Introduction
B. amyloliquefaciens is widely present in
soil, plants and some animals (1–6);
therefore, up to now, several strains of B.
amyloliquefaciens have been isolated. Although different
strains of B. amyloliquefaciens have a similar genome
sequence, they have marked differences in the variable part of the
genome (7); therefore, various
strains of B. amyloliquefaciens may possess different
abilities. Previous studies have demonstrated that numerous strains
of B. amyloliquefaciens exert antifungal (8–10)
and antibacterial activities (6,11–13).
However, these studies have also indicated that various strains
exert different antimicrobial activities. At present, several
studies have suggested that B. amyloliquefaciens may be used
as a potential therapeutic strategy that targets pathogens.
The major antimicrobial components of B.
amyloliquefaciens are cyclic lipopeptides (CLPs) (14–16).
At present, the three major CLPs that are known to possess
antimicrobial activities are surfactin, iturin and fengycin A.
Previous studies have demonstrated that surfactin, iturin and
fengycin A possess antifungal and antibacterial activities
(6,17–19).
Notably, CLPs have several advantages, including low toxicity, high
biodegradability and environmentally positive characteristics
(20–23). These studies indicated that CLPs
may be suitable for the treatment of pathogenic infections. In
addition, other non-CLP antimicrobial peptides exist, such as
amylolysin, which is a type of bacteriocin that is produced by
B. amyloliquefacien (24,25).
The majority of bacteriocins only inhibit gram-positive bacteria,
and certain non-CLPs are produced using the ribosomal synthesis
method with gene modification, which is not the same as in the
Bacillus genus (25).
Therefore, CLPs isolated from B. amyloliquefaciens may be
considered a better choice for the treatment of pathogenic
infections, compared with non-CLPs.
Listeria monocytogenes is a foodborne
pathogen, which induces listeriosis (26,27).
L. monocytogenes is present in several ready-to-eat foods,
including vegetables, poultry and beef, which have been found to
cause human listeriosis (28,29).
Due to clinical severity and the high mortality rates of L.
monocytogenes-induced listeriosis, the control of L.
monocytogenes food contamination is an important issue
(30,31). Previous studies have reported that
bacteriocins produced by B. amyloliquefaciens can inhibit
the growth of L. monocytogenes (32,33).
These studies indicated that products of B.
amyloliquefaciens may be useful to control growth of L.
monocytogenes; however, whether CLPs produced by B.
amyloliquefaciens may target L. monocytogenes remains
unclear.
Staphylococcus aureus can cause clinical
acute hematogenous, osteomyelitis, endocarditis, pulmonary
abscesses and spondylodiscitis (34–36).
Methicillin-resistant S. aureus (MRSA) is not inhibited by
treatment with several antibiotics (37,38).
MRSA is a major and serious pathogen, which is particularly
associated with nosocomial infection (39,40).
Since numerous antibiotics can not effectively inhibit MRSA, the
development of a novel anti-MRSA drug is important. A previous
study indicated that B. amyloliquefaciens produced
amylolysin, a ribosomally synthesized peptide, which can inhibit
MRSA (25). However, to the best
of our knowledge, whether CLPs produced by B.
amyloliquefaciens can target MRSA has not yet been
reported.
Based on the findings of previous studies, the
present study aimed to determine whether CLPs isolated from B.
amyloliquefaciens purified from chicken intestines can exert
antibacterial effects on MRSA and L. monocytogenes.
Materials and methods
Materials
Difco™ agar and Luria broth (LB) culture medium were
obtained from BD Biosciences (Franklin Lakes, NJ, USA). The
polymerase chain reaction (PCR) primers for 16S ribosomal (r)RNA
and DNA gyrase, subunit B (gyrB) genes were synthesized by MD Bio
(Taipei, Taiwan). The API® 50 CHB system assay kit was
obtained from bioMerieux (Marcy-l'Étoile, France). Bond
Elut® C18 was purchased from Agilent Technologies (Santa
Clara, CA, USA). The polytetrafluoroethylene membrane (JP020) was
obtained from Advantec Co., Ltd. (Tokyo, Japan). The indicator
strains Staphylococcus epidermidis and Bacillus
cereus were obtained from Bioresource Collection and Research
Center (Hsinchu, Taiwan). The indicator strains Streptococcus
pyogenes, Listeria monocytogenes, Clostridium tyrobutyricum,
Escherichia coli, Helicobacter pylori, Salmonella typhimurium,
Pseudomonas aeruginosa, Aspergillus flavus var. flavus,
Aspergillus niger and Penicillium pinophilum were
obtained from American Type Culture Collection (Manassas, VA, USA).
MRSA HCT20 was kindly provided by the Department of Pathology and
Laboratory Medicine, Taichung Veterans General Hospital (Taichung,
Taiwan). The iturin A and surfactin standards were purchased from
Sigma-Aldrich; Merck Millipore (Darmstadt, Germany).
Selection of potential bacterial
strains from chicken intestines
Potential bacterial strains with antimicrobial
activities isolated from chicken intestines (obtained from a local
market, Taichung, Taiwan) using microscopic examination and streak
plate methods were selected and determined using the modified
spot-on-the-lawn method, as previously described (41,42).
Briefly, 0.7% top agar containing 105 colony-forming
units (CFU)/ml indicator strains (A. niger or P.
pinophilum) was overlaid onto a potato dextrose agar (PDA)
plate, dried for 30 min, and subsequently spotted with 2 µl
(108 CFU/ml) of an overnight culture of the candidate
strains. Subsequently, the PDA plates were incubated at 25°C for 5
days. Zones of inhibition are found in the sites containing
potential strains with antimicrobial activities.
Identification of potential bacterial
strain
The B. amyloliquefaciens JN68 strain isolated
from chicken intestines was identified by analyzing data from the
API® 50 CHB system (43,44),
as well as the results of a 16S rRNA and gyrB sequence detection
(45–47). The results were analyzed on the
National Center for Biotechnology Information BLAST tool
(blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome).
According to morphological and physiological characteristics,
biological testing and comparison of the 16S rRNA sequence, the
strain was identified as B. amyloliquefaciens. Briefly,
phenotypic characterization and sugar fermentation pattern of the
candidate strain was analyzed according to standard methods, using
the API® 50 CHB system assay kit. The obtained sugar
fermentation pattern was analyzed using APILAB Plus software
version 3.2.2 (bioMerieux). Further strain identification was
confirmed by PCR detection of 16S rRNA and gyrB. Briefly, the
candidate strain was cultured in LB culture medium (30°C, 200 ×
g, 8–12 h) and the genomic DNA was extracted using the Blood
and Tissue Genomic DNA Extraction Miniprep (Viogene BioTek
Corporation, New Taipei City). The amplification of the 16S rRNA
and gyrB gene region of candidate strain was conducted in a 50 µl
reaction mixture containing 1 µl genomic DNA template, 5 µl PCR
reaction buffer [10 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2,
50 mM KCl and 0.1% Triton X-100] containing 200 µM of each
deoxynucleoside triphosphate (GE Healthcare Life Sciences,
Chalfont, UK), 4 µl MgCl2 (52 mM), 1 µM each primer
(Table I), 1 unit of Taq
polymerase (GE Healthcare Life Sciences) and 39 µl autoclaved
Milli-Q water (Merck Millipore). The amplification was performed
with thermocycling conditions as follows: Denaturation for 10 min
at 95°C; 35 cycles (as presented in Table I); termination at 72°C for 7 min.
