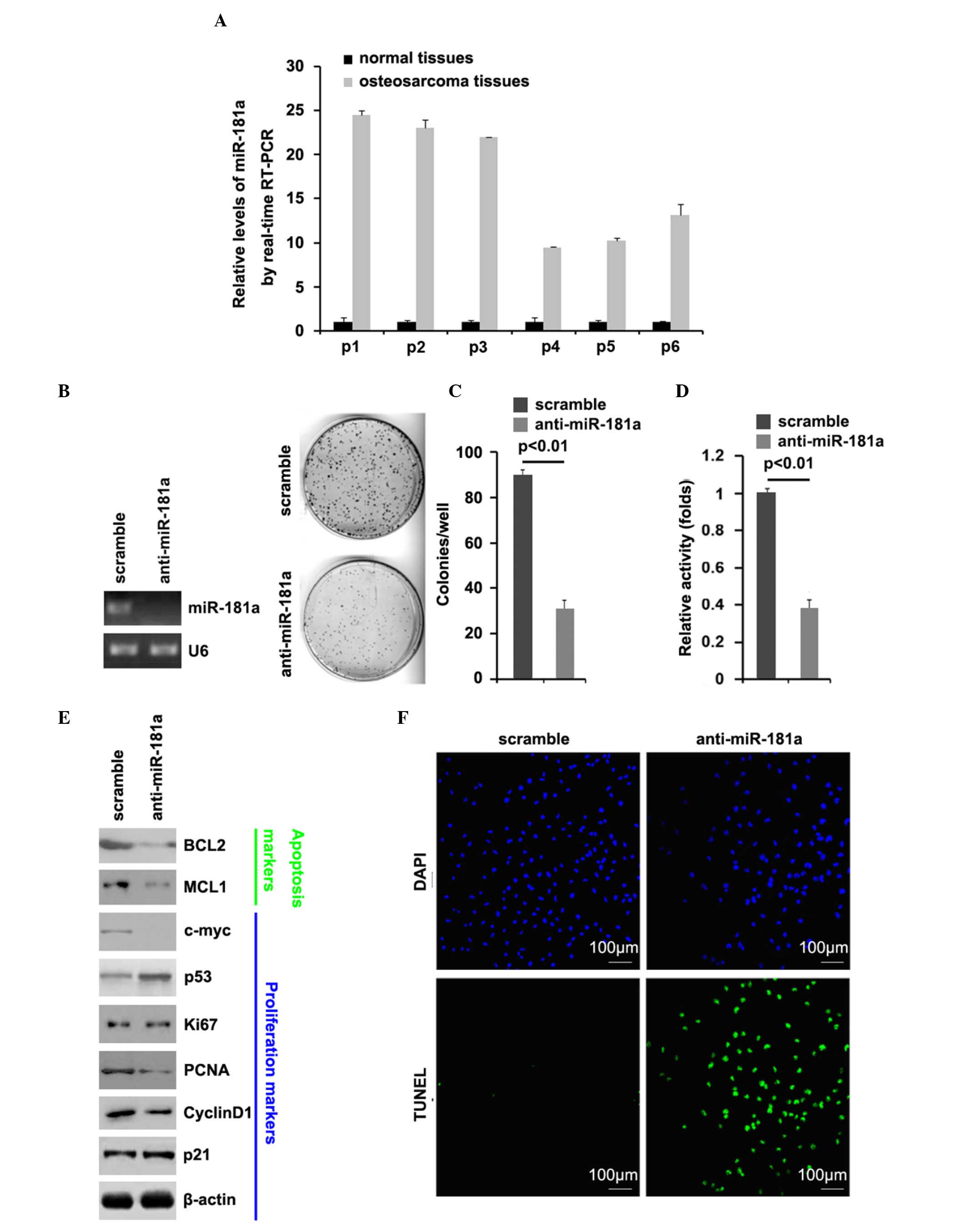 | Figure 1.Expression of miR-181a is upregulated
in osteosarcoma tissues, and its silencing inhibits proliferation
and promotes apoptosis. RT-qPCR analysis was used to compare
miR-181a between (A) osteosarcoma tissues and adjacent normal
tissues (n=6) and between (B) MG63 cells transfected with scramble
and anti-miR-181a (n=3). U6 was used as a loading control. (C)
Colony formation assay of MG63 cells transfected with scramble and
anti-miR-181a. Colonies of >50 cells were counted (n=3). (D)
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
assay (n=3) and (E) Western blot analysis for BCL2, MCL1, c-myc,
p53, Ki-67, PCNA, cyclin D1 and p21 in MG63 cells transfected with
scramble and anti-miR-181a. β-actin was used as a loading control
(n=3). (F) TUNEL assay (n=3) for MG63 cells transfected with
scramble and anti-miR-181a. miR, microRNA; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; BCL2, B cell
lymphoma 2; MCL1, myeloid cell leukemia 1; PCNA, proliferating cell
nuclear antigen. |
To further identify the role of miR-181a in
osteosarcoma cells, the MG63 osteosarcoma cells were transfected
with anti-miR-181a. Following stable transfection, the expression
of miR-181a was detected using RT-qPCR analysis, and colony
formation of the MG63 cells was assessed using a colony formation
assay. The results showed that exogenous anti-miR-181a
significantly downregulated the expression of miR-181a in the MG63
cells (Fig. 1B) and silencing
miR-181a significantly decreased the colony formation rate of the
MG63 cells (Fig. 1C). In addition,
MTT assays were performed to detect the proliferation of the MG63
cells transfected with anti-miR-181a and the scramble control. The
results showed that anti-miR-181a inhibited the proliferation of
