Introduction
Sepsis is a systemic inflammatory response syndrome
caused by infection, which may lead to multiple organ dysfunction.
It is the leading cause of mortality in critically ill patients
(1). The inflammatory response is
primarily mediated by white blood cells (WBCs), including
neutrophils (Neu), lymphocytes (Lym), and monocytes (Mon) (2,3).
Previous studies have suggested that the inflammatory response may
be regulated by histone methylation (4–9).
Histone methylation regulates transcription and
chromatin dynamics, and has roles in pathogenic development and the
maintenance of normal physiology (10,11).
Methylation occurs on lysine and arginine residues of histone
proteins (12). Each lysine may
undergo three different states of methylation, with one (mono), two
(di), or three (tri) potential methyl groups being covalently
attached to the amine group of the lysine side chain, and arginine
may be mono- or di-methylated symmetrically or asymmetrically
(13,14). Histone methylation may repress or
activate transcription, depending on the specific residues and
modification states. Histone 3 lysine 9 (H3K9), H3K27 and H4K20
methylation are associated with transcriptional repression, whereas
methylation of H3K4, H3K36 and H3K79 are associated with
transcriptional activation (15–17).
Histone methylation is a reversible process,
catalyzed by two antagonistic groups of enzymes, histone
methyltransferases (HMTs) and demethylases (HDMs), with the levels
of each of these determining the biological outcome (18–21).
Certain studies have revealed that the process of inflammation and
infection in inflammatory cells is regulated by histone
methylation, including H3K4, H3K9 and H3K27 methylation (4–9,22–24).
Specific HMTs or HDMs were revealed to link histone methylation to
transcriptional genes of inflammation/infection, including nuclear
factor κB (NF-κB), tumor necrosis factor-α and interleukin-6, when
inflammatory cells, particularly Mon or macrophages, were
investigated in vitro (4–9,22–24).
The majority of studies that have been performed examined H3K9
methylation, and H3K9 dimethylation (H3K9me2) is primarily
associated with the inflammatory response (4–9,22–24).
However, the specific histone methylation alterations that occur in
the peripheral WBCs of patients with critical illness and sepsis
remain to be elucidated.
The present study hypothesized that histone
methylation in peripheral WBCs from patients is important in sepsis
occurrence and progression. The current study established and
optimized an immunofluorescence staining method to investigate
H3K9me2 alterations in WBCs from critically ill patients. The pilot
data provided clinical evidence for the hypothesis of histone
methylation in WBCs, and suggested that the characteristic changes
of histone methylation are a potential diagnostic and prognostic
biomarker for septic patienthowever, further evaluation in a larger
cohort is required.
Materials and methods
Study design
The present study was approved by the Research
Ethics Board of Zhongshan Hospital, Fudan University (Shanghai,
China). Written informed consent was obtained from all recruited
patients. The majority of patients admitted to the surgical
intensive care unit (ICU) of Zhongshan Hospital were suffering from
postoperative trauma or esophageal cancer, and were frequently
affected by sepsitherefore, these two groups of patients became the
focus of the present study. To minimize individual differences,
each group comprised a similar number of patients within the same
age range. Only male patients were enrolled to avoid gender bias. A
total of 43 patients (23 trauma and 20 esophageal canceage range,
40–75 years) were enrolled in the present study between October
2012 and December 2014. The enrolled patients were divided into
four groups: i) Trauma, ii) trauma+sepsis, iii) cancer and iv)
cancer+sepsis. Exclusion criteria were: Patients with trauma and
cancer, endocrine disorders (including diabetes and
hyperthyroidism), obesity [body weight 20% greater than the ideal
standard, which is equal to height (cm)-105], or any other chronic
organ impairments. None of the cases had infectious diseases for
more than two weeks prior to the sample collection.
Traumatic insults resulted from road accidents,
accidental traumas and falls. The pathological diagnosis of all
enrolled esophageal cancer patients was squamous cell carcinoma,
grade II–III. Sepsis patients (12 trauma; 11 postoperative
esophageal cancer) were diagnosed according to the 2001
International Sepsis Definition Conference (25). The clinical and biochemical
parameters of the subjects were recorded, including number, age and
gender, Acute Physiology and Chronic Health Evaluation II (APACHE
II) score (26), Sequential Organ
Failure Assessment (SOFA) score (27), microbiological findings, site of
infection, length of ICU stay, mortality occurring in ICU,
C-reactive protein (CRP), procalcitonin and predominant variables
of routine blood tests.
Sample collection and preparation
Blood samples were collected within the first 24 h
following operations in nonseptic patients, and following diagnosis
of sepsis in septic patients. The blood was obtained using an
existing arterial or venous catheter and transferred into a sterile
heparin anticoagulatant vacutainer. Healthy volunteers donated 5 ml
blood samples via the elbow vein. The samples were processed within
2 h following collection.
Samples were centrifuged at 1,200 x g for 15
min at 4°C and the plasma supernatant was collected for storage.
Red blood cell lysis buffer (3 ml; Beyotime Institute of
Biotechnology, Haimen, China) was added, and the blood cell samples
were mixed at room temperature for 10 min, centrifuged at 1,000 x
g for 2 min at 4°C, and the supernatant discarded. WBC
samples were washed with PBS buffer 2–3 times. 293T cells (human
embryonic kidney cells, kindly granted by Prof. Charlie Degui Chen,
Shanghai Institutes for Biological Sciences, Chinese Academy of
Sciences, China) were grown in Iscove's DMEM containing in 10%
fetal calf serum medium for 48 h. An aliquot of WBCs and 293T cells
in a ratio of 4:1 were mixed and resuspended in PBS. Following
this, 10 µl of the mixed cell suspension (~1×104 cells)
was dropped vertically onto a slide. Following drying of the slide,
the cells were fixed in 4% paraformaldehyde for 10 min, and washed
with PBS 2–3 times. Finally, the slides were stored at −80°C until
measurement.