The PCR was conducted in a Robocycler® temperature
thermal cycler (Agilent Technologies, Inc., Santa Clara, CA, USA).
The PCR products were sequenced using an automatic sequencer (ABI
PRISM® 3730; Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Databases were screened for
similarities using Basic Local Alignment Search Tool (48) and the alignment of overlapping
fragments was performed using Vector NTI Advance 10 Contig Express
software (Thermo Fisher Scientific, Inc.).
 | Table I.Primer sequences and PCR conditions
used in the present study. |
Table I.
Primer sequences and PCR conditions
used in the present study.
| First author,
year | Target gene and
primers | Sequence (5′ to
3′) | PCR conditions | Product size | Refs. |
|---|
| Wang, 2007 | gyrB degenerate
primers |
| 94°C ×1 min/55°C
×30 sec/72°Cx1 min | 898 bp | (47) |
|
|
UP-1S |
CGTAGAGCCACTTGAGCG |
|
|
UP-2Sr |
CTGCCGTTACAGTTCCTT |
| Perea Vélez,
2007 | 16S universal
primer for bacteria |
| 94°C ×1 min/45°C
×30 sec/72°Cx1 min | ~1.5 kb | (51) |
|
|
Pro26 |
AGAGTTTGATCCTGGCTCAG |
|
|
Pro27 |
AAGGAGGTGATCCAGCCGCA |
Culture conditions and isolation of
the lipopeptide fraction
The B. amyloliquefaciens JN68 strain was
cultured and activated with 5 ml LB culture medium at 30°C.
Subsequently, 5% B. amyloliquefaciens JN68 was seeded into
200 ml number 3 medium (49).
After incubation under an agitation rate of ~200 rpm at 30°C for 4
days, the fermentation medium was centrifuged at 11,000 × g
for 20 min at 4°C. The supernatant was collected and filtered
through a polytetrafluoroethylene membrane (0.22 µM pore size). The
filtered supernatant was then treated with 6 N HCl (pH 2.0) at 4°C
overnight, and was further centrifuged at 11,000 × g for 20
min at 4°C. The sediment was washed with PBS and extracted three
times with methanol. The methanolic extracts were concentrated and
dissolved in methanol. Finally, the purified CLPs of the B.
amyloliquefaciens JN68 strain were obtained from the methanolic
extracts using the Bond Elut® C18 (5 g) mini column as
previously described (50).
Identification of CLP structure by
liquid chromatography-electrospray ionization-mass spectrometry
(LC-ESI-MS)/MS
Purified CLPs produced by B.
amyloliquefaciens JN68 were obtained from methanolic extracts
as aforementioned. Subsequently, the structure of the CLPs was
detected using LC and MS. For LC, an Agilent 1200 series system
(Agilent Technologies), including a binary pump and an autosampler,
was used for the chromatographic separation. The separation was
conducted using an Atlantis C18 column (150×2.1 mm, 3.5 µm; Waters
UK, Elstree, UK) at 30°C with a flow rate of 200 µl/min using the
following gradient: i) 50% B for 2 min; ii) a gradient of 50–100% B
for 10 min; iii) 100% B for 10 min; iv) re-equilibration with 50% B
for 10 min. Mobile phase A consisted of 10% methanol and mobile
phase B consisted of 100% methanol. The injection volume was set to
10 µl. For MS analysis an LTQ XL™ Linear Ion Trap MS was used
(Thermo Fisher Scientific, Inc.), which was equipped with an ESI
source. MS was operated in positive-ion mode with a spray voltage
of 5.0 KV, and a capillary voltage of 40 V. Capillary temperature
and sheath gas (N2) were set at 275°C and 20 arbitrary
units, respectively. The precursor ion width was set at 3 Da and
the normalized collision energy was set at 35%. For LC-ESI-MS/MS,
the MS was operated in the selected target precursor ions as
follows: m/z 1,036, surfactin; 1,043, iturin A; 1464, fengycin A,
using the Linear Ion Trap MS. The three major CLP components
containing surfactin, iturin A and fengycin A were analyzed in the
present study as previously described (52–54).
Antimicrobial profile assay
The antimicrobial activities of the purified CLPs
produced by B. amyloliquefaciens JN68 were determined using
an agar well diffusion assay, as previously described (41,55).
Briefly, small holes were dug into agar plates using a 200 µl
pipette tip. Each indicator microorganism (~106 CFU/ml
bacterium or 105 CFU/ml fungus) was spread onto an agar
plate. Subsequently, 50 µl B. amyloliquefaciens JN68
extracts were seeded into the holes on the agar plates for 2 h at
4°C. The agar plates were then incubated for 16–18 h at 37°C
(indicator bacteria) or at 30°C (indicator fungi). After a 16–18 h
incubation, the appearance of the zones of inhibition was
determined.
Results
Screening of potential antimicrobial
strains
Several bacterial strains were obtained from the
chicken intestines. In order to identify potential strains with
antimicrobial activities, the spot-on-the-lawn method was used
(41). In the present study,
bacterial strains isolated from chicken intestines were spotted
onto top agar plates containing indicator strains (A. niger
or P. pinophilum). Zones of inhibition are found in the
sites containing potential strains with antimicrobial activities.
