Introduction
A large number of newborns and infants are currently
treated with anesthetics for surgery in what is considered to be a
safe manner. However, accumulating evidence from rodent and
nonhuman primate studies, as well as from epidemiological studies
(1–5), suggest that exposure to anesthetics
during a susceptive neurodevelopmental period may lead to neuronal
apoptosis and subsequent learning difficulties (6,7). A
previous study demonstrated that sevoflurane exposure led to
neuronal apoptosis and pathological alterations in the hippocampus
of neonatal rats (8), which was
consistent with reports from additional previous studies (9,10).
However, the molecular mechanisms underlying sevoflurane-induced
neurodegeneration remain largely unknown.
A previous study provided evidence of an association
between long-term neurological impairment induced by sevoflurane
and alterations in the expression levels of numerous genes in the
developing brain (11). MicroRNAs
(miRNAs/miRs) are small non-coding RNAs that serve as regulators of
endogenous epigenetic gene expression, and serve important roles in
multiple physiological/pathological processes, such as cell death
and survival (12,13). A recent study demonstrated that
sevoflurane combined with propofol anesthesia may lead to
alterations in expression levels of miRNAs in the hippocampus of
rats (14). In addition, miR-34a
has been demonstrated to be involved in ketamine-induced
hippocampal apoptosis (15).
However, there is limited information regarding the identity of
miRNA targets in the hippocampus during and after
sevoflurane-induced neural apoptosis.
miR-34c belongs to the miR-34 family (miRNA-34
a/b/c), which is highly conserved among different species (16). miR-34c is highly expressed in the
hippocampus. Targets of miR-34c are considered to be involved in
neuronal processes and functions (17). A previous study indicated that
miR-34 s are direct transcriptional targets of the p53 tumor
suppressor (18). In response to
DNA damage, the transcription factor p53 binds to its consensus
binding sequence in promoter DNA, and regulates the expression of
miR-34 family genes to induce apoptosis (16). It has been hypothesized that
miR-34s complement p53 function through regulation of various
targets involved in apoptosis, such as B-cell lymphoma-2 (Bcl-2)
(19).
In the present study, the miRNA expression patterns
in the hippocampus of rats at 6 h following exposure to sevoflurane
for 6 h, when neural death began to occur, was investigated. In
order to determine the functional roles of target genes regulated
by altered miR-34c levels, bioinformatics analyses were performed.
The results suggested miR-34c may serve a key role in
sevoflurane-induced apoptosis, which was the result of p53
phosphorylation, downregulation of Bcl-2 expression and
upregulation of Bax.
Materials and methods
Animals
Experiments were performed using Sprague-Dawley rats
(male; age, 7 days; average weight 16–17 g) obtained from the
Experimental Animal Center of Sun Yat-sen University, (Guangzhou,
China). All interventions and post-anesthesia animal care were
performed in accordance with the Guide for the Care and Use of
Laboratory Animals (National Research Council, 1996) and the
Guidelines of the Institutional Animal Care and Use Committee at
Sun Yat-sen University (Guangzhou, China). The use of animals in
this study was approved by the Institutional Animal Care and Use
Committee at Sun Yat-Sen University (approval no. 2014A-073). All
efforts were made to minimize suffering and the number of animals
used. The rats were maintained under a 12-h light-dark cycle (light
between 07:00 and 19:00), and the room temperature was maintained
at 21±1°C. Rats had ad libitum access to water and food. A total of
19 litters, 48 control and 51 treated pups were used in the present
study.
Sevoflurane exposure
Rats were anesthetized using the protocol described
previously (8). Seven-day-old male
rats were randomly divided into air-treated control (Con group,
n=15) and sevoflurane-treated groups (Sevo group, n=15). Rats in
the Sevo group were placed in a plastic container and exposed to
2.3% sevoflurane for 6 h using air as a carrier, with a total gas
flow of 2 l/min. In order to maintain the body temperature of rats
during sevoflurane exposure, the chamber was heated to 38°C using
an external heating device (NPS-A3 heated device; Midea Group,
Beijiao, China) and an internal hot water bag. The levels of
sevoflurane, oxygen and carbon dioxide in the container were
monitored using a gas monitor (Detex Ohmeda, Inc., Louisville, CO,
USA). During exposure, the respiration frequency and the skin color
of the pups was monitored by an investigator. Rats were excluded
from the experiment if any signs of apnea or hypoxia were detected.
Sevoflurane exposure ceased following 6 h. Upon waking from
anesthesia, rats were returned to the maternal cage for 6 h before
they were sacrificed. The rats in the control group were placed
into the same plastic container as those for the sevoflurane group,
where they were exposed to air for 6 h.
Arterial blood gas analysis
Arterial blood analysis was performed on both groups
using the methods described previously (8). The blood samples were obtained from
rats immediately following removal from the maternal cage (0 h, n=3
in each subgroup) and at the end of exposure (6 h, n=3 in each
subgroup). Briefly, the arterial blood sample (50 µl) was rapidly
obtained from the left cardiac ventricle and transferred into a
heparinized glass capillary tube. Each sample was analyzed
immediately following collection with a blood gas analyzer (GEM
Premier 3000; Instrumentation Laboratory, Bedford, MA, USA), and
the analysis was repeated >3 times. The pH, arterial carbon
dioxide tension, arterial oxygen tension, and blood glucose levels
in arterial blood samples were analyzed in order to exclude other
factors, including acidosis, hypercapnia, hypoxemia and
hypoglycemia that may induce apoptosis.