Western blot analysis
Proteins from the prepared WBCs were extracted using
SDS lysis buffer (Beyotime Institute of Biotechnology). To examine
H3K9me2 levels in WBCs, a 10 µg extract of cell proteins from these
cells was separated on a 15% SDS-PAGE gel. The proteins were
transferred to a polyvinyl difluoride membrane in blocking reagent
[5% milk in Tris-buffered saline (TBST)] at room temperature for 30
min. The membrane was washed with 1X TBST three times. A mouse
histone H3K9me2-specific antibody (1:2,000; cat. no. ab1220; Abcam,
Cambridge, MA, USA) was used for western blot analysis, and a
rabbit histone H3 antibody (1:5,000; cat. no. ab1791; Abcam) served
as a loading control. The primary antibodies were incubated with
the membrane at room temperature for 90 min. The membrane was then
washed with 1X TBST three times, and incubated with goat anti-mouse
and goat anti-rabbit horseradish peroxidase-conjugated secondary
antibodies (1:10,000; cat. nos. 115-035-003 and 111-035-045;
Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) at
room temperature for 60 min. Enhanced chemiluminescence
(SuperSignal West Pico Chemiluminescent Substrate; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) was subsequently used to
visualize the blot.
Immunofluorescence staining
The stored leukocyte slides were defrosted at room
temperature for 20 min. Following washing with PBS, the cells were
permeabilized with cold PBS containing 0.2% Triton X-100 for 5 min.
Subsequent to blocking in 1% bovine serum albumin (Roche
Diagnostics, Basel, Switzerland) in PBS at room temperature for 30
min, the cells were incubated with a H3K9me2 antibody (1:800; cat.
no. ab1220; Abcam) in a humidified chamber at 37°C for 60 min, then
incubated with the Cy3-labeled goat anti-mouse IgG (1:1,000; cat.
no. 115-165-146; Jackson ImmunoResearch Laboratories, Inc.) at room
temperature for 60 min, and stained with DAPI, and mounted prior to
viewing. Fields with appropriate cell density were selected and
observed by fluorescence microscopy with the exposure time of 2
msec for H3K9me2 and 130 msec for DAPI. The total leukocyte cell
counts in each patient sample were all >100 in 5 different
fields.
The fluorescence intensity of different cells on the
slide was analyzed by Image-Pro Plus software version 6.0 (Media
Cybernetics, Inc., Rockville, MD, USA). The software allowed the
staining intensity and area of each cell to be presented in one
table. The quantitative intensity of the three WBC types in each
sample was calculated as the mean intensity of all observed cells.
All data were analyzed by two independent investigators, and the
intensity values quantified individually were averaged by
calculation of the mean to obtain the final intensity. Microsoft
Excel software 2007 (Microsoft Corporation, Redmond, WA, USA) was
used to calculate the fluorescence intensity of different
cells.
Statistical analysis
The data were analyzed using the unpaired Student's
t-test for comparison of two groups, and one way analysis of
variance for data from all four groups followed by the Bonferroni
post-hoc test. All analyses were performed using Stata software
version 10.0MP (StataCorp LP, College Station, TX, USA). Data are
presented as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient characteristics
Table I presents a
comparison of characteristics between non-septic and septic trauma
(n=23) and esophageal cancer (n=20) patients. Routine blood tests
revealed a normal WBC count in patients in the trauma group;
however, levels of Neu and Mon in the other three groups were
elevated, along with a reduced percentage of Lym. It was
demonstrated that there were significant differences in the
percentages of increased Neu and decreased Lym between sepsis and
non-sepsis trauma patients. Additionally, the level of CRP was
elevated in sepsis compared with non-sepsis grouphowever, this was
not the case for procalcitonin (Table
I).
 | Table I.Clinical and biochemical parameters of
study subjects. |
Table I.
Clinical and biochemical parameters of
study subjects.
| Variable | Trauma | Trauma+sepsis | Cancer | Cancer+sepsis |
|---|
| Primary etiology | Trauma | Esophageal squamous
cell carcinoma |
| Number | 11 | 12 | 9 | 11 |
| Gender | Male |
| Age (years) | 52.46±17.17 | 62.17±18.47 | 56.11±7.29 | 62.55±9.34 |
| APACHE II score | 2.64±2.11 |
16.58±6.58b | 3.56±1.24 |
15.27±7.07b |
| SOFA score | 0.45±0.69 |
7.33±4.89b | 1.33±0.87 |
8.36±5.90b |
| Infection |
|
|
|
|
|
Gram-positive bacteria | NA | 5 | NA | 4 |
|
Gram-negative bacteria | NA | 3 | NA | 6 |
| Site of
infection |
|
|
|
|
|
Pulmonary | NA | 8 | NA | 8 |
|
Abdominal | NA | 5 | NA | 3 |
|
Other | NA | 0 | NA | 1 |
| Patients with organ
insufficiency | 0 | 6 | 0 | 7 |
| Length of ICU stay
(days) | 1±0 |
21.33±14.25b | 9.11±7.36 | 15.63±9.23 |
| Mortality occurring
in ICU | 0 | 4 | 0 | 2 |
| WBC
(×109/l) | 7.31±2.03 |
12.27±4.67b | 13.63±3.52 | 14.84±4.96 |
| Neu
(%) | 62.75±7.15 |
87.27±5.27b | 83.13±2.30 |
88.78±3.39b |
| Lym
(%) | 28.51±7.91 |
7.62±4.56b | 7.49±3.59 | 5.82±1.87 |
| Mon
(%) | 6.53±1.96 | 5.20±2.04 | 7.91±1.08 |
4.63±2.18b |
| Procalcitonin
(µg/l) | 0.23±0.04 |
0.17±0.07a | 0.24±0.06 | 0.20±0.07 |
| C-reactive protein
(mg/l) | 9.75±21.23 |
154.07±109.64b | 40.95±27.84 |
91.74±77.18a |
Western blot analysis
The present study conducted a western blot analysis
of the total level of histone methylation in whole WBCs in sepsis
and non-sepsis patients. Notably, H3 from each sample contained
three bands, with the largest band corresponding to the intact H3
probed with histone H3K9me2 antibody (data not shown), suggesting
that the two smaller bands were derived from cleavage of histone H3
following K9 residues in WBCs. H3 cleavage following residues A21
to K27 in mouse embryonic stem cells has been reported previously
(28,29), suggesting that the cleaved
N-terminal fragments, including A1-A21, A1-A23 and A1-A27,
containing the target K9 residue, escaped detection. The present
study therefore failed to obtain the total level of histone H3K9me2
from whole WBCs by western blot analysis. The levels of histone
methylation from specific types of leukocytes were not identified
or compared.