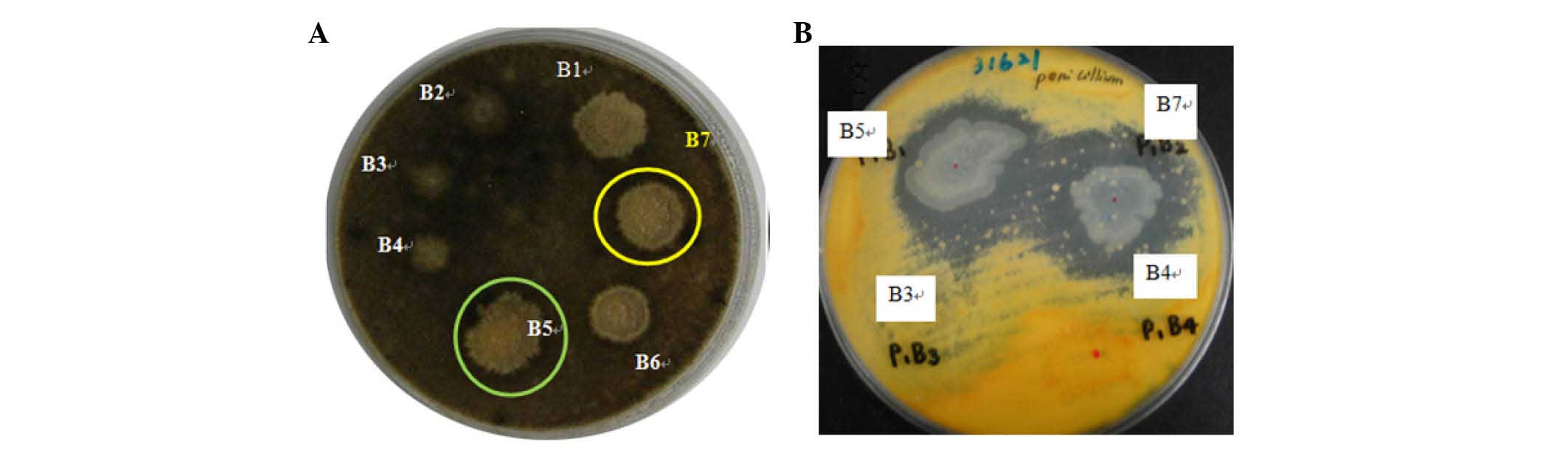
The results indicated that marked zones of inhibition appeared at
the B5 and B7 sites; these sites therefore contained potential
strains that can effectively inhibit the growth of A. niger
(Fig. 1A) and P. pinophilum
(Fig. 1B). Therefore, the strains
at the B5 and B7 sites were considered to possess antimicrobial
activities. In the present study, the potential bacterium at the B7
site was selected and analyzed (identical to B5).
Identification of the bacterial
strain
The potential bacteria at the B7 site was primarily
identified using the API® 50 CHB system (Table II). Subsequently, the potential
bacteria at the B7 site was further confirmed by 16S rRNA and gyrB
sequence detection. The PCR primer sequences of 16S rRNA and gyrB
used in the present study are presented in Table I. Analysis of the API®
50 CHB system and gene sequence detection indicated that the
potential bacteria at the B7 site belonged to the Bacillus
amyloliquefaciens family. In the present study, this potential
bacterial strain was named Bacillus amyloliquefaciens
JN68.
 | Table II.Sugar fermentation pattern of the
candidate strain was determined using the API 50 CHB system. |
Table II.
Sugar fermentation pattern of the
candidate strain was determined using the API 50 CHB system.
| Carbohydrate
fermentation |
|---|
|
|---|
| Active
ingredient | Response |
|---|
| Control | − |
| Glycerol | + |
| Erythritol | − |
| D-arabinose | − |
| L-arabinose | + |
| D-ribose | + |
| D-cylose | w |
| L-cylose | − |
| D-adonitol | − |
|
β-methyl-D-xylopyranoside | − |
| D-galactose | − |
| D-glucose | + |
| D-fructose | + |
| D-mannose | + |
| L-sorbose | − |
| L-rhamnose | − |
| Dulcitol | − |
| Inositol | + |
| D-mannitol | + |
| D-sorbitol | + |
| Methyl
α-D-mannopyranoside | − |
| Methyl
α-D-glucopyranoside | + |
| N-acetyl
glucosamine | − |
| Amygdalin |
|
| Arbutin | + |
| Esculin (ferric
citrate) | + |
| Salicin | + |
| D-cellobiose | + |
| D-maltose | + |
| D-lactose (bovine
origin) | w |
| D-melibiose | − |
| D-saccharose
(sucrose) | + |
| D-trehalose | + |
| Inulin | − |
| D-melezitose | − |
| D-raffinose | − |
| Amidon
(starch) | w |
| Glycogen | ± |
| Xylitol | − |
| Gentiobiose | w |
| D-turanose | − |
| D-lyxose | − |
| D-tagatose | − |
| D-fucose | − |
| L-fucose | − |
| D-Arabitol | − |
| L-arabitol | − |
| Potassium
giuconate | − |
| Potassium
2-ketogluconate | − |
| Potassium
5-ketogluconate | − |
Determination of CLP extract
components
LC-ESI-MS/MS analysis of the CLP extracts is
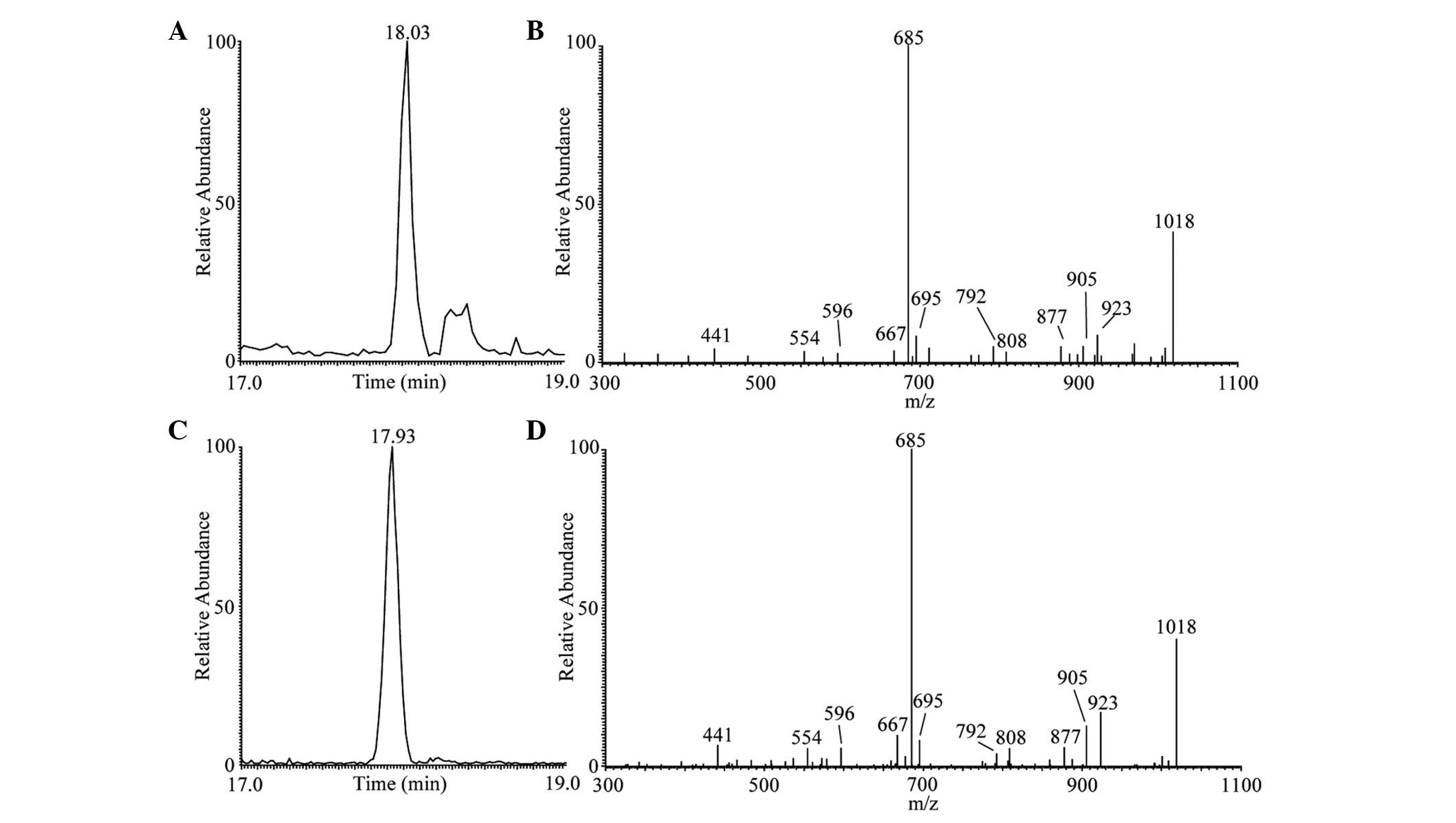
presented in Figs. 2–4. As shown in Fig. 2C, the main peak of surfactin
