Introduction
The myelin basic protein (MBP) gene is a genetic
complex containing three transcription start sites, which generate
two families of proteins. Golli proteins are generated from the
first transcription start site, while the classic MBPs are produced
from the second and third transcription start sites. The classic
MBPs are major constituents of the myelin sheath of
oligodendrocytes and Schwann cells in the central and peripheral
nervous system, respectively. The Golli-MBPs are structurally
associated with the classic MBPs, however are more ubiquitously
expressed, particularly in the immune system, including T
lymphocytes, B lymphocytes and macrophages (1–3). T
lymphocytes serve an important role in the immune system. Numerous
diseases of the immune system are associated with the proliferation
and apoptosis of T lymphocytes. The study of T lymphocytes may aid
in the clarification of the pathogenesis of immune diseases.
Golli-MBP is predominantly expressed in T cells within the immune
system, which can negatively regulate the proliferation and
activation of the T cells through a mechanism involving the
modulation of calcium homeostasis (4,5).
Thus, Golli-MBP may be an important pathogenetic factor associated
with immune diseases.
In the current study, a Golli-MBP-RNAi lentiviral
vector was constructed and it was used to transfect Jurkat cells.
DNA microarray techniques were applied to investigate alterations
in gene expression profiles. DNA microarrays measure the expression
levels of large numbers of genes simultaneously. With the extensive
use of microarrays, expression profiling has identified numerous
applications including the discovery of gene functions and pathway
dissection. In the current study, the Agilent human gene microarray
system was used, which may aid in determining the detailed
functional roles of numerous associated genes in the development of
autoimmune diseases.
Materials and methods
Materials
The T lymphocytic leukemia Jurkat cell line was
purchased from the cell bank of the Chinese Academy of Sciences
(Shanghai, China). Three samples from Golli-MBP knockdown Jurkat
cells and three samples from control Jurkat cells were included in
the study. The Agilent Human Gene Expression system (8*60K; design
ID: 039494; Agilent Technologies, Inc., Santa Clara, CA, USA) was
used in the current study, and data analysis of the 6 samples was
completed.
Lentiviral vector preparation and
transfection
RNAi was used for silencing Golli-MBP in the Jurkat
cells. The target sequence was 5′-GGGAGGACAACACCTTCAA-3′. The sense
strand of the oligonucleotide sequence was
5′-CCGGGAGGGAGGACAACACCTTCAACTCGAGTTGAAGGTGTTGTCCTCCCTCTTTTTG-3′
and the antisense strand was
5′-AATTCAAAAAGAGGGAGGACAACACCTTCAACTCGAGTTGAAGGTGTTGTCCTCCCTC-3′.
The Golli-MBP-RNAi lentiviral vector (GV248-KD-MBP) labeled with
green fluorescent protein was prepared by GeneChem Co., Ltd.
(Shanghai, China). Jurkat cells were cultured in RPMI-1640 medium
at 37°C with 5% CO2 and 30% saturated humidity. The
medium was supplemented with 10% fetal calf serum, 100 IU/ml
penicillin and 100 IU/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Jurkat cells in the
logarithmic growth phase were seeded at a density of
1.5×107 cells/well in six-well plates and were
transfected with GV248-KD-MBP, which also included a blank control
and a negative control. The multiplicity of infection ratio of 80
was selected, in addition, 8 µg/ml polybrene (Takara Bio, Inc.,
Otsu, Japan) was added to increase infection efficiencies.
Subsequent to 72 h incubation, Jurkat cells were observed under an
inverted fluorescence microscope and cells with >80%
transfection efficiency were considered for gene interference.
RNA extraction
Total RNA extraction was performed using a Qiagen
RNeasy Mini Kit (Qiagen China Co., Ltd., Shanghai, China) according
to the manufacturer's instructions. Total RNA was quantified using
NanoDrop ND-2000 (Thermo Fisher Scientific, Inc.) and the RNA
integrity was assessed using an Agilent Bioanalyzer 2100 (Agilent
Technologies, Inc.). RNA samples with RIN≥7.0 and 28S/18S>0.7
were used. All equipment (homogenizers, mortar, pestle etc.) was
pretreated with RNase and rinsed with diethyl pyrocarbonate-treated
water prior to use.