Immunofluorescence staining of
WBCs
To examine the level of histone methylation in
different WBCs, the present study used an immunofluorescence
staining method. Considering the complex cell components, wide
range of cell number, and short survival time in vitro of
peripheral WBCs, the leukocytes were directly separated within 2 h
following blood collection to obtain the maximum number of live
cells. Various suspension solvents and fixed slides were used to
analyze leukocytes, due to complications in fixing the suspended
WBCs onto a glass slide, and the suspension, blood smear and drop
methods were compared. It was concluded that use of fresh leukocyte
samples, PBS for cell suspension and the drop method enabled
optimal immunofluorescence staining of WBCs.
To control for the influence of different staining
operations and compare the immunofluorescence between samples, 293T
cells were co-stained alongside WBCs, and served as a reference.
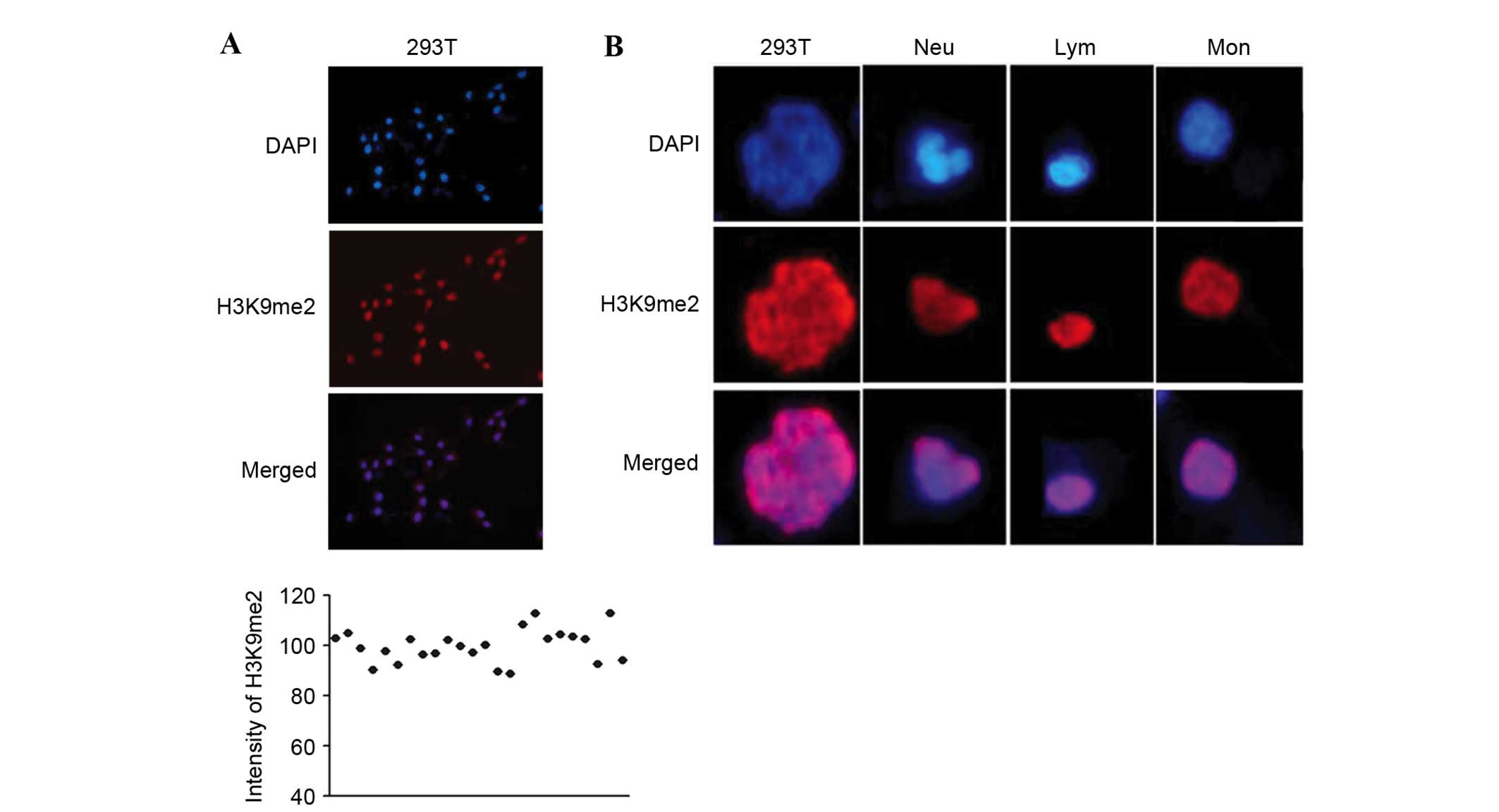
293T cells have stable and homogeneous staining intensity (Fig. 1A), and contain nuclei with a shape
distinct to the three primary types of WBCs, Neu, Lym and Mon. Neu
has a lobulated or rod-shaped nucleus with loose chromatin; Lym has
a round or oval nucleus with dense chromatin; and Mon is larger
than Lym, and has a kidney- or horseshoe-shaped nucleus with loose
chromatin. The nuclei of 293T cells are irregular, round and
significantly larger than those of WBCs (Fig. 1B).
Sepsis-induced alterations of H3K9me2
in WBCs from trauma and cancer patients
Immunostaining analysis detected different levels of
histone H3K9me2 in the three types of WBCs from trauma patients.
The staining intensity of Lym was stronger than that of Neu and Mon
(Fig. 2). The levels of H3K9me2
were elevated in different WBCs from cancer patients, and
demonstrated a similar level of intensity to that exhibited by 293T
cells (Fig. 3). In the trauma +
sepsis group, the levels of H3K9me2 in all of the three WBCs
increased ~20%, this was particularly observed for Neu (P=0.0005)
and Mon (P=0.0002; Figs. 2 and
4). Notably, the presence of
sepsis significantly reduced the elevation of H3K9me2 in cancer
patients by 20 to 40% (Figs. 3 and
4). The serum CRP values from
trauma and cancer patients were elevated when sepsis was present;
however, there was no statistical significance indicated in the
cancer patients (Fig. 4).