lipopeptide was revealed at retention time (Rt) 17.93 min
corresponding to protonated molecules [M+H]+ at m/z
1,036 in positive modality. The representative MS/MS spectrum is
shown in Fig. 2D. These results
were similar to those observed during surfactin standard
LC-ESI-MS/MS analysis (Fig. 2A and
B). This analysis confirmed the presence of one main surfactin
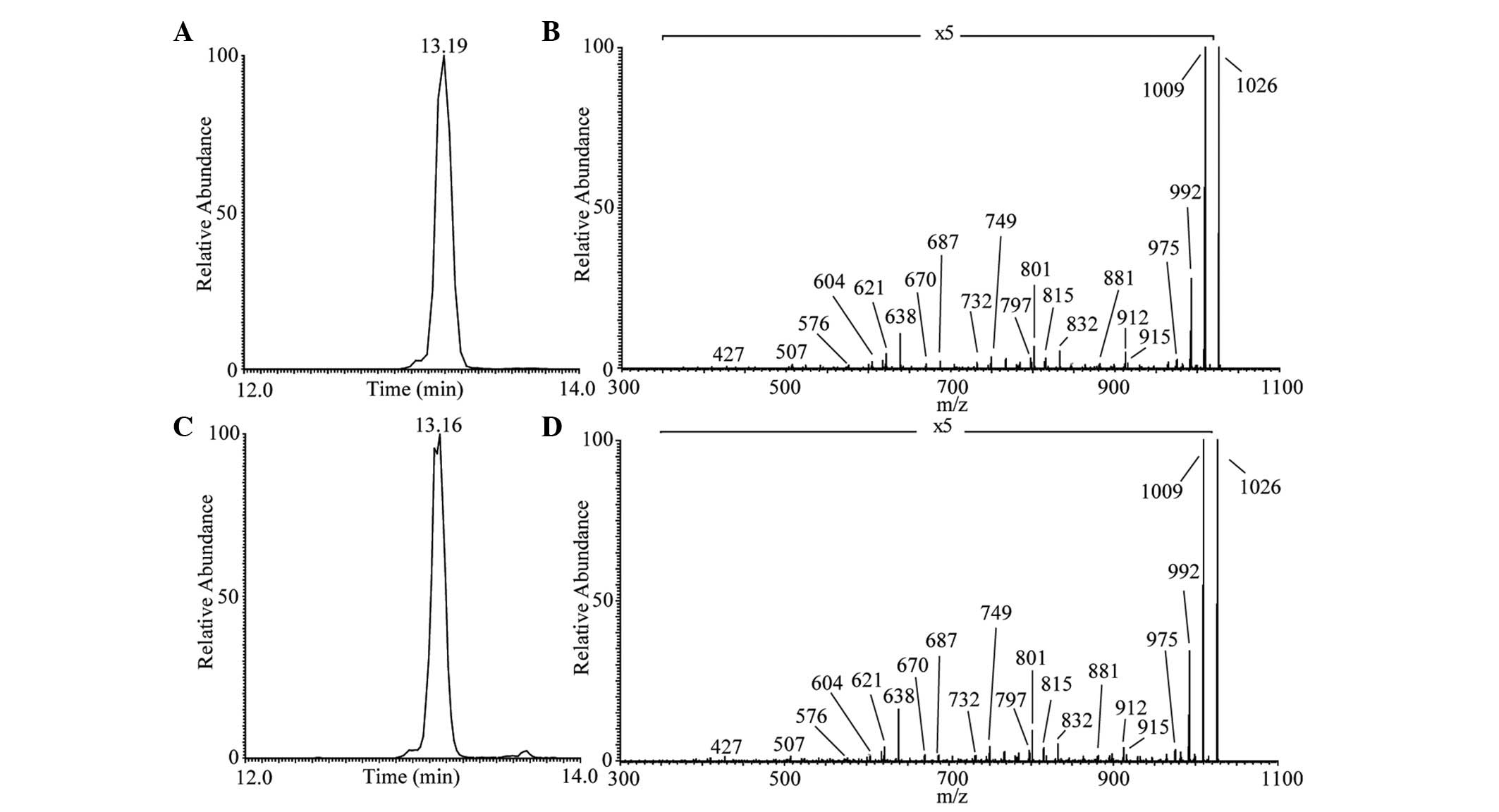
in the extracted CLPs. As shown in Fig. 3C, the one main peak of iturin A
lipopeptide was revealed at Rt 13.16 min corresponding to
the protonated molecules [M+H]+ at m/z 1,043 in positive
modality. The representative MS/MS spectrum is shown in Fig. 3D. The Rt and product ions
were similar to those obtained during iturin A standard
LC-ESI-MS/MS analysis (Fig. 3A and
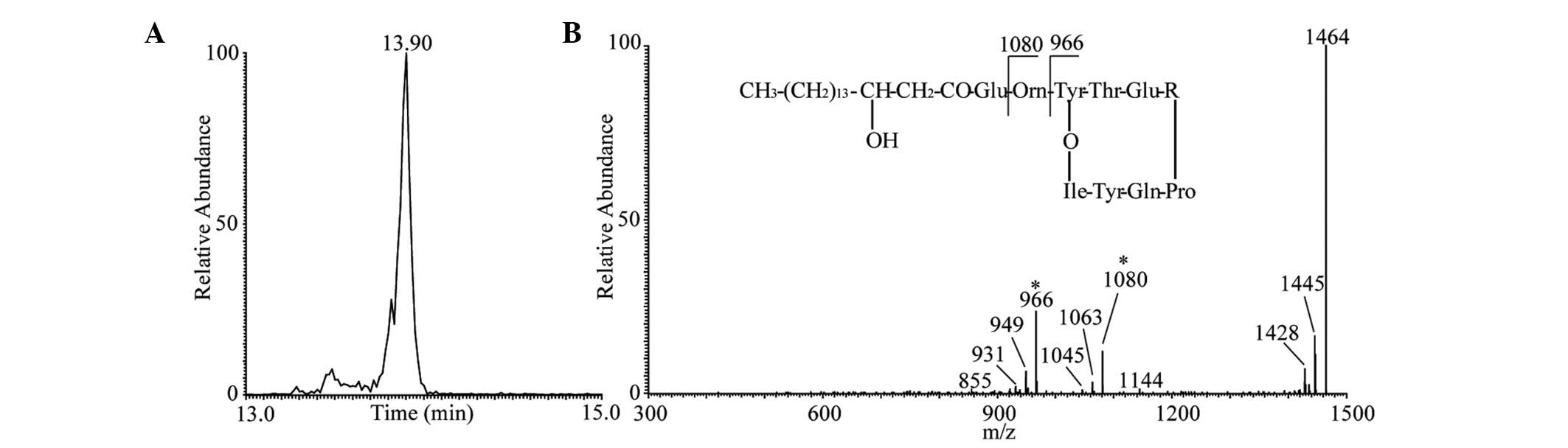
B). The results of the LC-ESI-MS/MS analysis of fengycin A
lipopeptide are presented in Fig.
4. The one main peak of fengycin A was observed at Rt
13.90 min corresponding to the protonated molecules
[M+H]+ at m/z 1,464 in positive modality (Fig. 4A). The representative MS/MS
spectrum is shown in Fig. 4B.
Although no fengycin A standard was used in the present study, the
product ions were consistent with those from previous studies,
which reported that product ions at m/z 1,080 and 966 were
representative ions present in the fengycin A MS/MS spectrum at m/z
1,464 (56,57). Taking into account the results
obtained by LC-MS/MS analysis of CLPs, there are at least three
lipopeptide groups within the whole B. amyloliquefaciens
extract, which included surfactant, iturin A and fengycin A.
Antimicrobial spectrum of the
potential strain
Previous studies have demonstrated that CLPs
isolated from various B. amyloliquefaciens strains possess
distinct antifungal (9,17,58,59)
and antibacterial activities (60–63).
However, to the best of our knowledge, no previous experiment has
demonstrated that CLPs isolated from B. amyloliquefaciens
can inhibit MRSA and L. monocytogenes. The present study
indicated that CLPs containing surfactin, iturin and fengycin A,
isolated from B. amyloliquefaciens JN68, can inhibit several
pathogenic fungal and bacterial strains (Table III). Notably, the present study
is the first, to the best of our knowledge, to indicate that CLPs
isolated from B. amyloliquefaciens JN68 can inhibit the
growth of MRSA and L. monocytogenes (Table III).
 | Table III.Antimicrobial spectrum of CLPs
produced by Bacillus amyloliquefacients JN68 using the agar
well diffusion method. |
Table III.
Antimicrobial spectrum of CLPs
produced by Bacillus amyloliquefacients JN68 using the agar
well diffusion method.
|
| Antimicrobial