the MG63 cells following 48 h of transfection, compared with the
scramble-transfected cells (Fig.
1D). In subsequent experiments, western blot analysis was used
to identify whether the proteins of proliferation-associated
markers were also affected by silencing miR-181a in the cells. The
results showed that the protein expression levels of c-myc, PCNA
and cyclin D1 were downregulated, and the expression of p53 was
upregulated by anti-miR-181a in the cells (Fig. 1E).
On demonstrating that silencing miR-181a inhibited
the proliferation of the MG63 cells, to provide further evidence
that miR-181a was involved in the regulation of MG63 cell
apoptosis, a TUNEL assay was performed to analyze whether silencing
miR-181a affected apoptosis in the MG63 cells. The TUNEL assay
revealed a change in the apoptotic rate of the MG63 cells
transfected with anti-miR-181a. Specifically, silencing miR-181a
promoted apoptosis in the MG63 cells (Fig. 1F).
Western blot analysis was also performed to identify
whether proteins of apoptosis-associated markers were also affected
by miR-181a in the cells. Bcl-2 and MCL1 are important
anti-apoptotic molecules (41,42).
The results showed that the expression levels of BCL2 and MCL1 were
downregulated by anti-miR-181a in the cells (Fig. 1E).
Silencing miR-181a restores the
protein expression of CFIm25 in osteosarcoma
miRNAs/miRs are a class of non-coding RNAs, which
are able to regulate gene expression at the post-transcriptional
level by binding to the 3′UTR of target mRNAs through partial
sequence homology; this inhibits translation and/or mRNA
degradation (24–26). The upregulation of specific miRNAs
can contribute to the downregulation of tumor suppressive genes
(31,43,44).
Thus, the present study hypothesized that miR-181a affected
proliferation and apoptosis by regulating target genes.
