Introduction
The liver, an organ with a complex structure and
function, is damaged by various factors, including viruses, drugs,
trauma, chemotoxicity and metabolic disorders. A reduction in liver
mass and ischemia-reperfusion caused by surgery are also causes of
liver injury in patients with diseases of the liver. The high
regenerative potential is the primary mechanism by which the liver
compensates for loss of weight and assists in recovery from injury.
Reasonable control of liver regeneration is of substantial value in
therapy for liver diseases.
There have been studies on liver regeneration for
over a century with notable progress. The mechanisms of liver
regeneration have been elucidated in detail and have benefited from
the contribution of partial hepatectomy (PH) models, first
established by Higgins and Anderson in 1931. The classic process of
liver regeneration, which involves numerous genes, consists of
three stages: Initiation, proliferation and termination (1). Hepatocytes leave their quiescent
state, resulting in DNA synthesis, with the stimulation of priming
signals, including interleukin (IL)-6, tumor necrosis factor-α
(TNF-α) and nitric oxide in the initiation step. The proliferation
step is characterized by hepatocytes entering the G1 phase of the
cell cycle, and mitogens, including hepatocyte growth factor (HGF),
transforming growth factor (TGF)-α and epidermal growth factor
(EGF), are essential in this step. When the remnant liver grows to
a suitable volume, similar to that of the original liver,
regeneration enters the termination step, during which stop and
differentiation signals are involved. Compared with the initiation
and proliferation steps, the termination step is less well
understood.
As is already known, the liver regeneration process
is a well-orchestrated process requiring a balance between
pro-proliferation and anti-proliferation factors. However, the
majority of studies on liver regeneration have been associated with
the regulation of initiation and proliferation-promoting signals,
with less known regarding termination and growth inhibitors. The
proliferation inhibitors act as a ‘brake’ and ‘steering wheel’ to
control the speed and direction of liver regeneration. The remnant
hepatocytes in the damaged liver do not stop growing to the correct
size and the proliferation is not suppressed. In extreme cases, the
proliferation of liver cells may lead to oncogenesis. For example,
loss of the expression of suppressor of cytokine signaling (SOCS)3
enhances liver regeneration but allows hepatocellular carcinoma
formation (2). Furthermore,
certain growth inhibitors can themselves function as tumor
suppressor genes, for example p53 and p21, which are essential for
the inhibition of carcinogenesis from liver damage by chronic
injury (3). The known
proliferation-inhibiting factors and associated pathways are
discussed in this review.
Cytokine-associated pathways
TGF-β
TGF-β is the most well-known hepatocyte
proliferation inhibitor and stop signal in the process of hepatic
regeneration. There have been investigations on the association
between TGF and liver regeneration for >30 years. Nakamura et
al reported in 1985 that TGF-β from platelets inhibited the DNA
synthesis of adult rat hepatocytes induced by epidermal growth
factor (EGF) in primary culture (4). The administration of platelet-derived
TGF-β has also been shown to suppress liver regeneration in a
two-thirds PH rat model (5). The
inhibition of TGF-β1 with monoclonal antibody has been shown to
enhance liver regeneration in a porcine model of partial portal
vein ligation (6). A previous
study also found that inhibiting the TGF-β pathway with the TGF-β
type I receptor kinase inhibitor increased hepatocyte proliferation
during acute liver damage (7).
Previous investigations demonstrated that TGF-β is
predominantly secreted by nonparenchymal cells, including hepatic
stellate cells (HSCs), Kupffer cells (KCs) and platelets, in liver
regeneration and modulates the proliferation of hepatocytes in a
paracrine manner (8,9). Subsequent studies reported that TGF-β
can also be expressed in regenerating hepatocytes (10,11).
The expression of TGF-β was found to increase at 4 h and reached a
peak 72 h following PH when DNA synthesis had stopped, suggesting
that TGF-β was involved in the inhibition and termination of liver
regeneration (8,12). Further evidence for the
anti-proliferation function of TGF-β1 includes the reduction in DNA
synthesis in transgenic mice with overexpressed TGF-β1 following PH
(13). Although several members
are included in the TGF-β superfamily, the β1 form is the most
important subtype in regeneration (10,12).
TGF-β modulates the proliferation of hepatocytes
through multiple mechanisms. The earliest mechanism to be
identified is the inhibition of EGF-induced DNA synthesis by TGF-β
following its binding to TGF-β receptors on the surface of
hepatocytes (8,11,14).