activitya |
|---|
|
|
|
|---|
| Indicator
strain | Growth medium | Unpurified CLP |
|---|
| Gram-positive |
|
|
| HCT20
MRSA | TSA | +++ |
|
Staphylococcus
epidermidis BCRC15245 | NA | ++ |
|
Streptococcus pyogenes
ATCC12344 | TSA with 5%
blood | +++ |
|
Listeria monocytogenes
ATCC15313 | BHI | +++ |
|
Clostridium
tyrobutyricum ATCC25755 | NA | + |
|
Bacillus cereus
BCRC10250 | NA | + |
| Gram-negative |
|
|
|
Escherichia coli
ATCC11775 (10675) | NA | ++ |
|
Helicobacter pylori
ATCC43526 | Brucella with 3%
blood | + |
|
Salmonella typhimurium
ATCC13311 | TSA | +++ |
|
Pseudomonas aeruginosa
ATCC9027 (11633) | NA | ++ |
| Mold |
|
|
|
Aspergillus flavus var.
flavusb
ATCC26770 | PDA | +++ |
|
Aspergillus niger
ATCC16404 | PDA | + |
|
Penicillium pinophilum
ATCC9644 | PDA | +++ |
Discussion
At present, various B. amyloliquefaciens
strains have been isolated from plants and soil; in addition, some
B. amyloliquefaciens strains have been found to be present
in animals (1–6). Several studies have reported that
extracts of various B. amyloliquefaciens strains exert
antimicrobial activities (12,64,65).
However, to the best of our knowledge, no previous studies have
reported that B. amyloliquefaciens strains isolated from
chickens possess antimicrobial activities. The present study
isolated the B. amyloliquefaciens JN68 strain from chicken
intestines, and confirmed it possessed antimicrobial activities. At
present, several components have been identified that exert
antimicrobial activities, including CLPs (15,66,67),
antifungal enzymes (5,68) and non-CLPs (24,25).
Among these components, CLPs exert more broad-spectrum antifungal
and antibacterial activities, compared with antifungal enzymes and
non-CLPs (17,64). In general, CLPs can exert
antifungal and antibacterial activities (against both gram-positive
and gram-negative bacteria) (64,67),
whereas antifungal enzymes only exert antifungal activities
(5,68) and non-CLPs exert only antibacterial
activities against gram-positive bacteria (24,25).
In the present study, the CLP extracts purified from the B.
amyloliquefaciens JN68 strain exerted antifungal and
antibacterial activities on gram-positive and gram-negative
bacteria.
Since CLPs produced by B. amyloliquefaciens
possess several advantages, including broad-spectrum antimicrobial
activities, low toxicity, high biodegradability and environmentally
positive characteristics (20–23,64),
CLPs may therefore be considered potential antimicrobials for food
and clinical application. Previous studies have reported that
surfactin, iturin and fengycin A are major CLP components that
exert antimicrobial activities (6,17–19).