In the present study, targeted genes of miR-181a
were screened using TargetScan (http://www.microrna.org/microrna/home.do), and a large
number of target genes were identified. The gene of interest,
CFIm25, was used as it has been regarded as a tumor suppressor gene
in malignant tumors (22). Target
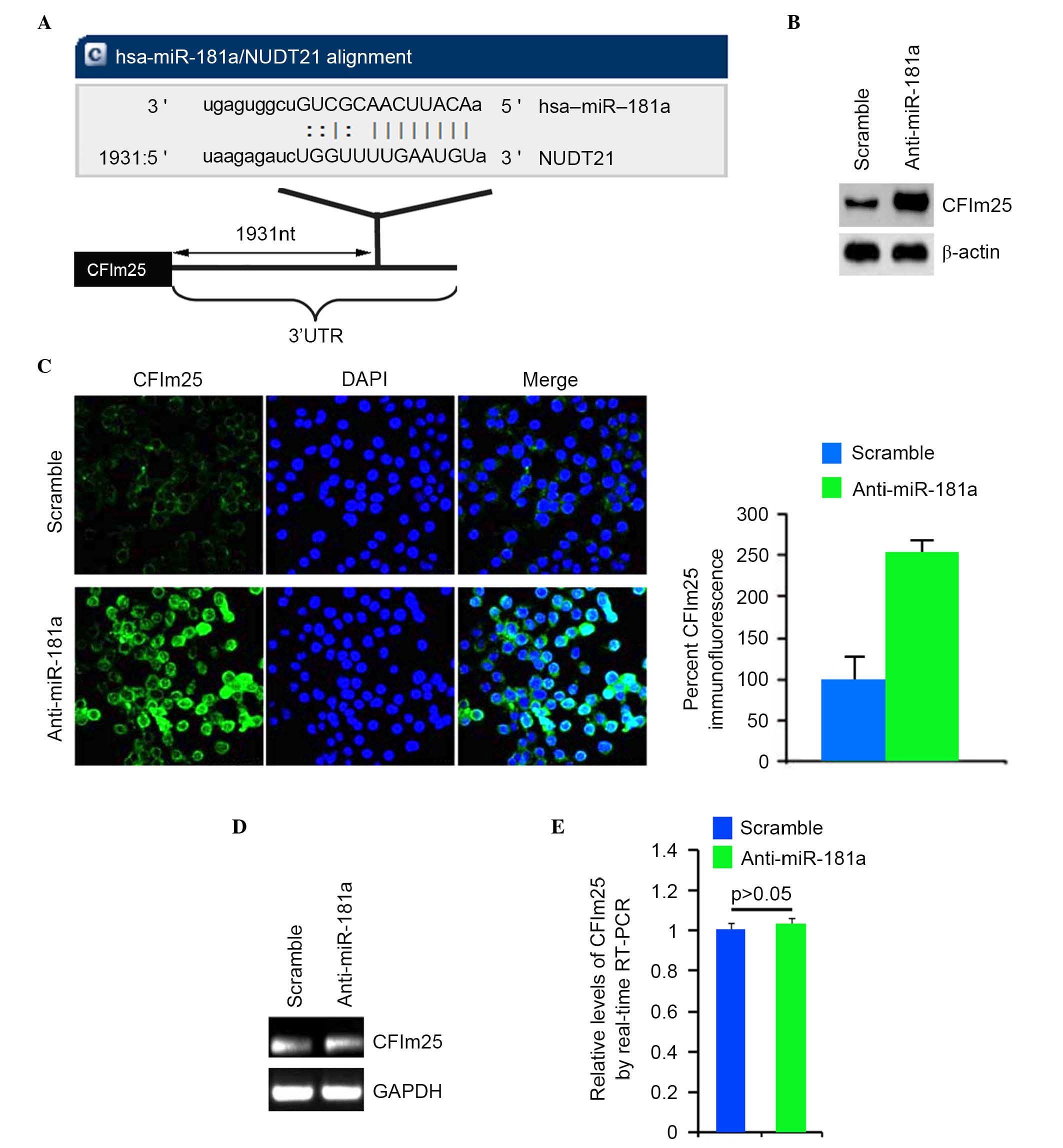
sites on the 3′UTR of CFIm25 are shown in Fig. 2A. Therefore, miR-181a may
downregulate the expression of CFIm25 by targeting its 3′UTR in
osteosarcoma cells.
Western blot analyses was performed on MG63 cells
transfected with anti-miR-181c or scramble. The results confirmed
that the protein level of CFIm25 was increased in the cells
transfected with anti-miR-181a (Fig.
2B). Consistent with the results of western blot analyses,
immunofluorescence analyses demonstrated that the protein level of
CFIm25 increased in MG63 cells transfected with anti-miR-181a,
compared with scramble-transfected groups (Fig. 2C). To identify whether CFIm25 mRNA
was increased by the silencing of miR-181a, RT-qPCR analysis was
performed to detect the mRNA expression of CFIm25 in the MG63 cells
transfected with anti-miR-181a or scramble. The results of the
RT-qPCR analysis demonstrated that silencing of miR-181a had no
effect on the mRNA expression of CFIm25 in the MG63 cells (Fig. 2D and E).