Immunofluorescence analysis
In order to determine alterations in the activity of
cleaved caspase-3 in the hippocampus following sevoflurane
exposure, rats from the Con and Sevo groups (n=3) were deeply
anesthetized with 10% chloral hydrate (350 mg/kg, Jianglai Biology,
Inc., Shanghai, China) intraperitoneally on P7 at 6 h following
exposure to sevoflurane or air for 6 h, before they were perfused
with 4% paraformaldehyde (Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany) in phosphate-buffered saline (PBS; pH 7.4)
through the left cardiac ventricle. The brains were dissected and
placed in 4% paraformaldehyde (Sigma-Aldrich; Merck Millipore) at
4°C overnight. Following fixation, the brains were soaked in 10, 20
and 30% sucrose for an additional 24 h. A sliding microtome
(Scientific Instruments, Inc., West Palm Beach, FL, USA) was used
to cut the brains into coronal sections (30 µm), which were blocked
with a solution containing 1% bovine serum albumin (Sigma-Aldrich;
Merck Millipore) and 0.4% Triton X-100 (Sigma-Aldrich; Merck
Millipore) for 2 h at room temperature. Tissue sections were
subsequently incubated with a rat anti-rabbit cleaved caspase-3
primary antibody (cat. no. 9661, dilution, 1:1,500; Cell Signaling
Technology, Inc., Danvers, MA, USA) at 4°C for 72 h. After washing
with PBS, the sections were then incubated with fluorescein
isothiocyanate-conjugated anti-rabbit IgG secondary antibody (cat.
no. 43413, dilution, 1:400; Sigma-Aldrich; Merck Millipore) in the
dark for 2 h at room temperature. Finally, the sections were washed
with PBS, mounted on gel-coated slides and observed under a
fluorescence microscope (Axio Imager Z1; Carl Zeiss AG, Oberkochen,
Germany). CA1 and CA3 hippocampal regions in both groups were
selected for analysis by fluorescence microscopy. The images were
analyzed using Image-Pro Plus software (version, 16.0; Media
Cybernetics, Inc., Rockville, MD, USA), and the integral optical
density (IOD) of part of the CA1 and CA3 regions in each photograph
was collected. A total of 3 sections were analyzed for each rat in
both experimental groups (n=3).
Total RNA extraction
Total RNA was isolated from the hippocampus using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), and the miRNeasy Mini kit (Qiagen GmbH, Hilden,
Germany) according to manufacturer's instructions. The RNA quality
and quantity were measured using the NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc., Wilmington, DE, USA), and RNA integrity was determined by gel
electrophoresis.
miRNA microarray assay and
analysis
The microarray assay and analysis of miRNA
expression were performed according to the procedures described
previously (20). Briefly, the RNA
samples from the Con and Sevo groups were labeled using the
miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon A/S, Vedbaek,
Denmark) and hybridized on the miRCURY LNA™ array (version, 18.0;
Exiqon A/S). Following three washes with the wash buffer kit
(Exiqon A/S), the slides were scanned using the Agilent G2505C
Microarray Scanner system (Agilent Technologies, Inc., Santa Clara,
CA, USA), and the images were imported into the Axon GenePix Pro
6.0 software program (Axon Instruments; Molecular Devices, LLC,
Sunnyvale, CA, USA) for grid alignment and data extraction. The
intensity of the green signal following subtraction of background
signal was calculated, and the mean intensity values for replicated
spots on the same slide were ascertained to calculate the median
intensity for each sample. The following median normalization
method was applied to obtain normalized data: (Foreground
signal-background signal)/median signal. The median signal was in
the 50th percentile of miRNA intensity, and was observed to be
>30 in all samples following background correction. The
threshold value for significance used to define upregulation or
downregulation of miRNA expression was considered to be a
fold-change of >1.5. Hierarchical clustering analysis was
performed to demonstrate distinguishable miRNA expression profiles
among the samples. A false discovery rate correction was applied
for multiple comparisons. miRNAs were selected for further
investigation in the present study based on demonstrating a
significant different expression pattern when compared with
controls (fold-change >1.5). Target genes of differentially
expressed miRNA sequences were identified using Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
In order to verify the accuracy of the microarray
analysis, RT-qPCR experiments were performed to determine the
expression of selected miRNAs using the procedures described in the
manufacturer's protocol (Takara Biotechnology, Co., Ltd., Dalian,
China). The primers (Invitrogen; Thermo Fisher Scientific, Inc.)
used are shown in Table I. The RNA
extracts were reverse transcribed into cDNA. In specific, the MMLV
reverse transcriptase enzyme (Takara Biotechnology, Co., Ltd.) and
RT primers of U6 and rno-miR-34c were used. The RT-qPCR reaction
was performed at 95°C for 10 min (pre-denaturation) followed by 42
cycles of 95°C for 15 sec, 58°C for 30 sec and 72°C for 30 sec.
Following the reaction, a melting curve analysis from 55 to 95°C
was applied to all reactions to ensure the consistency and
specificity of the amplified product. The data were analyzed using
the iQTM5 Optical System software (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The RT-qPCR data were normalized using U6 RNA.
The relative miRNA expression was determined by calculating the
mean difference between the cycle thresholds of the miRNA and the
U6 control for each sample [Δ quantification cycle
(ΔCq)] within each sample group, which was expressed as
-ΔCq for the relative change in expression. The
fold-change in miRNA expression was determined by calculating the
difference between the mean ΔCq of the Sevo and Con
groups at the same time point and hippocampal location
(ΔΔCq), and this value was expressed as the fold-change
(2-ΔΔCq) (21). The
difference in cycle threshold alterations (−ΔΔCq) was
determined using a Student's two-tailed t-test, and a P<0.05 was
considered to indicate a statistically significant difference.
 | Table I.Primers used for RT and RT-qPCR
analysis of the miRNA expression. |
Table I.
Primers used for RT and RT-qPCR
analysis of the miRNA expression.