In the present study, the extracts purified from the B.
amyloliquefaciens JN68 strain were confirmed to be surfactin,
iturin and fengycin A. Since various B. amyloliquefaciens
strains have similar genome sequences, with the exception of
differences in the variable region of the genome (7), there are several types and expression
levels of surfactin, iturin and fengycin A in the different strains
(69). This may be why CLPs
produced by different strains possess distinct antimicrobial
activities.
Previous studies have reported that CLPs produced by
some B. amyloliquefaciens strains possess antifungal
activities, including the CGMCC5569, NJN-6, Q-426 and PPCB004
strains (9,17,58,59).
Furthermore, CLPs produced by some B. amyloliquefaciens
strains possess antibacterial activities, including FZB42,
HR62, NJN-6 and B9601-Y2 strains (60–63).
In addition, CLPs produced by some B. amyloliquefaciens
strains possess antifungal and antibacterial activities, such
as the NJN-6 and B9601-Y2 strains (9,62,63).
Compared with previous studies, antifungal and antibacterial
activities were detected in the B. amyloliquefaciens JN68
strain isolated from chicken intestines in the present study. It is
well known that MRSA is an important bacterial species that causes
nosocomial infections (39,40),
whereas L. monocytogenes induces listeriosis, which has a
high mortality rate (30,31). No previous studies, to the best of
our knowledge, have demonstrated that CLPs produced by B.
amyloliquefaciens can inhibit the growth of MRSA and L.
monocytogenes. Therefore, the present study is the first to
indicate that the B. amyloliquefaciens JN68 strain can
inhibit MRSA and L. monocytogenes.
In conclusion, the present study isolated B.
amyloliquefaciens JN68 from chicken intestines, and confirmed
it possessed antifungal and antibacterial activities. In
particular, B. amyloliquefaciens JN68 was able to inhibit
the growth of MRSA and L. monocytogenes. These results
suggested that the B. amyloliquefaciens JN68 strain may be
applied to prevent the occurrence of foodborne diseases and
nosocomial infections.
Acknowledgements
The present study was supported by grants from the
Taipei Tzu Chi Hospital (grant nos. TCRD-TPE-104-34 and
TCRD-TPE-104-06).
References
|
1
|
Niazi A, Manzoor S, Bejai S, Meijer J and
Bongcam-Rudloff E: Complete genome sequence of a plant associated
bacterium Bacillus amyloliquefaciens subsp. plantarum UCMB5033.
Stand Genomic Sci. 9:718–725. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cai J, Liu F, Liao X and Zhang R: Complete
genome sequence of Bacillus amyloliquefaciens LFB112 isolated from
Chinese herbs, a strain of a broad inhibitory spectrum against
domestic animal pathogens. J Biotechnol. 175:63–64. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ait Kaki A, Chaouche N Kacem, Dehimat L,
Milet A, Youcef-Ali M, Ongena M and Thonart P: Biocontrol and plant
growth promotion characterization of Bacillus species isolated from
calendula officinalis rhizosphere. Indian J Microbiol. 53:447–452.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Geetha I, Aruna R and Manonmani AM:
Mosquitocidal Bacillus amyloliquefaciens: Dynamics of growth &
production of novel pupicidal biosurfactant. Indian J Med Res.
140:427–434. 2014.PubMed/NCBI
|
|
5
|
Han JH, Shim H, Shin JH and Kim KS:
Antagonistic activities of Bacillus spp. strains isolated from
tidal flat sediment towards anthracnose pathogens Colletotrichum
acutatum and C. gloeosporioides in South Korea. Plant Pathol J.
31:165–175. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu HM, Rong YJ, Zhao MX, Song B and Chi
ZM: Antibacterial activity of the lipopetides produced by Bacillus
amyloliquefaciens M1 against multidrug-resistant Vibrio spp.
isolated from diseased marine animals. Appl Microbiol Biotechnol.
98:127–136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ruckert C, Blom J, Chen X, Reva O and
Borriss R: Genome sequence of B. amyloliquefaciens type strain
DSM7(T) reveals differences to plant-associated B.
amyloliquefaciens FZB42. J Biotechnol. 155:78–85. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu Z, Shao J, Li B, Yan X, Shen Q and
Zhang R: Contribution of bacillomycin D in Bacillus
amyloliquefaciens SQR9 to antifungal activity and biofilm
formation. Appl Environ Microbiol. 79:808–815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yuan J, Raza W, Huang Q and Shen Q: The
ultrasound-assisted extraction and identification of antifungal
substances from B. amyloliquefaciens strain NJN-6 suppressing
Fusarium oxysporum. J Basic Microbiol. 52:721–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao P, Quan C, Jin L, Wang L, Wang J and
Fan S: Effects of critical medium components on the production of
antifungal lipopeptides from Bacillus amyloliquefaciens Q-426
exhibiting excellent biosurfactant properties. World J Microbiol
Biotechnol. 29:401–409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chi Z, Rong YJ, Li Y, Tang MJ and Chi ZM:
Biosurfactins production by Bacillus amyloliquefaciens R3 and their
antibacterial activity against multi-drug resistant pathogenic E.
coli. Bioprocess Biosyst Eng. 38:853–861. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kadaikunnan S, Rejiniemon T, Khaled JM,
Alharbi NS and Mothana R: In-vitro antibacterial, antifungal,
antioxidant and functional properties of Bacillus
amyloliquefaciens. Ann Clin Microbiol Antimicrob. 14:92015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao H, He S, Wei R, Diong M and Lu L:
Bacillus amyloliquefaciens G1: A potential antagonistic bacterium
against eel-pathogenic aeromonas hydrophila. Evid Based Complement
Alternat Med. 2011:8241042011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma Z, Hu J, Wang X and Wang S: NMR
spectroscopic and MS/MS spectrometric characterization of a new
lipopeptide antibiotic bacillopeptin B1 produced by a marine
sediment-derived Bacillus amyloliquefaciens SH-B74. J Antibiot
(Tokyo). 67:175–178. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mora I, Cabrefiga J and Montesinos E:
Cyclic lipopeptide biosynthetic genes and products, and inhibitory
activity of plant-associated Bacillus against phytopathogenic
bacteria. PLoS One. 10:e01277382015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nihorimbere V, Cawoy H, Seyer A, Brunelle
A, Thonart P and Ongena M: Impact of rhizosphere factors on cyclic
lipopeptide signature from the plant beneficial strain Bacillus
amyloliquefaciens S499. FEMS Microbiol Ecol. 79:176–191. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arrebola E, Jacobs R and Korsten L: Iturin
A is the principal inhibitor in the biocontrol activity of Bacillus
amyloliquefaciens PPCB004 against postharvest fungal pathogens. J
Appl Microbiol. 108:386–395. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arguelles-Arias A, Ongena M, Halimi B,
Lara Y, Brans A, Joris B and Fickers P: Bacillus amyloliquefaciens
GA1 as a source of potent antibiotics and other secondary
metabolites for biocontrol of plant pathogens. Microb Cell Fact.