Preparation of fluorescent cDNA
probes
Double-stranded cDNA was synthesized from total RNA
using the SuperScript Double-Stranded cDNA Synthesis kit
(Invitrogen; Thermo Fisher Scientific, Inc.). cRNA was then
obtained from the double-stranded cDNA using the MEGAscript T7 kit
(Ambion; Thermo Fisher Scientific, Inc.). Aliquots of each reaction
mixture were electrophoresed on a 1% agarose gel to determine the
cRNA yield. Subsequently, the cRNA was purified using the RNeasy
Mini protocol. Finally, fluorescent cDNA probes were obtained by
reverse transcription from the cRNA using the CyScribe First-Strand
cDNA labeling kit (GE Healthcare Life Sciences, Piscataway, NJ,
USA) with cyanine-3-cytidine triphosphate.
Gene expression profiling and data
analysis
The labeled cRNAs were hybridized onto the
microarray. Subsequent to washing, the arrays were scanned by the
Agilent Scanner G2505C (Agilent Technologies, Inc.). GeneSpring
feature extraction software (version 10.7.1.1; Agilent
Technologies, Inc.) was used to analyze array images for raw data.
GeneSpring was employed to finish the basic analysis with the raw
data. The raw data were normalized with the quantile algorithm. The
probes with at least 100% of the values in any 1 out of all
conditions with flags in ‘Detected’ were selected for further data
analysis. Differentially expressed genes were then identified
through fold changes in addition to the P-values, which were
calculated with the t-test. The threshold set for up- and
downregulated genes was a fold change ≥2.0 and P≤0.05.
Subsequently, gene ontology (GO) analysis and Kyoto Encyclopedia of
Genes and Genomes analysis were applied to determine the roles of
these differentially expressed mRNAs. Hierarchical clustering was
additionally conducted in order to display the gene expression
patterns among the samples.
Results
Microarray scan image
Microarray scan images broadly reflect the
microarray hybridization conditions. The clearer and more uniform
the images are, the better the hybridization conditions are
(Fig. 1).
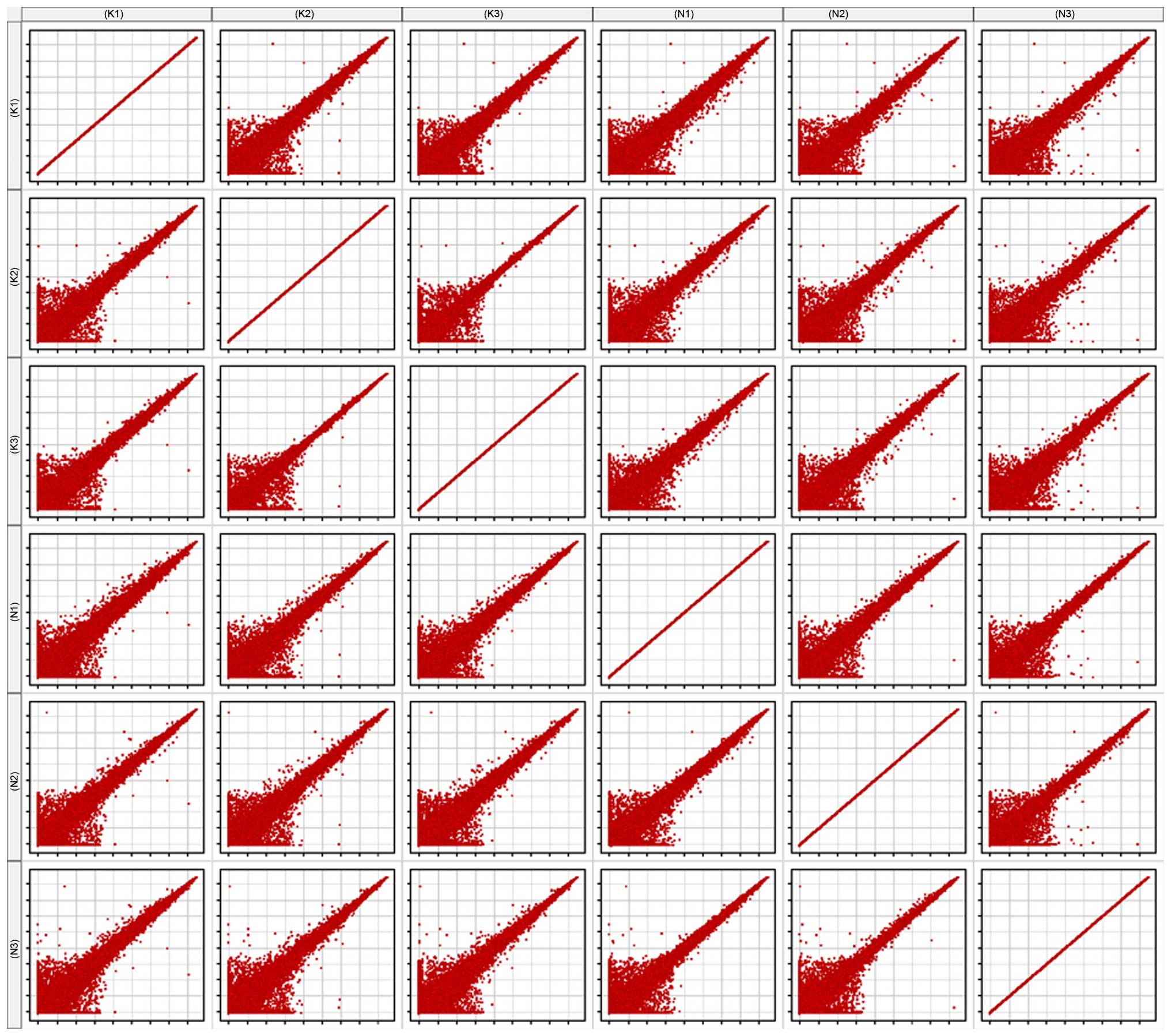
Scatter plot
The scatter plot is commonly used to assess the