| Procedure | Name | Sequence (5′-3′) |
|---|
| RT | U6 |
CGCTTCACGAATTTGCGTGTCAT |
|
| miR-3559-5p |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTCATGTAG |
|
| miR-92a-2-5p |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGTAATG |
|
| miR-214-5p |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAGACAC |
|
| miR-382-3p |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAAGTGT |
|
| miR-181b-1-3p |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTTGCAT |
|
| miR-101a-5p |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGCATCAG |
|
| miR-130b-5p |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGTAGTG |
|
| miR-188-3p |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGCAAAC |
|
| miR-34c-5p |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGCAATC |
| RT-qPCR | U6 | F:
GCTTCGGCAGCACATATACTAAAAT |
|
|
| R:
CGCTTCACGAATTTGCGTGTCAT |
|
| miR-3559-5p | GSP:
GGGGGGGTGACAGACTTAGTA |
|
|
| R:
GTGCGTGTCGTGGAGTCG |
|
| miR-92a-2-5p | GSP:
GGAAGGTGGGGATTAGTGC |
|
|
| R:
GTGCGTGTCGTGGAGTCG |
|
| miR-214-5p | GSP:
GGGGGGAAAGAGTTGTCATG |
|
|
| R:
GTGCGTGTCGTGGAGTCG |
|
| miR-382-3p | GSP:
GGGCAATCATTCACGGACA |
|
|
| R:
GTGCGTGTCGTGGAGTCG |
|
| miR-181b-1-3p | GSP:
GGGGGCTCACTGAACAATG |
|
|
| R:
GTGCGTGTCGTGGAGTCG |
|
| miR-101a-5p | GSP:
GGGGGGTCAGTTATCACAGTG |
|
|
| R:
GTGCGTGTCGTGGAGTCG |
|
| miR-130b-5p | GSP:
GCCACTCTTTCCCTGTTG |
|
|
| R:
CAGTGCGTGTCGTGGAG |
|
| miR-188-3p | GSP:
GGAATCTCCCACATGCAGG |
|
|
| R:
GTGCGTGTCGTGGAGTCG |
|
| miR-34c-5p | GSP:
GGGAGGCAGTGTAGTTAGC |
|
|
| R:
CAGTGCGTGTCGTGGAGT |
miRNA target gene prediction and
functional analysis
Using the Gene Ontology project (http://www.geneontology.org/), target genes that were
affected by the differentially expressed miRNAs in
sevoflurane-exposed rat pups were identified. The Gene Ontology
project was used to perform cluster analysis of the enriched
biological themes, and enrichment scores equal to -log10 (P-value)
were considered to indicate potential pathways. Higher enrichment
score values indicate a low P-value, which therefore indicates that
the cluster is significant.
Western blot analysis
At 6 h following exposure to sevoflurane or air for
6 h, 3 rats in each group were sacrificed by decapitation. The
brain was dissected, and the hippocampus was quickly removed. The
tissue samples were homogenized in 100 mg/ml
radioimmunoprecipitation assay lysis buffer (Shanghai Shenergy
Biocolor BioScience & Technology Company, Shanghai, China)
containing 1% (v/v) phenylmethylsulfonyl fluoride (Shanghai
Shenergy Biocolor BioScience & Technology Company) and
centrifuged at 13,000 × g for 20 min at 4°C. The supernatant was
separated, and the quantity of total protein extracted was measured
using a Bio-Rad Protein assay (Bio-Rad Laboratories, Inc.). The
samples were boiled, loaded at a concentration of 50 mg sample/lane
and electrophoresed on a 10% SDS-PAGE gel, and transferred onto
polyvinylidene fluoride membranes (Pall Life Sciences, Port
Washington, NY, USA). The blotted membranes were incubated with
rabbit polyclonal anti-p53 (dilution, 1:2,000; cat. no. Asp175;
Cell Signaling Technology, Inc.), rabbit polyclonal
anti-phosphorylated (p)-p53 (dilution, 1:3,000; cat. no. Asp175;
Cell Signaling Technology, Inc.), mouse monoclonal anti-Bcl-2
(dilution, 1:1,000; cat. no. 15071; Cell Signaling Technology,
Inc.), mouse monoclonal anti-Bax (dilution, 1:1,500; cat. no. 2772;
Cell Signaling Technology, Inc.), mouse monoclonal anti-β-actin
(dilution, 1:2,000; cat. no. sc-58673; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) or rabbit polyclonal anti-cleaved caspase-3
(dilution, 1:1,000; cat. no. 9664; Cell Signaling Technology, Inc.)
antibodies overnight at room temperature. The membranes were then
washed three times with PBS+0.05% Tween-20 (Sigma-Aldrich; Merck
Millipore) mixture, and incubated for 2 h at room temperature with
the anti-rabbit or anti-mouse IgG horseradish peroxidase-conjugated
secondary antibodies (dilution, 1:2,000; cat. no. sc-2372 or
sc-2377; Santa Cruz Biotechnology, Inc.). The protein expression
levels were examined using the Pierce ECL Plus Western Blotting
Substrate system (Pierce; Thermo Fisher Scientific, Inc.) and
photographed. The optical density was measured using ImageJ
software, V1.48u (National Institutes of Health, Bethesda, MD,
USA). Immunoblots were performed at least three times for each
hippocampus sample from each rat.
Statistical analysis
The data are presented as the mean ± standard
deviation. Statistical analysis was performed using the two-tailed
Student's t-test and the SPSS software program (version, 16.0;
SPSS, Inc., Chicago, IL, USA). All data in the study represent at
least three independent experiments for 3 samples in each
experiment. P<0.05 was considered to indicate a statistically
significant difference.
Results
Sevoflurane exposure did not alter the
parameters of arterial blood analysis
As shown in Table
II, no significant alterations were observed between the
control group and the sevoflurane group for pH, oxygen tension,
carbon dioxide tension and glucose levels. During sevoflurane and
air exposure, the skin color of the rat pups remained pink (data
not shown).
 | Table II.Arterial blood analysis. |
Table II.
Arterial blood analysis.