8:632009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vágvölgyi C, Sajben-Nagy E, Bóka B, Vörös
M, Berki A, Palágyi A, Krisch J, Skrbić B, Durišić-Mladenović N and
Manczinger L: Isolation and characterization of antagonistic
Bacillus strains capable to degrade ethylenethiourea. Curr
Microbiol. 66:243–250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim PI, Bai H, Bai D, Chae H, Chung S, Kim
Y, Park R and Chi YT: Purification and characterization of a
lipopeptide produced by Bacillus thuringiensis CMB26. J Appl
Microbiol. 97:942–949. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maget-Dana R and Peypoux F: Iturins, a
special class of pore-forming lipopeptides: Biological and
physicochemical properties. Toxicology. 87:151–174. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stein T: Bacillus subtilis antibiotics:
Structures, syntheses and specific functions. Mol Microbiol.
56:845–857. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoshida S, Hiradate S, Tsukamoto T,
Hatakeda K and Shirata A: Antimicrobial activity of culture
filtrate of Bacillus amyloliquefaciens RC-2 isolated from mulberry
leaves. Phytopathology. 91:181–187. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Benitez L, Correa A, Daroit D and
Brandelli A: Antimicrobial activity of Bacillus amyloliquefaciens
LBM 5006 is enhanced in the presence of Escherichia coli. Curr
Microbiol. 62:1017–1022. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Arias A Arguelles, Ongena M, Devreese B,
Terrak M, Joris B and Fickers P: Characterization of amylolysin, a
novel lantibiotic from Bacillus amyloliquefaciens GA1. PLoS One.
8:e830372013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salazar JK, Wu Z, McMullen PD, Luo Q,
Freitag NE, Tortorello ML, Hu S and Zhang W: PrfA-like
transcription factor gene lmo0753 contributes to L-rhamnose
utilization in Listeriamonocytogenes strains associated with human
food-borne infections. Appl Environ Microbiol. 79:5584–5592. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shalaby MA, Mohamed MS, Mansour MA and Abd
El-Haffiz AS: Comparison of polymerase chain reaction and
conventional methods for diagnosis of Listeria monocytogenes
isolated from different clinical specimens and food stuffs. Clin
Lab. 57:919–924. 2011.PubMed/NCBI
|
|
28
|
Neetoo H, Ye M and Chen H: Potential
antimicrobials to control Listeria monocytogenes in vacuum-packaged
cold-smoked salmon pâté and fillets. Int J Food Microbiol.
123:220–227. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pérez-Rodriguez F, van Asselt ED,
Garcia-Gimeno RM, Zurera G and Zwietering MH: Extracting additional
risk managers information from a risk assessment of Listeria
monocytogenes in deli meats. J Food Prot. 70:1137–1152. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Althaus D, Lehner A, Brisse S, Maury M,
Tasara T and Stephan R: Characterization of Listeria monocytogenes
strains isolated during 2011–2013 from human infections in
Switzerland. Foodborne Pathog Dis. 11:753–758. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pontello M, Guaita A, Sala G, Cipolla M,
Gattuso A, Sonnessa M and Gianfranceschi MV: Listeria monocytogenes
serotypes in human infections (Italy, 2000–2010). Ann Ist Super
Sanita. 48:146–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lisboa MP, Bonatto D, Bizani D, Henriques
JA and Brandelli A: Characterization of a bacteriocin-like
substance produced by Bacillus amyloliquefaciens isolated from the
Brazilian Atlantic forest. Int Microbiol. 9:111–118.
2006.PubMed/NCBI
|
|
33
|
Sağdiç O, Ozkan G, Ozcan M and Ozçelik S:
A study on inhibitory effects of Siğla tree (Liquidambar orientalis
Mill. var. orientalis) storax against several bacteria. Phytother
Res. 19:549–551. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Agarwal A and Aggarwal AN: Bone and joint
infections in children: Acute hematogenous osteomyelitis. Indian J
Pediatr. 2015.
|
|
35
|
Christiansen JG, Jensen HE, Johansen LK,
Kochl J, Koch J, Aalbaek B, Nielsen OL and Leifsson PS: Porcine
models of non-bacterial thrombotic endocarditis (NBTE) and
infective endocarditis (IE) caused by Staphylococcus aureus: A
preliminary study. J Heart Valve Dis. 22:368–376. 2013.PubMed/NCBI
|
|
36
|
Vos FJ, Kullberg BJ, Sturm PD, Krabbe PF,
van Dijk AP, Wanten GJ, Oyen WJ and Bleeker-Rovers CP: Metastatic
infectious disease and clinical outcome in Staphylococcus aureus
and Streptococcus species bacteremia. Medicine (Baltimore).
91:86–94. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hasanvand A, Ghafourian S, Taherikalani M,
Jalilian FA, Sadeghifard N and Pakzad I: Antiseptic resistance in
methicillin sensitive and methicillin resistant Staphylococcus
aureus isolates from some major hospitals, Iran. Recent Pat
Antiinfect Drug Discov. 10:105–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kaur DC and Chate SS: Study of antibiotic
resistance pattern in methicillin resistant Staphylococcus aureus
with special reference to newer antibiotic. J Glob Infect Dis.
7:78–84. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
McMaster J, Booth MG, Smith A and Hamilton
K: Meticillin-resistant Staphylococcus aureus in the intensive care
unit: Its effect on outcome and risk factors for acquisition. J
Hosp Infect. 90:327–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dancer SJ: Controlling hospital-acquired
infection: Focus on the role of the environment and new
technologies for decontamination. Clin Microbiol Rev. 27:665–690.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lima ET, Filho RL Andreatti, Okamoto AS,
Noujaim JC, Barros MR and Crocci AJ: Evaluation in vitro of the
antagonistic substances produced by Lactobacillus spp. isolated
from chickens. Can J Vet Res. 71:103–107. 2007.PubMed/NCBI
|
|
42
|
Chen H, Wang L, Su CX, Gong GH, Wang P and
Yu ZL: Isolation and characterization of lipopeptide antibiotics
produced by Bacillus subtilis. Lett Appl Microbiol. 47:180–186.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Østensvik Ø, From C, Heidenreich B,
O'Sullivan K and Granum PE: Cytotoxic Bacillus spp. belonging to
the B. cereus and B. subtilis groups in Norwegian surface waters. J
Appl Microbiol. 96:987–993. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zaghloul TI, Al-Bahra M and Al-Azmeh H:
Isolation, identification, and keratinolytic activity of several
feather-degrading bacterial isolates. Appl Biochem Biotechnol
70–72. 207–213. 1998. View Article : Google Scholar
|
|
45
|
Marroki A, Zúñiga M, Kihal M and
Pérez-Martinez G: Characterization of lactobacillus from algerian
goat's milk based on phenotypic, 16S rDNA sequencing and their
technological properties. Braz J Microbiol. 42:158–171. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tajbakhsh M, Nayer BN, Motavaze K,
Kharaziha P, Chiani M, Zali MR and Klena JD: Phylogenetic
relationship of Salmonella enterica strains in Tehran, Iran, using
16S rRNA and gyrB gene sequences. J Infect Dev Ctries. 5:465–472.