variation in RNA expression variation between two samples. Each
point of the scatter plot represents a probe on the chip, and the
position of the point in the two-dimensional plane is determined by
the X-axis and Y-axis coordinates. The signal values of six samples
were normalized (log2-scaled). All six samples were compared with
one another, to generate the matrix plot (Fig. 2).
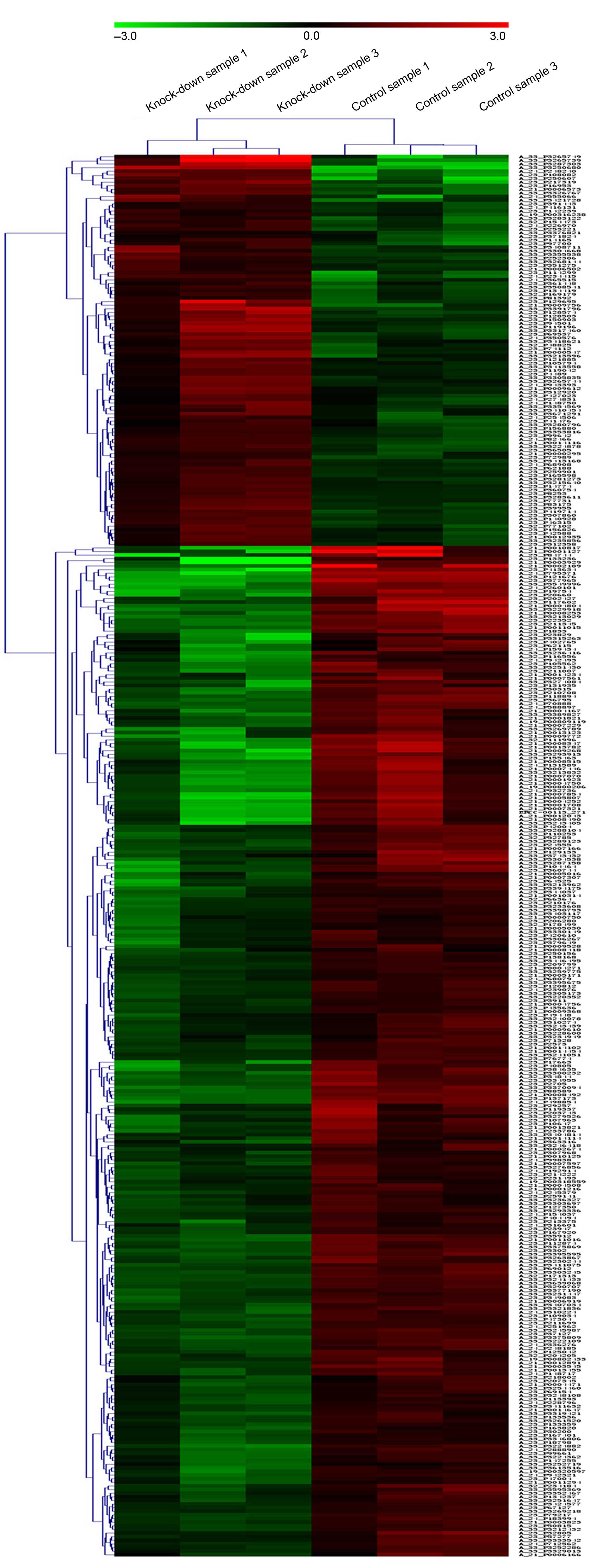
Gene expression alteration
Of the detected genes, 387 genes were differentially
expressed, including 108 upregulated genes and 279 downregulated
genes. Hierarchical clustering was performed to highlight the
differences in gene expression (Fig.
3).
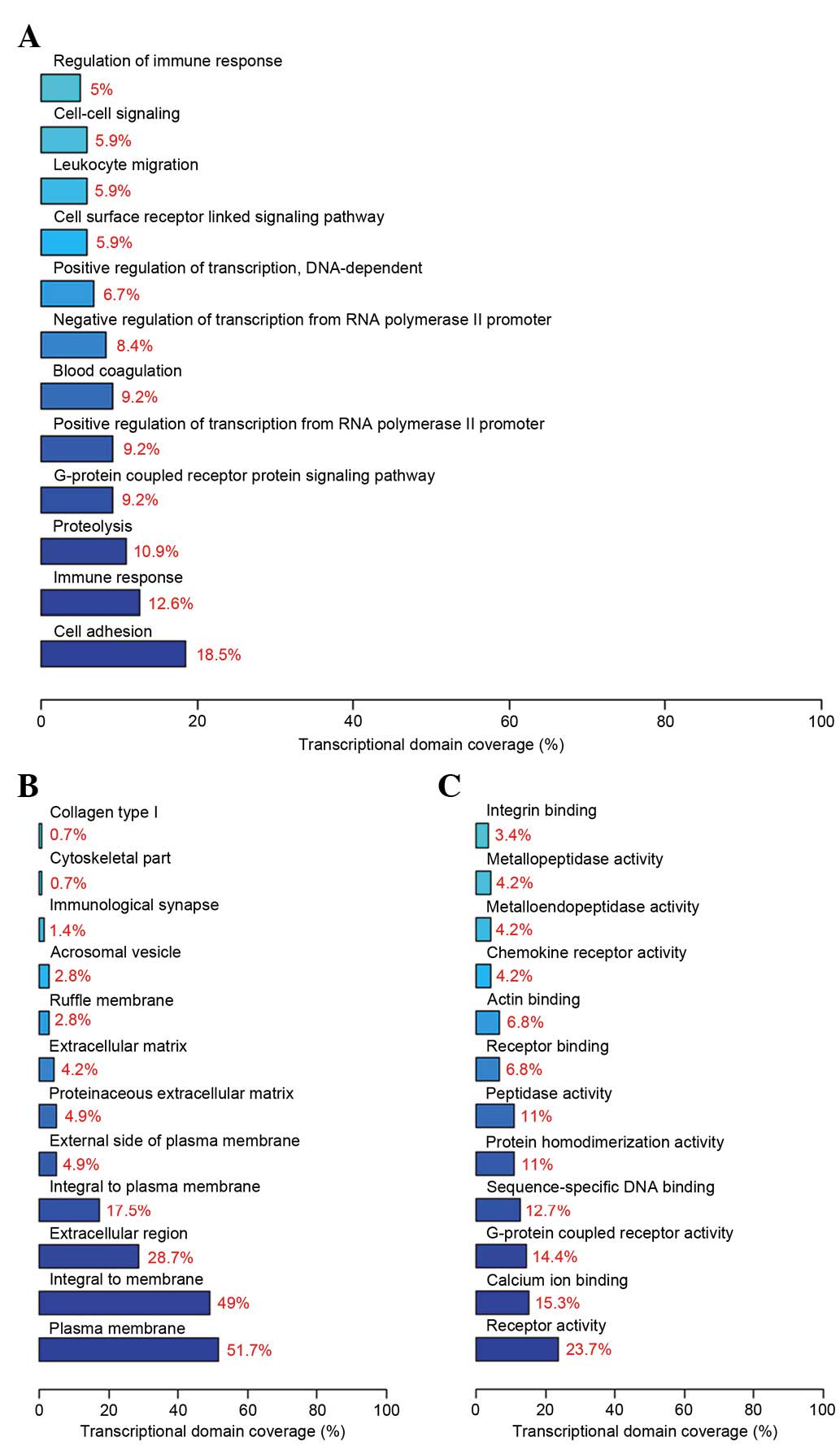
GO analysis and pathway analysis
GO analysis demonstrated that the differentially
expressed genes involve several biological processes, including
cell adhesion, the immune response and proteolysis. The components
of differential genes were predominantly located in the cell
membrane, and the molecular functions included receptor activity,
calcium ion binding and G-protein coupled receptor activity. The