Investigations found that TGF-β receptors were single-pass
transmembrane serine/threonine kinases. When the TGF-β ligand binds
to the complex of TGF-β receptors, the receptor-regulated
cytoplasmic small mothers against decpentaplegic (R-Smad) proteins
are phosphorylated and accumulate in the nucleus. The accumulated
Smads subsequently inhibit the transcription of several genes by
interacting with DNA binding proteins and transcriptional
regulators (15–18). Another study showed that the TGF-β
signaling may be inhibited by SnoN and Ski, Smad pathway
inhibitors, during the proliferative phase of liver regeneration
(19). Increased expression of
TGF-β and activation of Smad2 have also been found in a T
cell-mediated hepatitis model following PH, and were associated
with impaired hepatocellular proliferation (20). The activation of Smad2/3, which can
be inhibited by Smad7, is required for TGF-β to have an inhibitory
role in liver regeneration (21).
Only activated TGF-β has the ability to inhibit cell proliferation,
as reported by Schrum et al, who observed that activated
TGF-β, but not latent TGF-β, affected liver regeneration (22). A previous study found that the
activation of TGF-β required the involvement of thrombospondin-1
(TSP-1). Significantly reduced TGF-β/Smad signaling and accelerated
hepatocyte proliferation were observed in mice with TSP-1
deficiency following PH (23).
With the exception of EGF, other
proliferation-associated factors and signaling pathways can be
affected by TGF-β. TGF-β can inhibit the secretion of human
hepatocyte growth factor (HGF), which is known to be a complete
mitogen in the process of liver regeneration (24). DNA synthesis induced by TNF can
also be inhibited by TGF-β in cultured hepatocytes (25). The overexpression of TGF-β1 was
shown to inhibit the protein level of cdc25A, a cyclin-dependent
kinase-activating tyrosine phosphatase, by enhancing the binding of
histone deacetylase 1 to p130 in Alb-TGF-β1 transgenic mice
following PH (26). TGF-β1 was
found to inhibit DNA synthesis of the G1 stage in cultured
hepatocytes treated with HGF and heparin-binding epidermal growth
factor-like growth factor by decreasing the expression of cyclin E
without affecting the activity of c-Met, epidermal growth factor
receptor (EGFR) or mitogen-activated protein kinase (MAPK), or the
expression of cyclin D1 (27).
This indicated that TGF-β1 has the ability to restrain the growth
factor-induced signals between cyclin D1 and cyclin E (27).
Inducing cell apoptosis is important for TGF-β to
exert its anti-proliferative effect in liver regeneration. Studies
have indicated that liver cell regeneration and atypical bile duct
proliferation are associated with TGF-β in fulminant hepatitis
(28). The overexpression of
activated TGF-β by adenovirus vectors enhanced the apoptosis of
hepatocytes, resulting in the death of rats following two-thirds PH
(22). Enhanced apoptosis was also
found to be accompanied by the overexpression of TGF-β1 in
tetracycline-controlled TGF-β1 mice (29), whereas pro-apoptotic signals were
suppressed by insulin-like growth factor binding protein-1
(30). In experiments performed by
Samson et al, the overexpression of TGF-β1 resulted in
increased expression of c-Jun, a potential pro-apoptotic
transcription factor, which indicated that TGF-β1 induced
hepatocyte apoptosis through a c-Jun-independent mechanism
(31). The expression of TGF-β1
increases the generation of ROS, an essential mediator of
apoptosis, and induces the apoptosis of hepatocytes in liver
regeneration, which is associated with promoting the generation of
ROS from mitochondria and inducing cytochrome P450 1A1 (32).