| Group | Time (h) | n | pH | PaCO2
(Kpa) | PaO2
(Kpa) | Glucose
(mmol/l) |
|---|
| Control | 0 | 3 | 7.38±0.04 | 3.55±0.44 | 13.33±0.23 | 5.4±0.6 |
|
| 6 | 3 | 7.37±0.06 | 3.56±0.14 | 13.32±0.51 | 5.3±0.8 |
| Sevoflurane | 0 | 3 | 7.38±0.03 | 3.56±0.29 | 13.35±0.25 | 5.5±0.3 |
|
| 6 | 3 | 7.38±0.02 | 3.57±0.54 | 13.32±0.61 | 5.3±0.6 |
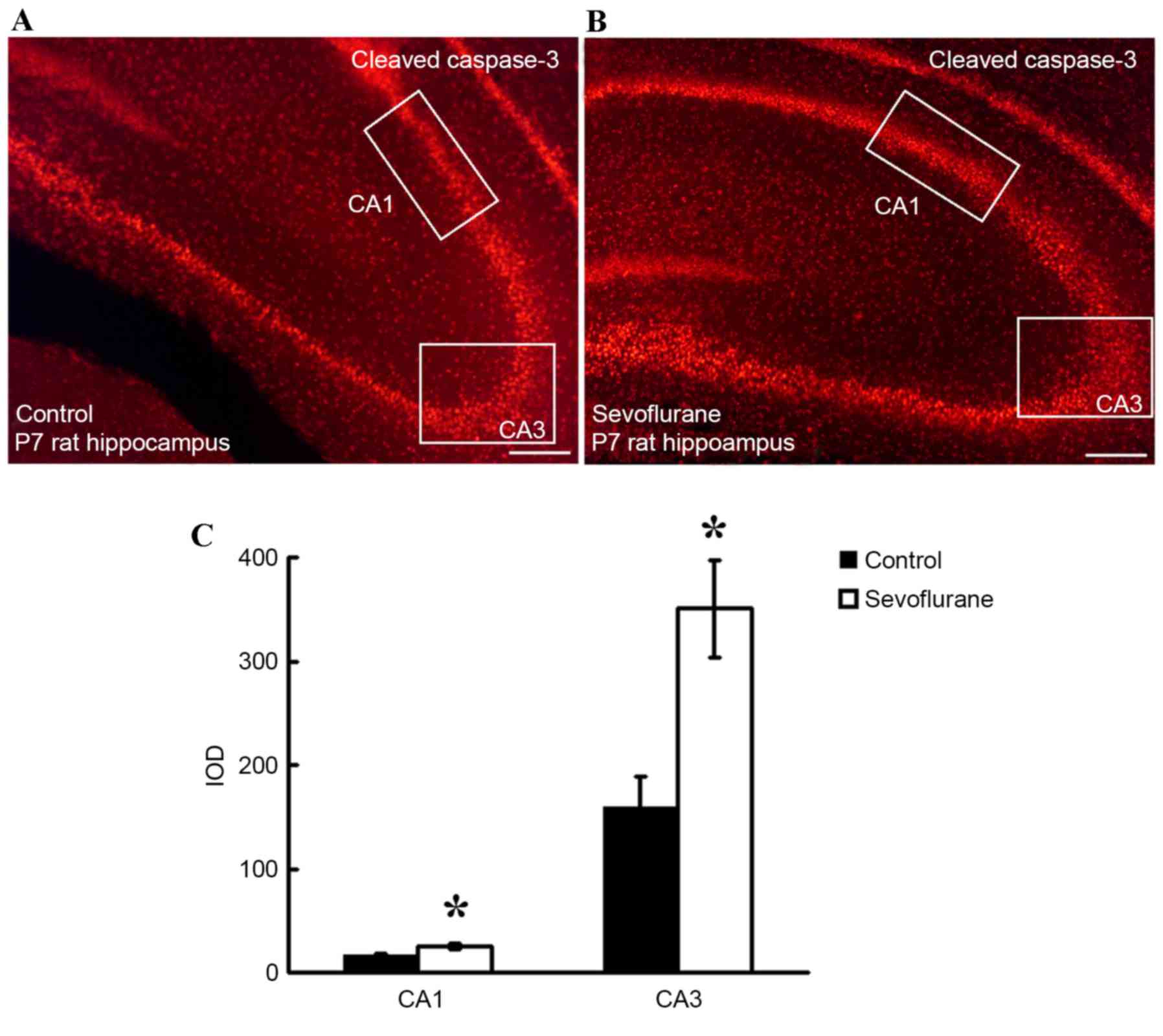
Sevoflurane exposure increased the
levels of cleaved caspase-3 in the hippocampus
In order to determine the effect of sevoflurane
exposure on apoptosis in hippocampal tissues, immunohistochemistry
assay analysis was performed to measure the level of cleaved
caspase-3 (red regions; Fig. 1A and
B) and the IOD of CA1 and CA3 regions. As illustrated in
Fig. 1C, the IODs in the CA1 and
CA3 regions were significantly higher in the sevoflurane group when
compared with the control group (P=0.001).
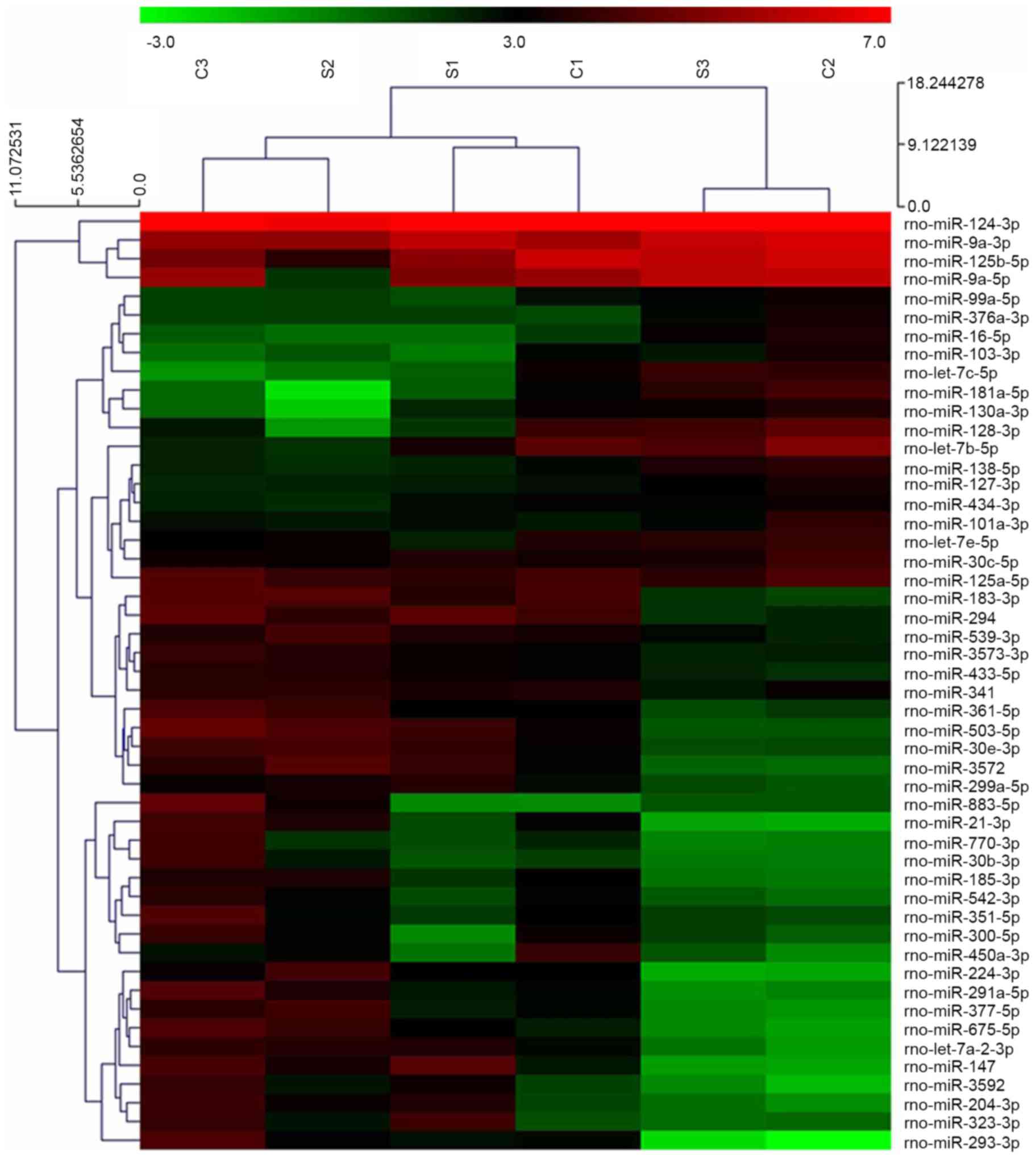
Sevoflurane exposure altered miRNA
expression in the hippocampus
Microarray analysis indicated that 688 miRNAs were
expressed in the hippocampus of rats at 7 days of age. Following
normalization of the signal intensities to the miRNA expression
levels, miR-124-3p, miR-9a-3p and miR-125b-5p were observed to be
expressed at the highest levels (Fig.