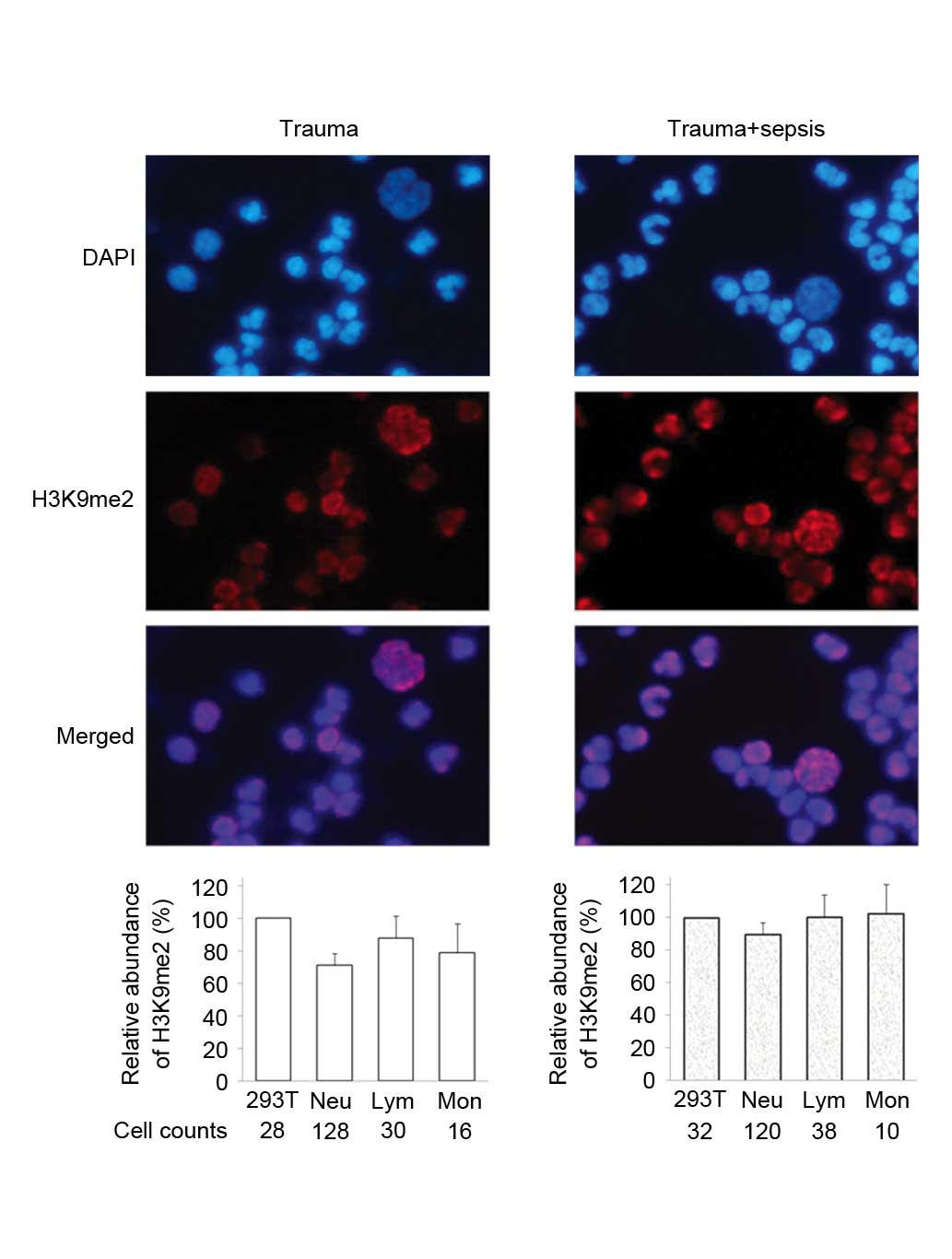 | Figure 2.Immunofluorescence staining of H3K9me2
in WBCs from trauma patients. Peripheral WBCs samples were obtained
from two patients, trauma and trauma+sepsis. For each sample, the
average intensity of H3K9me2 in 293T cells was defined as 100, and
the relative quantitative value of H3K9me2 in each leukocyte was
calculated. The relative fluorescence abundance of H3K9me2 in
counted Neu, Lym and Mon cells is presented as the mean ± standard
deviation. H3K9me2, histone 3 lysine 9 dimethylation; WBCs, white
blood cells; Neu, neutrophils; Lym, lymphocytes, Mon,
monocytes. |
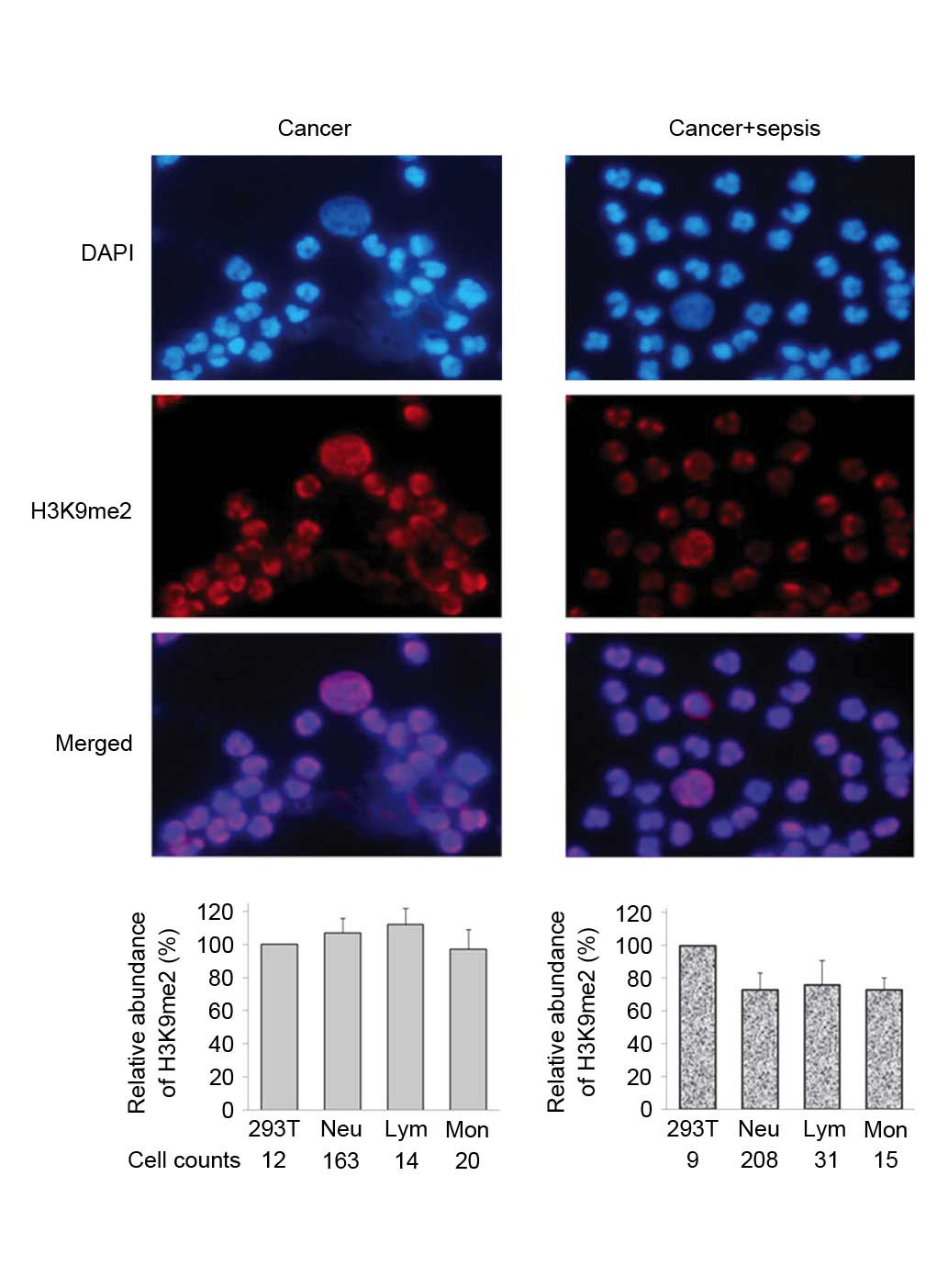 | Figure 3.Immunofluorescence staining of H3K9me2
in WBCs from cancer patients. Peripheral WBCs samples were obtained
from two patients, cancer and cancer+sepsis. For each sample, the
average intensity of H3K9me2 in 293T cells was defined as 100, and
the relative quantitative value of H3K9me2 in each leukocyte was
calculated. The relative fluorescence abundance of H3K9me2 in
counted Neu, Lym and Mon cells is presented as the mean ± standard
deviation. H3K9me2, histone 3 lysine 9 dimethylation; WBCs, white