CFIm25 protein inhibits proliferation
and promotes apoptosis in MG63 osteosarcoma cells
The observation that silencing miR-181a inhibited
proliferation, promoted apoptosis and upregulated the expression of
CFIm25 in the MG63 cells cells, indicated that anti-miR-181a
inhibited proliferation and promoted apoptosis via upregulating the
expression of CFIm25. Thus, the present study investigated the
roles of CFIm25 in osteosarcoma.
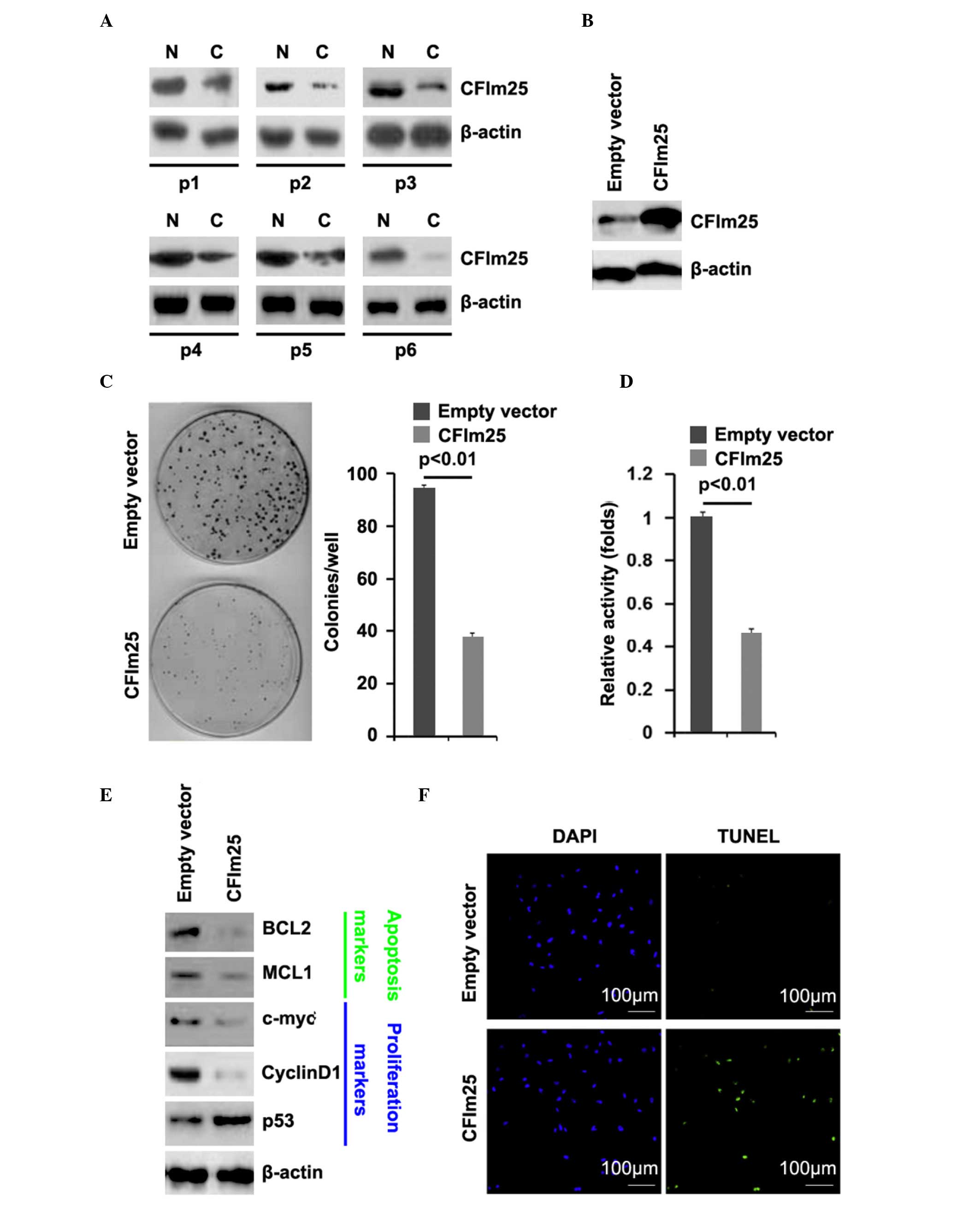
To assess the expression of CFIm25 in osteosarcoma,
western blot analysis was performed on six pairs of osteosarcoma
tissues and matched adjacent normal tissue samples. The expression
of CFIm25 was consistently lower in the osteosarcoma tissues,
compared with the normal tissues (Fig.