TGF-β is also the target of other proliferation
inhibiting factors in liver regeneration. Cation-independent
mannose 6-phosphate receptor (CIMPR), an indirect negative
regulator of hepatocyte growth, which has progressively
overexpressed in liver regeneration 8 h after PH, mediates the
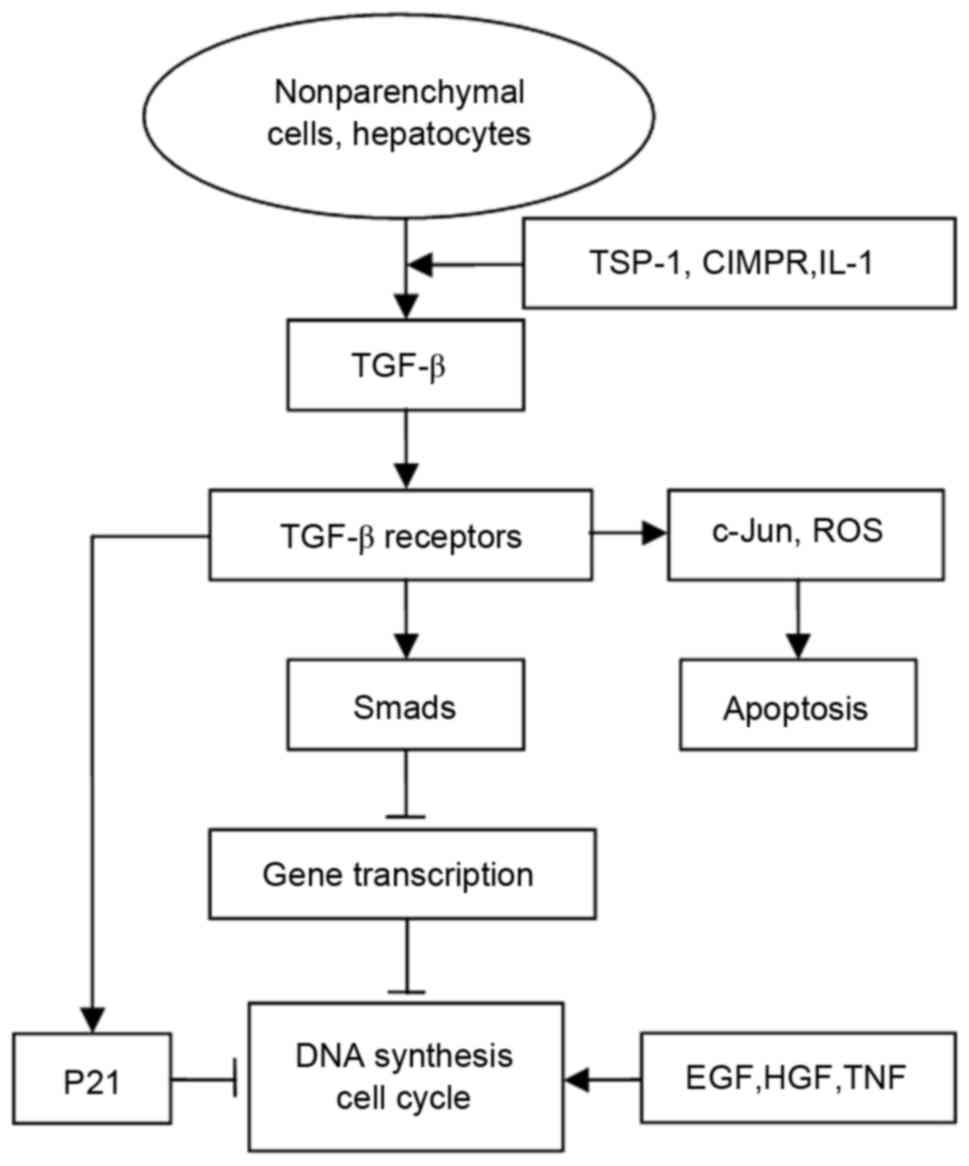
activation of latent pro-TGF-β (33). A schematic diagram of the TGF-β
signaling pathway is shown in Fig.
1.
Activin, another member of the TGF-β superfamily,
which is structurally related to TGF-β, also has an inhibitory
effect on hepatocyte proliferation and liver regeneration. Activin
A, with a similar ability to that of TGF-β, inhibits EGF-induced
DNA synthesis in an autocrine manner without competing with TGF-β
(34). A study by Schwal et
al indicated that activin induced the apoptosis of hepatocytes
in a different manner from that of TGF-β (35). The administration of follistatin,
an activin-binding protein that can inhibit the activity of
activin, enhances liver regeneration following PH (35–37).
The inhibitory effect of activin may also be inhibited by increased
expression of SnoN and Ski, which are Smad pathway inhibitors, in
early liver regeneration (19).
The impairment of activin type I receptors has also been shown to
increase DNA synthesis in hepatocytes, providing further evidence
of the anti-proliferative function of activin (38).