blood cells; Neu, neutrophils; Lym, lymphocytes, Mon,
monocytes. |
Differential H3K9me2 expression in
WBCs
The intensity of H3K9me2 in WBCs between non-septic
patients with different underlying diseases, trauma and cancer, was
compared. An increase of histone methylation in cancer patients was
demonstrated, in the absence or presence of sepsis. However, the
elevated histone methylation in the three WBCs in cancer and septic
trauma varied. The H3K9me2 levels of Lym and Mon cells in cancer
patients were similar compared with trauma patients with sepsis,
whereas the levels in Neu cells were greater (Fig. 4).
These results indicated that immunofluorescence
staining may be used to compare histone methylation in different
WBCs. Sepsis in trauma and cancer patients had opposing effects on
histone methylation; sepsis increased H3K9me2 levels in trauma
patients and decreased them in cancer.
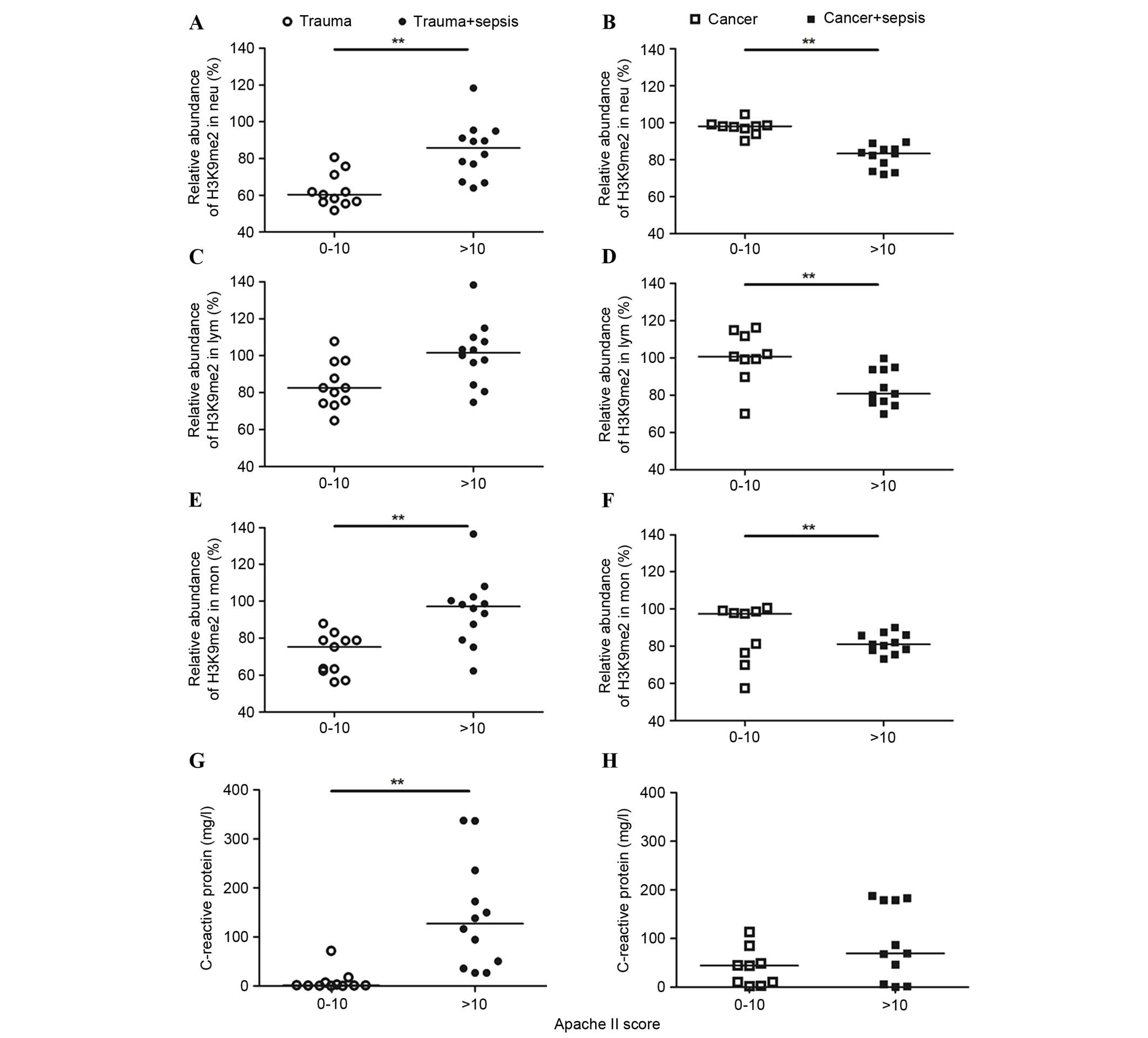
Diagnostic significance of H3K9me2 in
WBCs for sepsis
Furthermore, the H3K9me2 levels were used as a
diagnostic marker for sepsis using a receiver operating
characteristic analysis (Fig. 5).