top 12 significant biological processes (Fig. 4A), cell components (Fig. 4B) and molecular functions (Fig. 4C) are presented in Fig. 4.
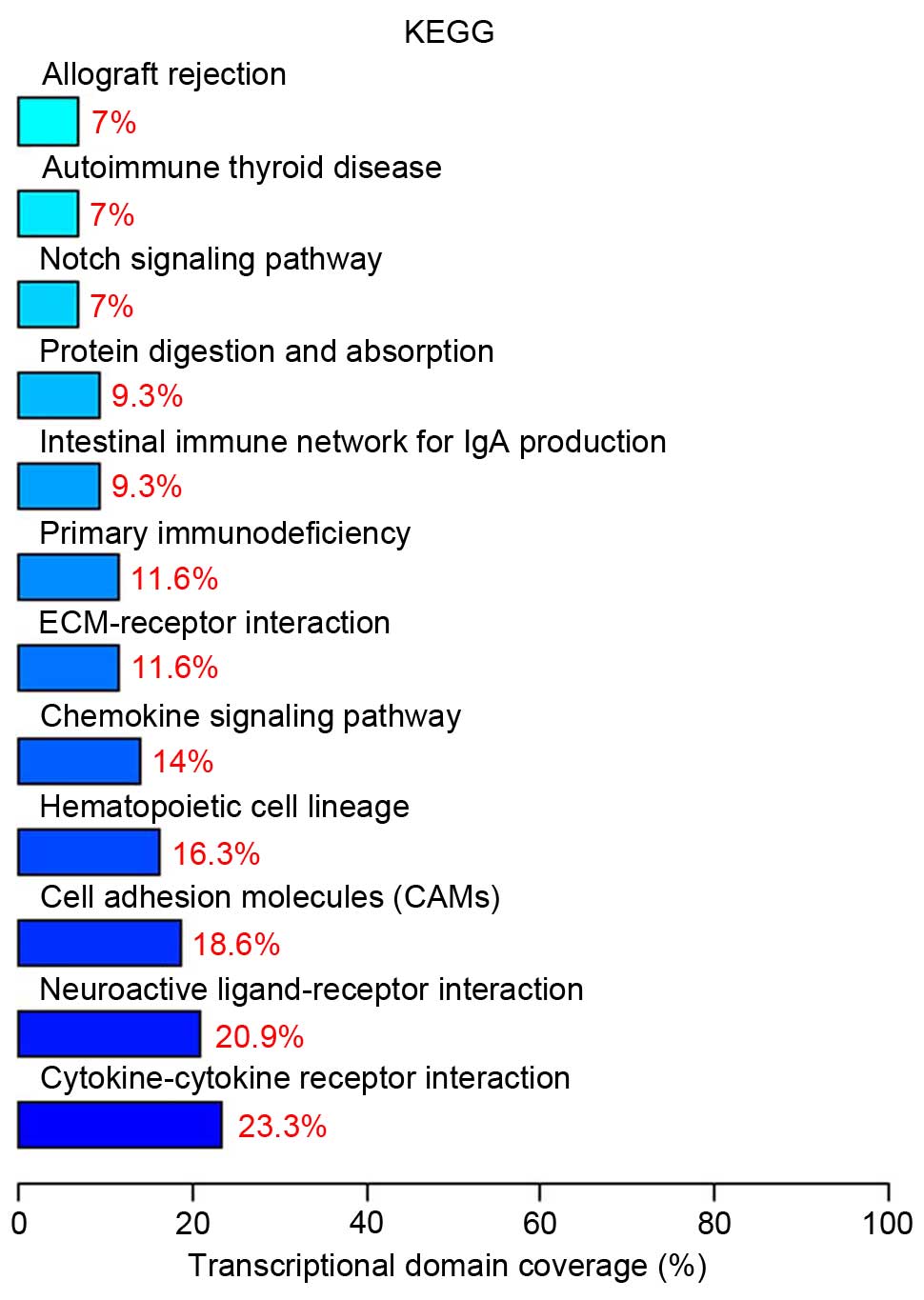
Pathway analysis indicated that 10 differentially
expressed genes (23.3%) are involved in cytokine-cytokine receptor
interaction (Table I). The top
twelve significant signaling pathways are presented in Fig. 5.
 | Table I.Genes involved in the
‘Cytokine-cytokine receptor interaction’ pathway with associated
P-values acquired from the microarray result. |
Table I.
Genes involved in the
‘Cytokine-cytokine receptor interaction’ pathway with associated
P-values acquired from the microarray result.
| Gene name | P-value | Regulation |
|---|
| V-kit Hardy-Zuckerman
4 feline sarcoma viral oncogene homolog | 0.001781 | Down |
| Interleukin 7
receptor | 0.002597 | Down |
| Interleukin 28
receptor α(interferon, λ receptor) | 0.013984 | Up |
| Interleukin 26 | 0.007562 | Up |
| Chemokine (C-X-C
motif) receptor 3 | 0.022956 | Up |
| Chemokine (C-C motif)
receptor 9 | 3.26E-04 | Down |
| Chemokine (C-C motif)
receptor 8 | 6.58E-04 | Down |
| Chemokine (C-C motif)
receptor 4 | 0.003523 | Up |
| Chemokine (C-C motif)
receptor 1 | 0.004038 | Down |
| CD40 ligand | 2.68E-05 | Up |
Discussion
The Golli-MBP protein is distributed in the myelin
sheath of the nervous system, and additionally in T cells,
macrophages and B cells (3). Upon
further study, understanding of the structure and function of
proteins is increased. Golli expression has been previously
observed to be increased in the lymph nodes of mice with relapsing
experimental autoimmune encephalomyelitis (EAE) (6). In heterozygous Golli-MBP knockout
mice, EAE relapses were observed to be reduced (7). Previous studies have identifed that
in addition to acting as an autoantigen in the pathogenesis of
autoimmune diseases, Golli-MBP can be combined with stromal
interaction molecule 1 to negatively regulate the proliferation and
activation of T cells, via a mechanism involving the modulation of
calcium homeostasis, which is known to serve an important role in
the immune system (4,5).