IL-1
IL-1 is another significant negative regulator of
cell proliferation in the process of liver regeneration. In 1988,
Takahashi et al first reported that IL-1 may be involved in
the suppression of liver regeneration on examining syngeneic spleen
cells treated with poly I:C in a mouse PH model (39). Another study by Nakamura et
al showed that IL-1β markedly inhibited DNA synthesis induced
by insulin and EGF in cultured rat hepatocytes (40). Similar inhibition of cultured
hepatocytes has also been found in other studies (41–43),
and enhanced liver regeneration by FK506 can be inhibited by IL-1
(44). The increased expression of
IL-1 impairs regeneration via an indirect mechanism in liver injury
induced by endotoxin following hepatectomy (45). The mRNA expression of IL-1α in the
whole rat liver was found to be downregulated at 10 h, and
upregulated 24 and 48 h following PH, suggesting an association
between the expression of IL-1 and liver regeneration (46). Further experiments have revealed
that sensitivity of hepatocytes isolated from the liver 24 h
following PH was increased to the inhibition of IL-1, and the
administration of exogenous IL-1β suppressed DNA synthesis at 0 and
12 h post-PH (46). Long-term
examination of the expression of IL-1 in the liver in liver
regeneration showed that the mRNA levels of IL-1 increased slowly
and marginally following 30 and 80% hepatectomy (47). The increased expression of IL-1β
was has also been observed in a shrinking liver lobe of a rat
portal vein ligation model, indicating that IL-1β was involved in
the process of cellular atrophy (48,49).
The expression of IL-1β was also increased in the regenerating
impaired liver caused by deficiency in the expression of peroxisome
proliferator-activated receptor α (50). The suppression of IL-1β by
FR167653, a selective inhibitor, was shown to promote liver
regeneration in rat models of classic PH (70%) and extended
hepatectomy (90%) (51,52). Hepatocyte proliferation was also
found to be enhanced by recombinant human interleukin 1 receptor
antagonist in a mouse model of acute liver injury induced by
CCl4 (53).
IL-1 is secreted by nonparenchymal cells (NPCs) in
the regenerating liver. Goss et al found that IL-1 is
produced by KCs and regulated by prostaglandin E2 (PGE2) in the rat
liver following PH (54,55). Heparin and PGE1 also suppress the
expression of IL-1 in KCs following PH (45). Increased mRNA levels of IL-1α and
β, which are associated with KCs and the spleen, were reported in
liver NPCs between 30 min and 1 h following PH (56). Medium conditioned by nonparenchymal
cells isolated from the regenerating liver inhibit DNA synthesis in
primary rat hepatocytes induced by HGF, EGF and TGF-α. This
suppression is inhibited by either IL-1 receptor antagonist or by
IL-1 α and β antibodies (46).
Although studies on the association between IL-1 and
liver regeneration have been performed, the inhibitory mechanism of
IL-1 on liver regeneration remains to be fully elucidated. Several
studies have shown that IL-1 receptor antagonist (IL-1Ra), a
competitive inhibitor of IL-1α and IL-1β, and anti-inflammatory
protein, inhibited the inhibition of IL-1 and facilitated liver
regeneration, suggesting that IL-1R was required for the inhibitory
action of IL-1 (53,57–59).
IL-1 was found to contribute to the impairment of liver
regeneration by reducing HGF and promoting TGF-β release in
reduced-size orthotopic liver transplantation models (59). A previous study showed that IL-1β
inhibits the fibroblast growth factor (FGF)19 signaling pathway,
which regulates cell growth and metabolism of hepatocytes in liver
regeneration (60). IL-1β also
inhibits the expression of β-Klotho, a co-receptor of FGF receptor
4 (FGFR4), with involvement of the c-Jun N-terminal kinase (JNK)
and nuclear factor (NF)-κB pathways. The activation of
extracellular signal-regulated kinase (Erk)1/2 and hepatocyte
proliferation induced by FGF19 is also suppressed by IL-1β.
IL-6
IL-6 is a multifunctional cytokine, which has been
investigated extensively. It is a known classical triggering signal
of liver regeneration following PH. The essential role of IL-6 and
its mechanisms in the process of liver regeneration have been
confirmed (61). IL-6 has been
shown to act as a positive regulator of liver regeneration in the
majority of studies, whereas it has also been shown that IL-6
impairs liver regeneration, which conflicts with the mainstream
findings. IL-6 inhibits mature hepatocyte proliferation and DNA
synthesis stimulated by TNF and EGF in primary culture (25,42).