2). Compared with the control group, the expression levels of
266 miRNAs in the hippocampus of the sevoflurane group were
altered. Among these 266 miRNAs, 8 miRNAs (miR-3559-5p,
miR-92a-2-5p, miR-214-5p, miR-382-3p, miR-181b-1-3p, miR-101a-5p,
miR-130b-5p, and miR-188-3p) were significantly upregulated, and
one miRNA (miR34-c) was significantly downregulated following
sevoflurane exposure (P=0.001; Fig.
3A).
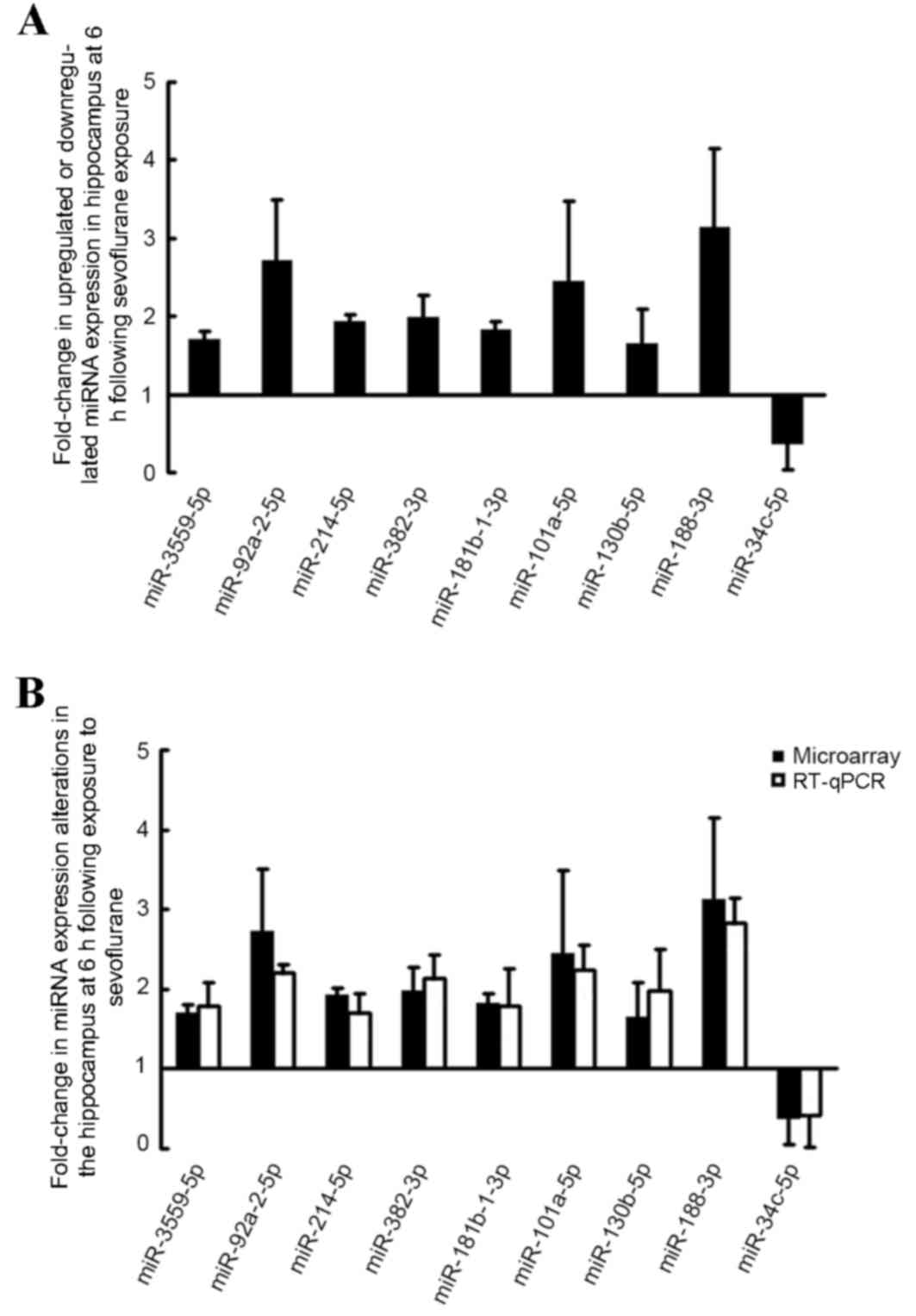
Sevoflurane exposure induced
alterations in miRNA expression as determined by identified by
RT-qPCR analysis
In order to validate the microarray results, RT-qPCR
analysis was performed to confirm the expression of the nine
identified miRNAs. As demonstrated in Fig. 3B, RT-qPCR analysis confirmed that,
miR-3559-5p, miR-92a-2-5p, miR-214-5p, miR-382-3p, miR-181b-1-3p,
miR-101a-5p, miR-130b-5p and miR-188-3p expression levels were
increased, whereas miR34-c expression was significantly reduced
following sevoflurane exposure.