It has been identified that the destruction of MBP
correlates with some autoimmune diseases. The cleavage of MBP has
been demonstrated to lead to the generation of immunogenic
fragments and demyelination in EAE in addition to human multiple
sclerosis (8,9). Previous studies have observed that
Golli-MBP exhibited significantly increased expression in oral
lichen planus (OLP) than in normal controls, A strong and negative
correlation between Golli-MBP and T-helper (Th) 1/Th2 gene
expression was observed in peripheral blood mononuclear cells
(PBMCs) of patients with OLP (10). This implies that Golli-MBP may
influence cytokine gene expression in PBMCs. However, it remains a
challenge to explain the association between the abnormal
expression of Golli-MBP and the disruption of the equilibrium of
Th1/Th2 in patients with OLP. Therefore, the study of alterations
in gene expression in the Jurkat T cell line following
downregulation of the Golli-MBP gene may aid in further
investigation of whether other genes are involved in influencing T
lymphocyte function together with Golli-MBP.
In the current study, DNA microarray was applied and
the results indicated that 387 genes were differentially expressed,
including 108 upregulated genes and 279 downregulated genes. GO
analysis indicated that the differentially expressed genes involved
several biological processes, including cell adhesion, the immune
response and the G-protein coupled receptor protein signaling
pathway. Pathway analysis demonstrated that the majority of the
differentially expressed genes served a role in cytokine-cytokine
receptor interaction, the chemokine signaling pathway and cell
adhesion molecules. Chemokine receptors were involved in several
signaling pathways, and it was hypothesized that Golli-MBP may
serve a role in the T lymphocyte signal transduction pathway via a
chemokine network, which affects autoimmune diseases.
The current study identified that chemokine (C-X-C
motif) receptor (CCR)1, CCR8 and CCR9 were downregulated, while
CXCR3 and CCR4 were upregulated. Chemokines are a large family of
small proteins and act on the G-protein coupled receptor
superfamily. Traditionally, chemokines and their receptors are
divided into four families (CXC, CC, C and CX3C) based on the
pattern of cysteine residues in the ligands (11). It has been previously reported that
chemokine and chemokine receptors are predominantly associated with
the chemotaxis of inflammatory cells and can additionally regulate
the activation of immune cells, the secretion of cytokines, cell
adhesion, cytotoxic effects, hematopoietic progenitor cell growth
and angiogenesis (12). Numerous
previous studies have confirmed that the chemokine receptor is
closely associated wtih the pathogenesis of human immune-associated
diseases, including multiple sclerosis (13), systemic lupus erythematosus
(14), asthma (15) and OLP, which is a chronic
inflammatory disease with small potential to become malignant
(16). The etiology of OLP remains
unclear, however it has been previously reported to be a T
cell-mediated autoimmune reaction (17). Hu et al (18) reported that serum levels of CCL5
and the percentage of CCR5+CD4+T cells was
increased in patients with OLP. Chemokine (C-C motif) ligand 5 and
CCR1 have been additionally observed to be expressed in OLP and are
associated with mast cell migration (19). The chemokine system is a complex
immune regulatory network, and further research into the chemokines
and their associations is required in order to better understand
the role of chemokines in human immune-associated diseases.
Subsequent to knockdown of Golli-MBP, the mechanism
regulating the alterations in the biological characteristics in
Jurkat cells appeared complex and remains unclear. It is suggested
that this may involve numerous types of functional proteins, and
metabolic and signaling pathways. However, chemokine receptors were
significantly correlated factors that may aid in the investigation
of the molecular and biological mechanisms of Golli-MBP in the
signal transduction pathway of T-cell-associated autoimmune
diseases.
Acknowledgements
The current study was supported by the National
Natural Science Foundation of China (grant nos. 81170961 and
81470748) and a project funded by the Priority Academic Program
Development of Jiangsu Higher Education Institutions (grant no.
PAPD 2014-37).
References
|
1
|
Feng JM: Minireview: Expression and
function of golli protein in immune system. Neurochem Res.