3A). Subsequently, CFIm25-expressing plasmids were used, and
the roles of CFIm25 in MG63 cell were investigated. In order to
demonstrate that CFIm25-expressing plasmids stably upregulated the
protein expression of CFIm25, western blot analysis was performed
on MG63 cells transfected with CFIm25-expressing plasmids. The
results showed that the protein expression of CFIm25 was
significantly increased by the CFIm25-expressing plasmids in the
cells (Fig. 3B). To determine the
roles of CFIm25 in colony formation ability and proliferation of
the MG63 cells, colony formation and MTT assays were performed. The
results of the colony formation assay demonstrated that CFIm25
suppressed colony formation in the MG63 cells (Fig. 3C) and, consistent with the colony
formation assay, the MTT assay showed that CFIm25 inhibited the
proliferation of the cells (Fig.
3D). In subsequent experiment, we performed western blot
analysis to identify whether the proteins of
proliferation-associated markers were also affected by CFIm25 in
the cells. The results of the present study showed that the
expression of cyclin D1 was downregulated the expression of p53,
were upregulated by CFIm25 in the cells (Fig. 3E).
Having demonstrated that overexpressing CFIm25
inhibited the proliferation in MG63 cells, to provide further
evidence that CFIm25 was involved in the regulation of MG63 cell
apoptosis, a TUNEL assay was performed to analyze whether
overexpressing CFIm25 affected apoptosis in MG63 cells. The TUNEL
assay revealed a change in the apoptotic rate of the MG63 cells
transfected with CFIm25. Specifically, overexpressing CFIm25
promoted apoptosis in the MG63 cells (Fig. 3F).
Western blot analysis was also performed to identify
whether the protein levels of apoptosis-associated markers were
also affected by CFIm25 in the cells. The results of the western
blot analysis showed that the expression levels of BCL2 and MCL1
were downregulated in the CFIm25-overexpressing cells (Fig. 3E).
Discussion
Osteosarcoma is the most common primary sarcoma of
bone and is a leading cause of cancer-associated mortality among
adolescents and young adults (3).
The cellular events, which initiate and propagate
osteosarcomagenesis remain to be fully elucidated (45). miRNAs are short noncoding RNAs,
which post-transcriptionally modify gene expression in eukaryotic
cells (46). The expression of a
single miRNA can silence a large number of genes, enabling these
molecules to have extensive control over several cellular functions
(46). Knowledge of individual
miRNAs affecting developmental biology, cellular differentiation
programs and oncogenesis continues to increase. miR-181a is
upregulated in osteosarcoma, can facilitate proliferation and
invasion, and suppress apoptosis in osteosarcoma cells (47). Consistent with a previous report
(47), the present study found
that the expression of miR-181a is upregulated in osteosarcoma
tissues, and that silencing it can inhibit proliferation and
promote apoptosis in osteosarcoma cells.
The global shortening of mRNAs through APA, which
occurs during enhanced cellular proliferation, represents a vital
mechanism of regulated gene expression, which remains to be fully
elucidated (12,23). The 3′UTR truncation of
growth-promoting mRNA transcripts, which can relieve intrinsic
miRNA-mediated repression has been observed to correlate with
cellular transformation (13).
CFIm25, is a broad repressor of proximal poly(A) site usage and,
when depleted, increases cell proliferation (47). Marked increases in the expression
of several known oncogenes, including cyclin D1, are observed as a
consequence of CFIm25 depletion (47). Consistent with this previous
report, the present study found that overexpressing CFIm25
suppressed proliferation, promoted apoptosis and inhibited the
expression of cyclin D1 in osteosarcoma cells. It was also
demonstrated that the protein expression of CFIm25 was restored by
silencing miR-181a in the osteosarcoma cells.
Elucidating the mechanism by which silencing
miR-181a inhibits proliferation and promotes apoptosis by restoring
CFIm25 improves understanding of the molecular mechanisms
underlying proliferation and apoptosis in osteosarcoma. The
suppression of miR-181a may represent a promising therapeutic
strategy to restore the CFIm25-mediated regulation of proliferation
and apoptosis. However, the roles of miR-181a and CFIm25 require
further confirmation in vivo.