Bioinformatics analyses of the miRNA
targets
Target genes of differentially expression miRNA
sequences were identified using KEGG pathway analysis (http://www.genome.jp/kegg/). All pathways that
demonstrated a P-value of <0.05 are listed in Table III. The identified pathways
included mineral absorption, tuberculosis, the p53 signaling
pathway, glycolysis/gluconeogenesis, drug metabolism-cytochrome
P450, metabolism of cytochrome by cytochrome P450, antigen
processing and presentation, and pyrimidine metabolism associated
genes. Among these pathways, only miR-34c was directly associated
with the p53 signaling pathway. This suggests that the miR-34c and
the p53 pathway may be involved in mediating sevoflurane-induced
apoptosis in the hippocampus of developing rat brains.
 | Table III.Signaling pathways identified by
Kyoto Encyclopedia of Genes and Genomes pathway analysis in rats at
6 h following exposure to sevoflurane for 6 h. |
Table III.
Signaling pathways identified by
Kyoto Encyclopedia of Genes and Genomes pathway analysis in rats at
6 h following exposure to sevoflurane for 6 h.
| Pathway | Count | P-value |
|---|
| Mineral
absorption | 46 | 0.008190094 |
| Tuberculosis | 193 | 0.01902255 |
| p53 signaling
pathway | 75 | 0.02084327 |
|
Glycolysis/gluconeogenesis | 79 | 0.02297618 |
| Drug
metabolism-cytochrome P450 | 85 | 0.02633797 |
| Metabolism of
xenobiotics by cytochrome P450 | 89 | 0.0286843 |
| Antigen processing
and presentation | 94 | 0.03173175 |
| Pyrimidine
metabolism | 100 | 0.03555134 |
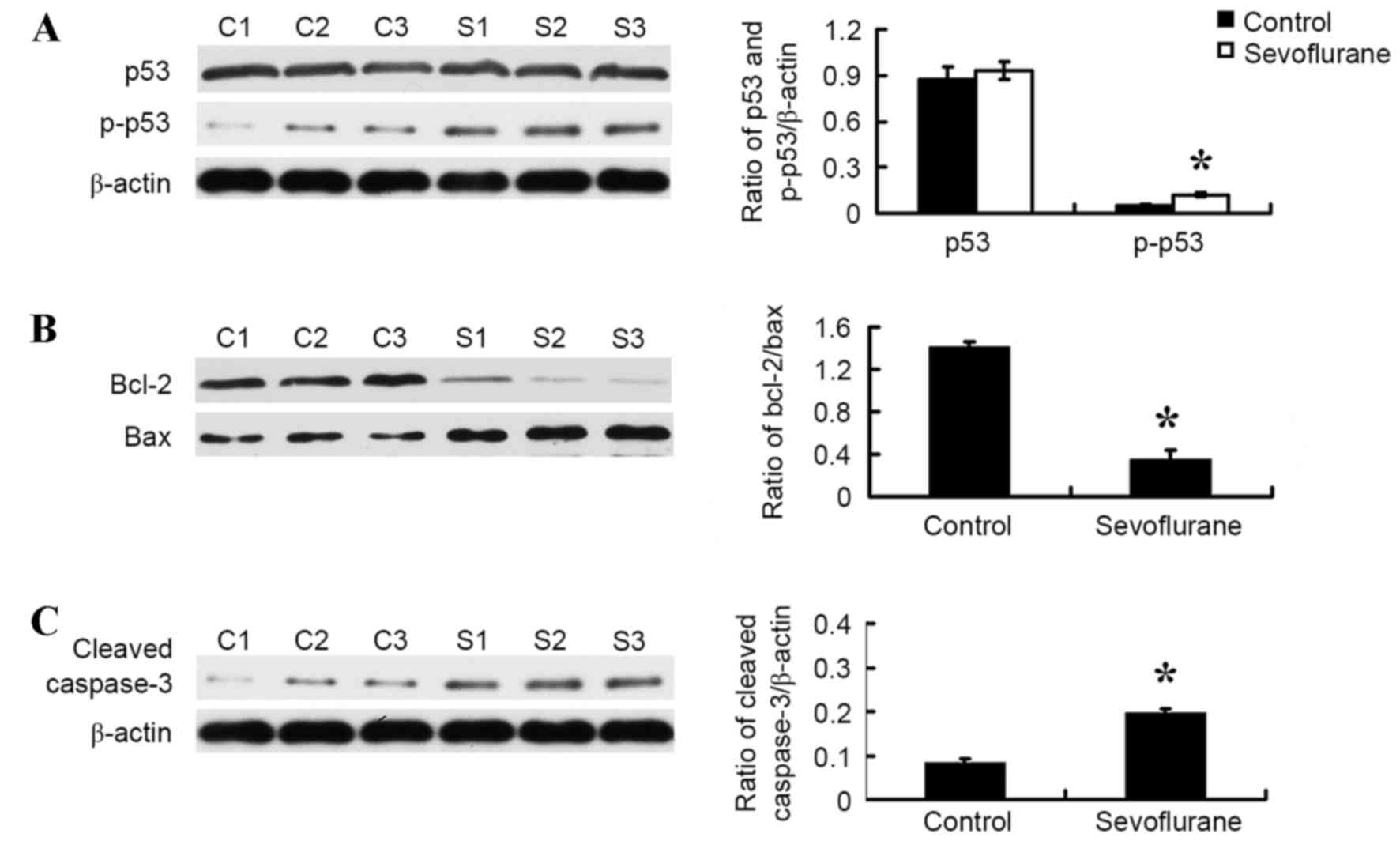
Sevoflurane altered the levels of p53,
Bcl-2, Bax and cleaved caspase-3 in the hippocampus of the neonate
rat brain
Western blot analysis was performed to assess the
effect of sevoflurane on the targets of the p53 signaling pathway
in the hippocampus. As shown in Fig.
4, sevoflurane exposure significantly increased the level of
the p-p53 protein (P=0.03; Fig.
4A) and Bax (P=0.001; Fig.
4B), and significantly decreased the level of Bcl-2 protein
(P=0.001; Fig. 4B) in the
hippocampus of neonate rat brains when compared with the control
rat brains. In addition, sevoflurane exposure significantly
increased the expression of cleaved caspase-3 when compared with
the control group (P<0.05; Fig.