32:273–278. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Campagnoni AT, Pribyl TM, Campagnoni CW,
Kampf K, Amur-Umarjee S, Landry CF, Handley VW, Newman SL, Garbay B
and Kitamura K: Structure and developmental regulation of
Golli-mbp, a 105-kilobase gene that encompasses the myelin basic
protein gene and is expressed in cells in the oligodendrocyte
lineage in the brain. J Biol Chem. 268:4930–4938. 1993.PubMed/NCBI
|
|
3
|
Pribyl TM, Campagnoni CW, Kampf K, Kashima
T, Handley VW, McMahon J and Campagnoni AT: The human myelin basic
protein gene is included within a 179-kilobase transcription unit:
Expression in the immune and central nervous systems. Proc Natl
Acad Sci USA. 90:10695–10699. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Feng JM, Fernandes AO, Campagnoni CW, Hu
YH and Campagnoni AT: The golli-myelin basic protein negatively
regulates signal transduction in T lymphocytes. J Neuroimmunol.
152:57–66. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feng JM, Hu YK, Xie LH, Colwell CS, Shao
XM, Sun XP, Chen B, Tang H and Campagnoni AT: Golli protein
negatively regulates store depletion-induced calcium influx in T
cells. Immunity. 24:717–727. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Slavin AJ, Maron R and Weiner HL: Mucosal
administration of IL-10 enhances oral tolerance in autoimmune
encephalomyelitis and diabetes. Int Immunol. 13:825–833. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Voskuhl RR, Pribyl TM, Kampf K, Handley V,
Liu HB, Feng J, Campagnoni CW, Soldan SS, Messing A and Campagnoni
AT: Experimental autoimmune encephalomyelitis relapses are reduced
in heterozygous golli MBP knockout mice. J Neuroimmunol. 139:44–50.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagasato K, Farris RW, Dubois-Dalcq M and
Voskuhl RR: Exon 2 containing myelin basic protein (MBP)
transcripts are expressed in lesions of experimental allergic
encephalomyelitis (EAE). J Neuroimmunol. 72:21–25. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shiryaev SA, Savinov AY, Cieplak P,
Ratnikov BI, Motamedchaboki K, Smith JW and Strongin AY: Matrix
metalloproteinase proteolysis of the myelin basic protein isoforms
is a source of immunogenic peptides in autoimmune multiple
sclerosis. PloS One. 4:e49522009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ding M, Zeng J, Sroussi H, Yu J, Xu J,
Cheng X and Fan Y: Interactions between Golli-MBP and Th1/Th2
cytokines in patients with oral lichen planus. Oral Dis.
20:205–211. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Allen SJ, Crown SE and Handel TM:
Chemokine: Receptor structure, interactions and antagonism. Annu
Rev Immunol. 25:787–820. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bennett LD, Fox JM and Signoret N:
Mechanisms regulating chemokine receptor activity. Immunology.
134:246–256. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cheng W and Chen G: Chemokines and
chemokine receptors in multiple sclerosis. Mediators Inflamm.
2014:6592062014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu SL, Kuan WP, Wong CK, Li EK and Tam LS:
Immunopathological roles of cytokines, chemokines, signaling
molecules and pattern-recognition receptors in systemic lupus
erythematosus. Clin Dev Immunol. 2012:7151902012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Siddiqui S, Secor ER Jr and Silbart LK:
Broncho-alveolar macrophages express chemokines associated with
leukocyte migration in a mouse model of asthma. Cell Immunol.
281:159–169. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Eisen D: The clinical features, malignant
potential and systemic associations of oral lichen planus: A study
of 723 patients. J Am Acad Dermatol. 46:207–214. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mattila R, Ahlfors E and Syrjänen S: CD27
and CD38 lymphocytes are detected in oral lichen planus lesions.
Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 111:211–217.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu JY, Zhang J, Cui JL, Liang XY, Lu R, Du
GF, Xu XY and Zhou G: Increasing CCL5/CCR5 on CD4+ T
cells in peripheral blood of oral lichen planus. Cytokine.
62:141–145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao ZZ, Sugerman PB, Walsh LJ and Savage
NW: Expression of RANTES and CCR1 in oral lichen planus and
association with mast cell migration. J Oral Pathol Med.
31:158–162. 2002. View Article : Google Scholar : PubMed/NCBI
|