The diagnostic significance was evaluated by the area under the
curve (AUC). The AUC values of H3K9me2 in the three WBCs from
trauma patients were 0.70–0.73; these were reduced compared with
the 0.98 demonstrated by CRP, indicating a poor predictive value of
H3K9me2 for the occurrence of sepsis in trauma patients. By
contrast, in cancer patients, the AUC values of H3K9me2 in the
three WBCs were 0.88 for Neu, 0.84 for Lym and 0.80 for Mon, all
greater than the 0.67 demonstrated by CRP (Fig. 5). A cutoff of 93.79 in Neu yielded
100% sensitivity and 66% specificity, 97.15 in Lym demonstrated 90%
sensitivity and 77% specificity, and 86.05 in Mon demonstrated 81%
sensitivity and 66% specificity for the occurrence of sepsis in
cancer patients. Considering the percentage of Neu of total WBCs,
H3K9me2 in WBCs exhibited a unique diagnostic significance for the
occurrence of sepsis in cancer patients compared with the commonly
used indicator.
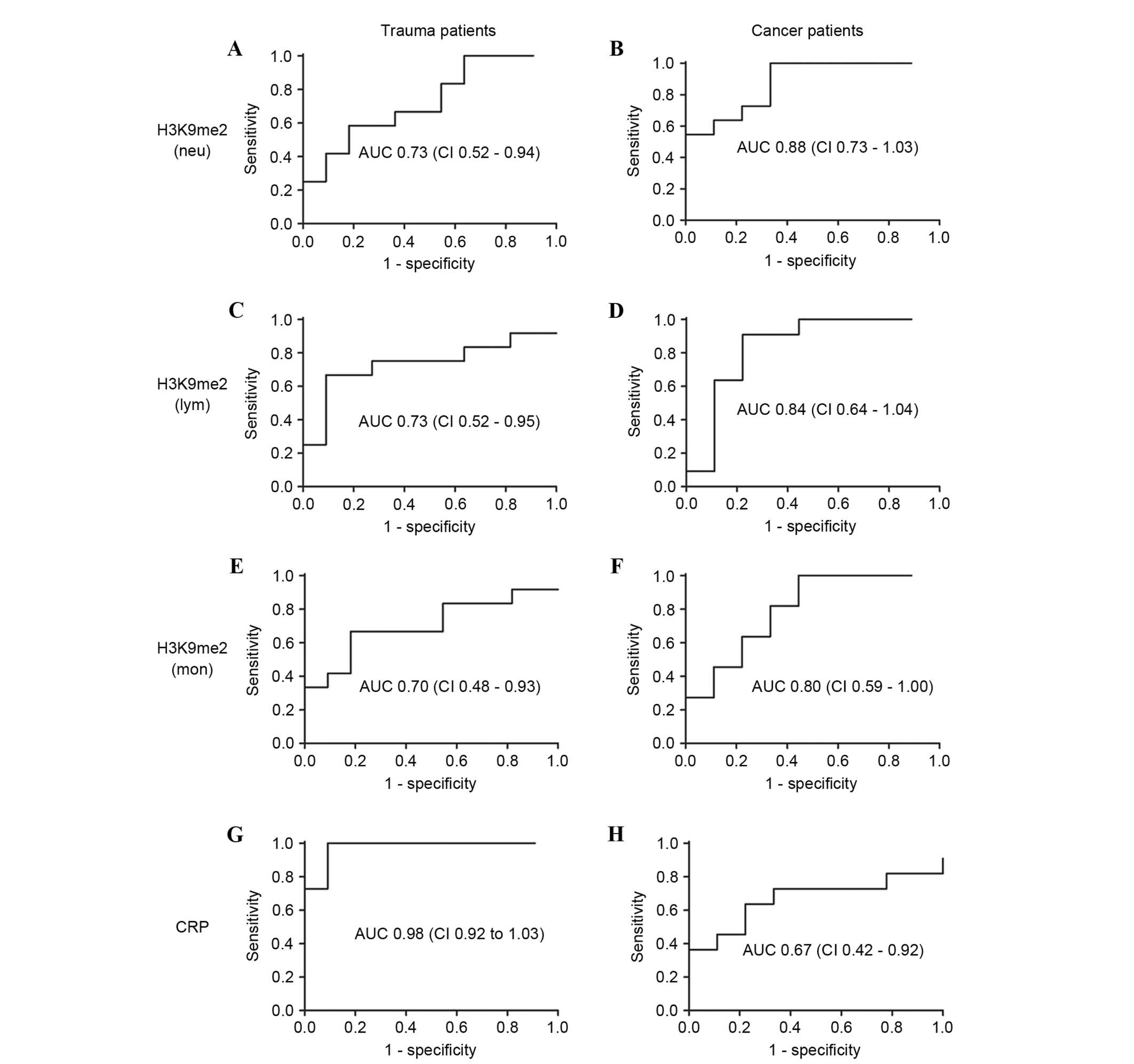 | Figure 5.Receiver operating characteristic AUC
of H3K9me2 and CRP levels in trauma and cancer patients. The AUC
values were calculated for H3K9me2 levels in (A and B) Neu, (C and
D) Lym and (E and F) Mon, and (G and H) CRP levels in patients with
trauma and cancer. H3K9me2 levels had a poor predictive value for
the occurrence of sepsis in trauma patients, but is a possible
diagnostic marker for the occurrence of sepsis in cancer patients.