4C).
Discussion
In the present study, arterial blood analysis was
first performed in rats exposed to sevoflurane or air, which
confirmed that none of the pups in either group had suffered from
apnea or hypoxia during treatment. The level of apoptosis in the
hippocampus of neonatal rats at 6 h following sevoflurane exposure
for 6 h was then examined. Although apoptosis is known to occur in
the developing brain (22), it was
more evident following exposure to 2.3% sevoflurane for 6 h, as
indicated by the significant increase in the level of cleaved
caspase-3; a marker of the execution phase of cell apoptosis
(23). This result is consistent
with studies conducted in our lab previously (8,24).
Additional studies involving a variety of adolescent animal models
have demonstrated that sevoflurane induces apoptosis, which may
contribute to the deficits in hippocampus-dependent learning and
memory (9,10,25).
However, the detailed mechanisms underlying the harmful effects of
sevoflurane in the developing brain are not fully understood.
miRNAs serve key roles in a wide range of biological
processes, including cell death and survival, as well as regulation
of gene expression, which occurs primarily at the translational
level (12,13). The results of the current study
provide novel evidence to suggest that exposure to 2.3% sevoflurane
for 6 h affects miRNA expression patterns in the hippocampus.
Hierarchical clustering analysis, presented as a heat map, was
performed to visualize the semantic similarities and expression
levels of the miRNAs. The results demonstrated that miR-124-3p,
miR-9a-3p and miR-125b-5p were expressed at the highest levels.
Following sevoflurane exposure, 8 miRNAs were significantly
upregulated, while only miR-34c was significantly downregulated.
Therefore, miR34c was selected for further investigation. Notably,
Goto et al (14) reported a
contrasting result. In this previous study, 4 miRNAs were observed
to be significantly increased, while 12 miRNAs were significantly
reduced in the rat hippocampus following sevoflurane anesthesia.
The reasons underlying the difference between their results and
those of the present study remain to be elucidated. However, one
notable difference is that Goto et al (14) conducted experiments on 6-week-old
male Wistar rats, whereas 7-day-old Sprague-Dawley rats were
employed in the current study. Previous studies have demonstrated
that ketamine leads to neuroapoptosis in the developing brain
(26), and that miR-34a negatively
regulates ketamine-induced apoptosis in the hippocampus (15). This is particularly notable, as
miR-34a and miR-34c belong to the miR-34 family. In addition,
previous studies have indicated that the miR-34 family is
associated with additional biological processes including cancer
(27); however its role in the
developing brain has been rarely addressed. It has been reported
that miR-34 family genes are involved in apoptosis (19), and the results of the present study
suggest that miR-34c may be involved in apoptosis induced by
sevoflurane.
miR-34 family members are direct transcriptional
targets of the p53 tumor suppressor (18). Previous studies have demonstrated
that p53 regulates the expression of miR-34c (16,28).
KEGG is a compendium of databases containing annotated genomes and
protein interaction networks for all organisms that have undergone
genome sequencing, as well as a compilation of manually verified
pathway maps displaying molecular interactions and biochemical
reactions (29). Using KEGG
pathway analysis in the current study, several pathways were
observed to be significantly correlated to the present condition
(sevoflurane-treated P7 rats), including the p53 pathway, and p53
phosphorylation was significantly increased. The authors
hypothesize that sevoflurane may lead to activation of p53 and the
downstream target, miR-34c. Among all the predicted pathways
identified, p53 regulates apoptosis, in part, by inducing intrinsic
cell death (30). The intrinsic,
mitochondrial pathway is regulated by Bcl-2 family proteins,
including the anti-apoptotic factor Bcl-2 and the pro-apoptotic
factor Bax (31). Recent studies
have indicated that a number of miRNAs directly target Bcl-2 family
proteins including miRNA-34 (32).
The results of the present study indicated that Bax protein levels
were significantly increased, while Bcl-2 protein levels were
significantly decreased. These results suggest that miR-34c may be
regulated by activated p53, and is involved in sevoflurane-induced
apoptosis in the developing brain potentially via the mitochondrial
pathway. These results are consistent with a previous study, which
demonstrated that sevoflurane induces caspase-dependent,
mitochondria-mediated apoptosis in human T lymphocytes in
vitro (33).
There are several limitations of the current study.
Firstly, the expression of additional miRNAs, aside from miR-34c,
was observed to be significantly altered following sevoflurane
exposure. It remains formally possible that these miRNAs may
contribute to the apoptosis induced by sevoflurane. Secondly,
miRNA-34c is transactivated by p53; however, there is insufficient
evidence to exclude the possibility that additional miRNAs may be
involved in the p53 signaling pathway. Further research will be
designed and performed to address these issues. For example, by
conducting interference of miR-34 expression in vivo or
in vitro, in order to identify the precise role of miR-34c
in sevoflurane-induced apoptosis. Furthermore, additional pathways
may be involved in sevoflurane-induced apoptosis, and these
mechanisms warrant further investigation.
In conclusion, exposure to 2.3% sevoflurane for 6 h
led to apoptosis in the developing rat hippocampus. The results of
the present study suggest that miR-34c may be regulated by p53, and
may be involved in sevoflurane-induced neural apoptosis.
Sevoflurane affects Bax and Bcl-2 levels and induces apoptosis
potentially via the mitochondrial pathway in the hippocampus of the
developing rat brains.
Acknowledgements
The present study was supported by the National
Science Foundation Council of China (81571032).
References
|
1
|
Wilder RT, Flick RP, Sprung J, Katusic SK,
Barbaresi WJ, Mickelson C, Gleich SJ, Schroeder DR, Weaver AL and
Warner DO: Early exposure to anesthesia and learning disabilities
in a population-based birth cohort. Anesthesiology. 110:796–804.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DiMaggio C, Sun LS and Li G: Early
childhood exposure to anesthesia and risk of developmental and
behavioral disorders in a sibling birth cohort. Anesth Analg.
113:1143–1151. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ing C, DiMaggio C, Whitehouse A, Hegarty
MK, Brady J, von U, ngern-Sternberg BS, Davidson A, Wood AJ, Li G
and Sun LS: Long-term differences in language and cognitive
function after childhood exposure to anesthesia. Pediatrics.