References
|
1
|
Sweetnam R: Osteosarcoma. Br J Hosp Med.
28(112): 116–121. 1982.
|
|
2
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Damron TA, Ward WG and Stewart A:
Osteosarcoma, chondrosarcoma, and Ewing's sarcoma. National Cancer
Data Base report. Clin Orthop Relat Res. 459:40–47. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dorfman HD and Czerniak B: Bone cancers.
Cancer. 75:(1 Suppl). S203–S210. 1995. View Article : Google Scholar
|
|
6
|
Geller DS and Gorlick R: Osteosarcoma: A
review of diagnosis, management, and treatment strategies. Clin Adv
Hematol Oncol. 8:705–718. 2010.PubMed/NCBI
|
|
7
|
Bielack SS, Marina N, Ferrari S, Helman
LJ, Smeland S, Whelan JS and Reaman GH: Osteosarcoma: The same old
drugs or more? J Clin Oncol. 26:3102–3103; author reply 3104–3105.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chou AJ, Geller DS and Gorlick R: Therapy
for osteosarcoma: Where do we go from here? Paediatr Drugs.
10:315–327. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
O'Day K and Gorlick R: Novel therapeutic
agents for osteosarcoma. Expert Rev Anticancer Ther. 9:511–523.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bernthal NM, Federman N, Eilber FR, Nelson
SD, Eckardt JJ, Eilber FC and Tap WD: Long-term results (>25
years) of a randomized, prospective clinical trial evaluating
chemotherapy in patients with high-grade, operable osteosarcoma.
Cancer. 118:5888–5893. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Link MP, Goorin AM, Miser AW, Green AA,
Pratt CB, Belasco JB, Pritchard J, Malpas JS, Baker AR, Kirkpatrick
JA, et al: The effect of adjuvant chemotherapy on relapse-free
survival in patients with osteosarcoma of the extremity. N Engl J
Med. 314:1600–1606. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sandberg R, Neilson JR, Sarma A, Sharp PA
and Burge CB: Proliferating cells express mRNAs withshortened 3′
untranslated regions and fewer microRNA target sites. Science.
320:1643–1647. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mayr C and Bartel DP: Widespread
shortening of 3′UTRs by alternative cleavage and polyadenylation
activates oncogenes in cancer cells. Cell. 138:673–684. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ji Z, Lee JY, Pan Z, Jiang B and Tian B:
Progressive lengthening of 3′ untranslated regions of mRNAs by
alternative polyadenylation during mouse embryonic development.
Proc Natl Acad Sci USA. 106:7028–7033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mangone M, Manoharan AP, Thierry-Mieg D,
Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR,
Khivansara V, et al: The landscape of C. elegans 3′UTRs. Science.
329:432–435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jacobson A and Peltz SW:
Interrelationships of the pathways of mRNA decay and translation in
eukaryotic cells. Annu Rev Biochem. 65:693–739. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wickens M, Anderson P and Jackson RJ: Life
and death in the cytoplasm: Messages from the 3′ end. Curr Opin
Genet Dev. 7:220–232. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Garneau NL, Wilusz J and Wilusz CJ: The
highways and byways of mRNA decay. Nat Rev Mol Cell Biol.
8:113–126. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
McCracken S, Fong N, Rosonina E, Yankulov
K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S and
Bentley DL: 5′-Capping enzymes are targeted to pre-mRNA by binding
to the phosphorylated carboxy-terminal domain of RNA polymerase II.
Genes Dev. 11:3306–3318. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hirose Y and Manley JL: RNA polymerase II
is an essential mRNA polyadenylation factor. Nature. 395:93–96.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi Y, Di Giammartino DC, Taylor D,
Sarkeshik A, Rice WJ, Yates JR III, Frank J and Manley JL:
Molecular architecture of the human pre-mRNA 3′ processing complex.
Mol Cell. 33:365–376. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Masamha CP, Xia Z, Yang J, Albrecht TR, Li
M, Shyu AB, Li W and Wagner EJ: CFIm25 links alternative
polyadenylation to glioblastoma tumour suppression. Nature.
510:412–416. 2014.PubMed/NCBI
|
|
23
|
Elkon R, Drost J, van Haaften G, Jenal M,
Schrier M, Vrielink JA Oude and Agami R: E2F mediates enhanced
alternative polyadenylation in proliferation. Genome Biol.