AUC, area under the curve; CI, 95% confidence interval; H3K9me2,
histone 3 lysine 9 dimethylation; Neu, neutrophils; Lym,
lymphocytes; Mon, monocytes; CRP, C-reactive protein. |
Discussion
Previous studies have suggested that the
inflammatory response in sepsis is regulated by histone
methylation, including H3K4, H3K9 and H3K27 methylation (4,9).
However, clinical evidence is lacking. The present study verified
that the levels of histone methylation of peripheral WBCs were
altered in critically ill patients.
The present study established and optimized an
immunofluorescence staining method for WBCs. Using this method,
immunofluorescence intensity in different leukocytes was
simultaneously determined, and the levels of histone methylation
from different patient samples were compared, with co-stained 293T
cells serving as a reference. The results of the present study
indicated that this method is applicable to a wider range of
cellular targets. The data suggested that sepsis increased the
levels of H3K9me2 in all three types of WBCs from trauma patients.
The gene-specific silencing methylation is a key feature of the
endotoxin tolerant phenotype demonstrated in a THP-1 human Mon
model of severe systemic inflammation (SSI) and in blood
polymorphonuclear leukocytes of patients with SSI (6,8,24,30,31).
A previous study suggested that the NF-κB component RelB was
induced by activation of endotoxin, whereas RelB induced the
formation of heterochromatin via a direct effect on the histone
H3K9 methyltransferase G9a, thereby inhibiting the expression of
acute proinflammatory genes to generate epigenetic silencing in
endotoxin tolerance (6).
Additionally, the study suggested that histone H3K9 methylation was
elevated during SSI, which is consistent with the results from the
present study in trauma patients with sepsis.
By contrast, the occurrence of sepsis significantly
downregulated H3K9 methylation in the three WBC types from
esophageal cancer patients, which was elevated in the cancer group
by 20–40% compared with the trauma group. The current study
compared the levels of H3K9me2 in WBCs from three healthy
volunteers and trauma patients by immunostaining analyses, and no
marked difference was observed. These results suggest that H3K9
methylation in WBCs of cancer patients was upregulated prior to
sepsis, and differently regulated in sepsis compared with trauma
patients. Further research is required to elucidate an underlying
mechanism.
Sepsis induced opposing effects on histone
methylation in trauma and cancer patients, suggesting that
differences in primary diseases should be considered. Inflammation
in sepsis may not be similar in different primary diseases, with
particular differences occurring in acute vs. chronic, and benign
vs. malignant cases. Additionally, the potential abnormality may
alter the background of the inflammatory response. Although CRP is
a marker that is commonly used to detect inflammation or infection
(25), the disordered inflammatory
state associated with cancer existed prior to sepsis, reflected by
the greater percentage of Neu and elevated CRP levels in the cancer
group compared with the trauma group. Pre-existing inflammation
reduced the predictive power of CRP for sepsis. By contrast,
H3K9me2 levels in WBCs exhibited a unique diagnostic significance
for sepsis in cancer patients.
The present study observed altered H3K9me2 levels in
peripheral blood leukocytes from critically ill patients via
immunofluorescence analysis, with the opposite effect induced by
sepsis in trauma and cancer patients. Further studies on the
differential effects of sepsis on histone methylation are currently
being performed. The observation that the level of H3K9me2 is
altered in sepsis is of interest, and other histone methylations,
including H3K4, H3K27, H3K36, H3K79, H4K20 and arginine
methylation, will be investigated in the future. It may be
important to determine other potential causes of sepsis that have a
similar effect on histone methylation. Establishing a unique
profile of histone methylation in sepsis may enable the development
of novel strategies to manage sepsis.
In conclusion, the present study analyzed histone
methylation in peripheral WBCs via immunofluorescence. The H3K9me2
levels in peripheral WBCs from trauma and cancer patients were
different. Altered histone methylation in trauma and cancer
patients with sepsis exhibited opposite trends, and may represent a
potential diagnostic and prognostic biomarker for septic
patients.
Acknowledgements
The present study was supported, in part, by the
National Natural Science Foundation of China (grant no. 81101404),
the Ministry of Health of China for Clinical Key Discipline in 2012
and the Grant to Young Scholarship of Fudan University (grant no.
20520133394).
Glossary
Abbreviations
Abbreviations:
|
H3K9me2
|
histone 3 lysine 9 dimethylation
|
|
WBCs
|
white blood cells
|
|
Neu
|
neutrophils
|
|
Lym
|
lymphocytes
|
|
Mon
|
monocytes
|
|
CRP
|
C-reactive protein
|
|
APACHE II
|
Acute Physiology and Chronic Health
Evaluation II
|
|
SOFA
|
Sequential Organ Failure
Assessment
|
|
AUC
|
area under the curve
|
References
|
1
|
Soong J and Soni N: Sepsis: Recognition
and treatment. Clin Med (Lond). 12:276–280. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
An G, Namas RA and Vodovotz Y: Sepsis:
From pattern to mechanism and back. Crit Rev Biomed Eng.