130:e476–e485. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hansen TG, Pedersen JK, Henneberg SW,
Pedersen DA, Murray JC, Morton NS and Christensen K: Academic
performance in adolescence after inguinal hernia repair in infancy:
A nationwide cohort study. Anesthesiology. 114:1076–1085. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun L: Early childhood general anaesthesia
exposure and neurocognitive development. Br J Anaesth. 105 Suppl
1:i61–i68. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Deng M, Hofacer RD, Jiang C, Joseph B,
Hughes EA, Jia B, Danzer SC and Loepke AW: Brain regional
vulnerability to anaesthesia-induced neuroapoptosis shifts with age
at exposure and extends into adulthood for some regions. Br J
Anaesth. 113:443–451. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zou X, Liu F, Zhang X, Patterson TA,
Callicott R, Liu S, Hanig JP, Paule MG, Slikker W Jr and Wang C:
Inhalation anesthetic-induced neuronal damage in the developing
rhesus monkey. Neurotoxicol Teratol. 33:592–597. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Feng X, Liu JJ, Zhou X, Song FH, Yang XY,
Chen XS, Huang WQ, Zhou LH and Ye JH: Single sevoflurane exposure
decreases neuronal nitric oxide synthase levels in the hippocampus
of developing rats. Br J Anaesth. 109:225–233. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang X, Xue Z and Sun A: Subclinical
concentration of sevoflurane potentiates neuronal apoptosis in the
developing C57BL/6 mouse brain. Neurosci Lett. 447:109–114. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Satomoto M, Satoh Y, Terui K, Miyao H,
Takishima K, Ito M and Imaki J: Neonatal exposure to sevoflurane
induces abnormal social behaviors and deficits in fear conditioning
in mice. Anesthesiology. 110:628–637. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan ZQ, Lu XF, Shao C, Zhang C, Yang J, Ma
T, Zhang LC and Cao JL: The effects of sevoflurane anesthesia on
rat hippocampus: A genomic expression analysis. Brain Res.
1381:124–133. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moreno-Moya JM, Vilella F and Simón C:
MicroRNA: Key gene expression regulators. Fertil Steril.
101:1516–1523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goto G, Hori Y, Ishikawa M, Tanaka S and
Sakamoto A: Changes in the gene expression levels of microRNAs in
the rat hippocampus by sevoflurane and propofol anesthesia. Mol Med
Rep. 9:1715–1722. 2014.PubMed/NCBI
|
|
15
|
Jiang XL, Du BX, Chen J, Liu L, Shao WB
and Song J: MicroRNA-34a negatively regulates anesthesia-induced
hippocampal apoptosis and memory impairment through FGFR1. Int J
Clin Exp Pathol. 7:6760–6767. 2014.PubMed/NCBI
|
|
16
|
He L, He X, Lim LP, De Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zovoilis A, Agbemenyah HY, Agis-Balboa RC,
Stilling RM, Edbauer D, Rao P, Farinelli L, Delalle I, Schmitt A,
Falkai P, et al: microRNA-34c is a novel target to treat dementias.
Embo J. 30:4299–4308. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cha YH, Kim NH, Park C, Lee I, Kim HS and
Yook JI: MiRNA-34 intrinsically links p53 tumor suppressor and Wnt
signaling. Cell Cycle. 11:1273–1281. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Suzuki HI and Miyazono K: Dynamics of
microRNA biogenesis: Crosstalk between p53 network and microRNA
processing pathway. J Mol Med (Berl). 88:1085–1094. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang Y, Ling ZM, Fu R, Li YQ, Cheng X,
Song FH, Luo HX and Zhou LH: Time-specific microRNA changes during
spinal motoneuron degeneration in adult rats following unilateral
brachial plexus root avulsion: Ipsilateral vs. contralateral
changes. BMC Neurosci. 15:922014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yudkowitz FS: Anesthetics and the
developing brain. Semin Cardiothorac Vasc Anesth. 14:44–45. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lemkuil BP, Head BP, Pearn ML, Patel HH,
Drummond JC and Patel PM: Isoflurane neurotoxicity is mediated by
p75 (NTR)-RhoA activation and actin depolymerization.
Anesthesiology. 114:49–57. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou X, Song FH, He W, Yang XY, Zhou ZB,
Feng X and Zhou LH: Neonatal exposure to sevoflurane causes
apoptosis and reduces nNOS protein expression in rat hippocampus.
Mol Med Rep. 6:543–546. 2012.PubMed/NCBI
|
|
25
|
Haseneder R, Kratzer S, von Meyer L, Eder
M, Kochs E and Rammes G: Isoflurane and sevoflurane
dose-dependently impair hippocampal long-term potentiation. Eur J
Pharmacol. 623:47–51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brambrink AM, Evers AS, Avidan MS, Farber
NB, Smith DJ, Martin LD, Dissen GA, Creeley CE and Olney JW:
Ketamine-induced neuroapoptosis in the fetal and neonatal rhesus
macaque brain. Anesthesiology. 116:372–384. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Maroof H, Salajegheh A, Smith RA and Lam
AK: Role of microRNA-34 family in cancer with particular reference
to cancer angiogenesis. Exp Mol Pathol. 97:298–304. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Corney DC, Flesken-Nikitin A, Godwin AK,
Wang W and Nikitin AY: MicroRNA-34b and MicroRNA-34c are targets of
p53 and cooperate in control of cell proliferation and
adhesion-independent growth. Cancer Res. 67:8433–8438. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40(Database
issue): D109–D114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Panduri V, Surapureddi S, Soberanes S,
Weitzman SA, Chandel N and Kamp DW: P53 mediates amosite
asbestos-induced alveolar epithelial cell mitochondria-regulated
apoptosis. Am J Respir Cell Mol Biol. 34:443–452. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cory S and Adams JM: The Bcl2 family:
Regulators of the cellular life-or-death switch. Nat Rev Cancer.
2:647–656. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ouyang YB and Giffard RG: MicroRNAs affect
BCL-2 family proteins in the setting of cerebral ischemia.
Neurochem Int. 77:2–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Loop T, Dovi-Akue D, Frick M, Roesslein M,
Egger L, Humar M, Hoetzel A, Schmidt R, Borner C, Pahl HL, et al:
Volatile anesthetics induce caspase-dependent,
mitochondria-mediated apoptosis in human T lymphocytes in vitro.
Anesthesiology. 102:1147–1157. 2005. View Article : Google Scholar : PubMed/NCBI
|