13:R592012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pasquinelli AE, Reinhart BJ, Slack F,
Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B,
Müller P, et al: Conservation of the sequence and temporal
expression of let-7 heterochronic regulatory RNA. Nature.
408:86–89. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kobayashi E, Satow R, Ono M, Masuda M,
Honda K, Sakuma T, Kawai A, Morioka H, Toyama Y and Yamada T:
MicroRNA expression and functional profiles of osteosarcoma.
Oncology. 86:94–103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Osaki M, Takeshita F, Sugimoto Y, Kosaka
N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T,
et al: MicroRNA-143 regulates human osteosarcoma metastasis by
regulating matrix metalloprotease-13 expression. Mol Ther.
19:1123–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duan Z, Choy E, Harmon D, Liu X, Susa M,
Mankin H and Hornicek F: MicroRNA-199a-3p is downregulated in human
osteosarcoma and regulates cell proliferation and migration. Mol
Cancer Ther. 10:1337–1345. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jovanovic M and Hengartner MO: miRNAs and
apoptosis: RNAs to die for. Oncogene. 25:6176–6187. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Deng G, Hu C, Zhu L, Huang F, Huang W, Xu
H and Nie W: Downregulation of ROS-FIG inhibits cell proliferation,
colony-formation, cell cycle progression, migration and invasion,
while inducing apoptosis in intrahepatic cholangiocarcinoma cells.
Int J Mol Med. 34:661–668. 2014.PubMed/NCBI
|
|
37
|
Zhang ZG, Niu XY, He XJ and Shu J:
Ginsenoside Rg1 reduces toxicity of fine particulate matter on
human alveolar epithelial cells: A preliminary observation. Mol Med
Rep. 9:989–992. 2014.PubMed/NCBI
|
|
38
|
Tsujimoto M, Doi T, Kuroyanagi G, Yamamoto
N, Matsushima-Nishiwaki R, Iida Y, Enomoto Y, Iida H, Ogura S,
Otsuka T, et al: αB-crystallin reduces ristocetin-induced soluble
CD40 ligand release in human platelets: Suppression of thromboxane
A2 generation. Mol Med Rep. 12:357–362. 2015.PubMed/NCBI
|
|
39
|
Zhang F, Li ZL, Xu XM, Hu Y, Yao JH, Xu W,
Jing HR, Wang S, Ning SL and Tian XF: Protective effects of
icariin-mediated SIRT1/FOXO3 signaling pathway on intestinal
ischemia/reperfusion-induced acute lung injury. Mol Med Rep.
11:269–276. 2015.PubMed/NCBI
|
|
40
|
Bekker-Méndez C, Guzmán-Aguilar RM,
Hernández-Cueto MA, Huerta-Yepez S, Jarillo-Luna RA,
González-Veyrand E and González-Bonilla CR: TUNEL-positive cells in
the surgical border of an amputation due to infected diabetic foot.
Mol Med Rep. 5:363–372. 2012.PubMed/NCBI
|
|
41
|
Ji F, Zhang H, Wang Y, Li M, Xu W, Kang Y,
Wang Z, Wang Z, Cheng P, Tong D, et al: MicroRNA-133a,
downregulated in osteosarcoma, suppresses proliferation and
promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 56:220–226.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang H, Cai X, Wang Y, Tang H, Tong D and
Ji F: microRNA-143, down-regulated in osteosarcoma, promotes
apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol
Rep. 24:1363–1369. 2010.PubMed/NCBI
|
|
43
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gorlick R: Current concepts on the
molecular biology of osteosarcoma. Cancer Treat Res. 152:467–478.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bacci G, Bertoni F, Longhi A, Ferrari S,
Forni C, Biagini R, Bacchini P, Donati D, Manfrini M, Bernini G and
Lari S: Neoadjuvant chemotherapy for high-grade central
osteosarcoma of the extremity. Histologic response to preoperative
chemotherapy correlates with histologic subtype of the tumor.
Cancer. 97:3068–3075. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jianwei Z, Fan L, Xiancheng L, Enzhong B,
Shuai L and Can L: MicroRNA 181a improves proliferation and
invasion, suppresses apoptosis of osteosarcoma cell. Tumour Biol.
34:3331–3337. 2013. View Article : Google Scholar : PubMed/NCBI
|