40:341–351. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cavaillon JM, Adrie C, Fitting C and
Adib-Conquy M: Reprogramming of circulatory cells in sepsis and
SIRS. J Endotoxin Res. 11:311–320. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen S, Ma J, Wu F, Xiong LJ, Ma H, Xu W,
Lv R, Li X, Villen J, Gygi SP, et al: The histone H3 Lys 27
demethylase JMJD3 regulates gene expression by impacting
transcriptional elongation. Genes Dev. 26:1364–1375. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
De Santa F, Narang V, Yap ZH, Tusi BK,
Burgold T, Austenaa L, Bucci G, Caganova M, Notarbartolo S, Casola
S, et al: Jmjd3 contributes to the control of gene expression in
LPS-activated macrophages. EMBO J. 28:3341–3352. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen X, El Gazzar M, Yoza BK and McCall
CE: The NF-kappaB factor RelB and histone H3 lysine
methyltransferase G9a directly interact to generate epigenetic
silencing in endotoxin tolerance. J Biol Chem. 284:27857–27865.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
De Santa F, Totaro MG, Prosperini E,
Notarbartolo S, Testa G and Natoli G: The histone H3 lysine-27
demethylase Jmjd3 links inflammation to inhibition of
polycomb-mediated gene silencing. Cell. 130:1083–1094. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chan C, Li L, McCall CE and Yoza BK:
Endotoxin tolerance disrupts chromatin remodeling and NF-kappaB
transactivation at the IL-1beta promoter. J Immunol. 175:461–468.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Foster SL, Hargreaves DC and Medzhitov R:
Gene-specific control of inflammation by TLR-induced chromatin
modifications. Nature. 447:972–978. 2007.PubMed/NCBI
|
|
10
|
Greer EL and Shi Y: Histone methylation: A
dynamic mark in health, disease and inheritance. Nat Rev Genet.
13:343–357. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bannister AJ and Kouzarides T: Reversing
histone methylation. Nature. 436:1103–1106. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Black JC, Van Rechem C and Whetstine JR:
Histone lysine methylation dynamics: Establishment, regulation, and
biological impact. Mol Cell. 48:491–507. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bedford MT and Richard S: Arginine
methylation an emerging regulator of protein function. Mol Cell.
18:263–272. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mikkelsen TS, Ku M, Jaffe DB, Issac B,
Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP,
et al: Genome-wide maps of chromatin state in pluripotent and
lineage-committed cells. Nature. 448:553–560. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vakoc CR, Mandat SA, Olenchock BA and
Blobel GA: Histone H3 lysine 9 methylation and HP1gamma are
associated with transcription elongation through mammalian
chromatin. Mol Cell. 19:381–391. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Carrozza MJ, Li B, Florens L, Suganuma T,
Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP and
Workman JL: Histone H3 methylation by Set2 directs deacetylation of
coding regions by Rpd3S to suppress spurious intragenic
transcription. Cell. 123:581–592. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cloos PA, Christensen J, Agger K, Maiolica
A, Rappsilber J, Antal T, Hansen KH and Helin K: The putative
oncogene GASC1 demethylates tri- and dimethylated lysine 9 on
histone H3. Nature. 442:307–311. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Klose RJ, Yamane K, Bae Y, Zhang D,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: The
transcriptional repressor JHDM3A demethylates trimethyl histone H3
lysine 9 and lysine 36. Nature. 442:312–316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi Y, Lan F, Matson C, Mulligan P,
Whetstine JR, Cole PA and Casero RA: Histone demethylation mediated
by the nuclear amine oxidase homolog LSD1. Cell. 119:941–953. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiang S, Callaghan MM, Watson KG and Tong
L: A different mechanism for the inhibition of the
carboxyltransferase domain of acetyl-coenzyme A carboxylase by
tepraloxydim. Proc Natl Acad Sci USA. 106:20723–20727. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Reddy MA, Miao F, Shanmugam N, Yee
JK, Hawkins D, Ren B and Natarajan R: Role of the histone H3 lysine
4 methyltransferase, SET7/9, in the regulation of
NF-kappaB-dependent inflammatory genes. Relevance to diabetes and
inflammation. J Biol Chem. 283:26771–26781. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Levy D, Kuo AJ, Chang Y, Schaefer U,
Kitson C, Cheung P, Espejo A, Zee BM, Liu CL, Tangsombatvisit S, et
al: Lysine methylation of the NF-κB subunit RelA by SETD6 couples
activity of the histone methyltransferase GLP at chromatin to tonic
repression of NF-κB signaling. Nat Immunol. 12:29–36. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
El Gazzar M, Yoza BK, Chen X, Hu J,
Hawkins GA and McCall CE: G9a and HP1 couple histone and DNA
methylation to TNFalpha transcription silencing during endotoxin
tolerance. J Biol Chem. 283:32198–32208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Levy MM, Fink MP, Marshall JC, Abraham E,
Angus D, Cook D, Cohen J, Opal SM, Vincent JL and Ramsay G:
SCCM/ESICM/ACCP/ATS/SIS: 2001 SCCM/ESICM/ACCP/ATS/SIS International
Sepsis Definitions Conference. Crit Care Med 2003. 31:1250–1256.
2003. View Article : Google Scholar
|
|
26
|
Knaus WA, Draper EA, Wagner DP and
Zimmerman JE: APACHE II: A severity of disease classification
system. Crit Care Med. 13:818–829. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dominguez TE and Portnoy JD: Scoring for
multiple organ dysfunction: Multiple organ dysfunction score,
logistic organ dysfunction, or sequential organ failure assessment.
Crit Care Med. 30:1913–1914. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Duncan EM, Muratore-Schroeder TL, Cook RG,
Garcia BA, Shabanowitz J, Hunt DF and Allis CD: Cathepsin L
proteolytically processes histone H3 during mouse embryonic stem
cell differentiation. Cell. 135:284–294. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Adams-Cioaba MA, Krupa JC, Xu C, Mort JS
and Min J: Structural basis for the recognition and cleavage of
histone H3 by cathepsin L. Nat Commun. 2:1972011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
El Gazzar M, Yoza BK, Hu JY, Cousart SL
and McCall CE: Epigenetic silencing of tumor necrosis factor alpha
during endotoxin tolerance. J Biol Chem. 282:26857–26864. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
El Gazzar M, Yoza BK, Chen X, Garcia BA,
Young NL and McCall CE: Chromatin-specific remodeling by HMGB1 and
linker histone H1 silences proinflammatory genes during endotoxin
tolerance. Mol Cell Biol. 29:1959–1971. 2009. View Article : Google Scholar : PubMed/NCBI
|