2011.PubMed/NCBI
|
|
47
|
Wang LT, Lee FL, Tai CJ and Kasai H:
Comparison of gyrB gene sequences, 16S rRNA gene sequences and
DNA-DNA hybridization in the Bacillus subtilis group. Int J Syst
Evol Microbiol. 57:1846–1850. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Altschul SF, Madden TL, Schäffer AA, Zhang
J, Zhang Z, Miller W and Lipman DJ: Gapped BLAST and PSI-BLAST: A
new generation of protein database search programs. Nucleic Acids
Res. 25:3389–3402. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tsuge K, Akiyama T and Shoda M: Cloning,
sequencing, and characterization of the iturin A operon. J
Bacteriol. 183:6265–6273. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Razafindralambo H, Paquot M, Hbid C,
Jacques P, Destain J and Thonart P: Purification of antifungal
lipopeptides by reversed-phase high-performance liquid
chromatography. J Chromatogr. 639:81–85. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Perea Vélez M, Hermans K, Verhoeven TL,
Lebeer SE, Vanderleyden J and De Keersmaecker SC: Identification
and characterization of starter lactic acid bacteria and probiotics
from Columbian dairyproducts. J Appl Microbiol. 103:666–674. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zeriouh H, de Vicente A, Pérez-Garcia A
and Romero D: Surfactin triggers biofilm formation of Bacillus
subtilis in melon phylloplane and contributes to the biocontrol
activity. Environ Microbiol. 16:2196–2211. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhao X, Han Y, Tan XQ, Wang J and Zhou ZJ:
Optimization of antifungal lipopeptide production from Bacillus sp.
BH072 by response surface methodology. J Microbiol. 52:324–332.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang J, Liu J, Wang X, Yao J and Yu Z:
Application of electrospray ionization mass spectrometry in rapid
typing of fengycin homologues produced by Bacillus subtilis. Lett
Appl Microbiol. 39:98–102. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tagg JR and McGiven AR: Assay system for
bacteriocins. Appl Microbiol. 21:9431971.PubMed/NCBI
|
|
56
|
Pecci Y, Rivardo F, Martinotti MG and
Allegrone G: LC/ESI-MS/MS characterisation of lipopeptide
biosurfactants produced by the Bacillus licheniformis V9T14 strain.
J Mass Spectrom. 45:772–778. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Vanittanakom N, Loeffler W, Koch U and
Jung G: Fengycin-a novel antifungal lipopeptide antibiotic produced
by Bacillus subtilis F-29-3. J Antibiot (Tokyo). 39:888–901. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhao P, Quan C, Wang Y, Wang J and Fan S:
Bacillus amyloliquefaciens Q-426 as a potential biocontrol agent
against Fusarium oxysporum f. sp. spinaciae. J Basic Microbiol.
54:448–456. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yuan B, Wang Z, Qin S, Zhao GH, Feng YJ,
Wei LH and Jiang JH: Study of the anti-sapstain fungus activity of
Bacillus amyloliquefaciens CGMCC 5569 associated with Ginkgo biloba
and identification of its active components. Bioresour Technol.
114:536–541. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kröber M, Wibberg D, Grosch R, Eikmeyer F,
Verwaaijen B, Chowdhury SP, Hartmann A, Pühler A and Schlüter A:
Effect of the strain Bacillus amyloliquefaciens FZB42 on the
microbial community in the rhizosphere of lettuce under field
conditions analyzed by whole metagenome sequencing. Front
Microbiol. 5:2522014.PubMed/NCBI
|
|
61
|
Huang J, Wei Z, Tan S, Mei X, Shen Q and
Xu Y: Suppression of bacterial wilt of tomato by bioorganic
fertilizer made from the antibacterial compound producing strain
Bacillus amyloliquefaciens HR62. J Agric Food Chem. 62:10708–10716.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yuan J, Zhang F, Wu Y, Zhang J, Raza W,
Shen Q and Huang Q: Recovery of several cell pellet-associated
antibiotics produced by Bacillus amyloliquefaciens NJN-6. Lett Appl
Microbiol. 59:169–176. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
He P, Hao K, Blom J, Rückert C, Vater J,
Mao Z, Wu Y, Hou M, He P, He Y and Borriss R: Genome sequence of
the plant growth promoting strain Bacillus amyloliquefaciens subsp.
plantarum B9601-Y2 and expression of mersacidin and other secondary
metabolites. J Biotechnol. 164:281–291. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Compaoré CS, Nielsen DS, Sawadogo-Lingani
H, Berner TS, Nielsen KF, Adimpong DB, Diawara B, Ouédraogo GA,
Jakobsen M and Thorsen L: Bacillus amyloliquefaciens ssp. plantarum
strains as potential protective starter cultures for the production
of Bikalga, an alkaline fermented food. J Appl Microbiol.
115:133–146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hajji S, Ghorbel-Bellaaj O, Younes I,
Jellouli K and Nasri M: Chitin extraction from crab shells by
Bacillus bacteria. Biological activities of fermented crab
supernatants. Int J Biol Macromol. 79:167–173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Alvarez F, Castro M, Principe A, Borioli
G, Fischer S, Mori G and Jofré E: The plant-associated Bacillus
amyloliquefaciens strains MEP2 18 and ARP2 3 capable of producing
the cyclic lipopeptides iturin or surfactin and fengycin are
effective in biocontrol of sclerotinia stem rot disease. J Appl
Microbiol. 112:159–174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hsieh FC, Lin TC, Meng M and Kao SS:
Comparing methods for identifying Bacillus strains capable of
producing the antifungal lipopeptide iturin A. Curr Microbiol.
56:1–5. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wang SL, Shih IL, Liang TW and Wang CH:
Purification and characterization of two antifungal chitinases
extracellularly produced by Bacillus amyloliquefaciens V656 in a
shrimp and crab shell powder medium. J Agric Food Chem.
50:2241–2248. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ongena M and Jacques P: Bacillus
lipopeptides: Versatile weapons for plant disease biocontrol.
Trends Microbiol. 16:115–125. 2008. View Article : Google Scholar : PubMed/NCBI
|