Beyer and Theologides showed that IL-6 suppressed the DNA synthesis
of hepatocytes at 3 days in vitro (43). The inhibitory effect of IL-6 in
hepatocyte proliferation may be associated with induction of the
expression of p21 in vitro (62).
Upregulated levels of IL-6 have been observed in a
number of abnormal liver regeneration models. Increased blood
levels of IL-6, which induce the expression of protein inhibitor of
activated Stat-3 and the suppressor of cytokine signaling (SOCS)-1,
were found to be associated with impairment of liver regeneration
in a model of hepatic failure (63). Hyperstimulation with IL-6 also
suppressed hepatocyte proliferation by inducing the expression of
p21 in transgenic mice overexpressing the human soluble IL-6
receptor/gp80 following PH (64).
The overexpression of STAT3, a downstream molecule of IL-6, was
found to impair hepatocyte proliferation in fatty liver following
PH (65). In addition, the
suppression of hepatocyte proliferation was accompanied by
increased levels of IL-6 in IL-1Ra-knockout mice following PH
(57). The impairment of liver
regeneration by hepatitis B virus X protein (HBx) via upregulating
the expression of IL-6 was observed in an HBx-overexpressed
transgenic mouse model (66). In
addition to the data obtained in animal models, elevated levels of
IL-6 have also been found in patients with chronic liver disease
(67–70).
Although the mechanisms of IL-6 in liver
regeneration are controversial, the majority of associated studies
support the positive role of IL-6, with IL-6 affecting hepatocyte
proliferation in a dose- and time-dependent manner (71,72).
The studies mentioned above also indicated that the inhibitory
effects of IL-6 in liver regeneration, particulary on a background
of liver disease, may be associated with its hyperstimulation of
liver cells.
SOCS
SOCS is a family of proteins, which negatively
regulate various cytokine signals (2). SOCS inhibits the activity of STAT3, a
downstream signal of cytokines and growth factors in hepatic
regeneration. SOCS3, a feedback inhibitor of the IL6/JAK/STAT3
pathway, is an important member of the SOCS family, which has
received the most attention in investigations of liver
regeneration. SOCS3 is transiently upregulated in the livers of
mice following PH and is involved in the inactivation of STAT3
signaling (73–75). SOCS3 can inhibit the
phosphorylation of gp130, a component of the IL-6 receptor, JAKs
and STATs (76). SOCS3
hepatocyte-specific-knockout mice showed increased capability of
cell proliferation and prolonged STAT3 phosphorylation following
PH, with enhanced activation of ERK1/2 also observed (77). SOCS3 can also negatively control
the proliferative responses mediated by HGF and EGF (78). The overexpression of SOCS3
suppresses IL-22 signaling, another cytokine positively correlated
with liver regeneration, via eliminating the IL-22-induced
activation of STAT (79). The
expression of SOCS3 is primarily induced by IL-6 and is also
upregulated by other factors, including TNFα, IL-1, IL-22 and
ZIP14, a zinc transporter (73,79–81).
SOCS1, another member of the SOCS family, has a
similar but weaker function to that of SOCS3 (74,78).
Unlike SOCS3, SOCS1 does not affect the growth signaling mediated
by EGFR (78). A previous study
indicated that SOCS1 negatively regulated the hepatocyte
proliferation induced by HGF by inhibiting c-Met signaling
(82).
IFN-γ
IFN-γ can downregulate hepatocyte proliferation by
activating STAT1 and its downstream genes, including RF-1, p21 and
SOCS1, in liver regeneration (83). The inhibitory effect of invariant
natural killer T (iNKT) cells, a major lymphocyte in the liver, was
also associated with IFN-γ (84).
iNKT cells affect liver regeneration to a lesser degree. The
deficiency of iNKT cells (CD1d and Jα281 cells) does not alter
mouse liver regeneration following PH. However, treatment with
α-galactosylceramide, an inducer of iNKT cell activation, impairs
hepatocyte proliferation via upregulating the expression of IFN-γ
in wild-type mice. Experiments also indicated that IL-4 is the
primary mediator in the activation of iNKT cells and induction of
IFN-γ (84).
Other cytokines
There are other cytokines, which negatively regulate
liver regeneration. TNF-α, another pro-proliferation factor in
liver regeneration, also has the ability to induce the apoptosis of
adenovirus-infected hepatocytes in a coupled TGF-α-IL-1α/β-IL-1ra
autocrine cascade manner (85).
The administration of α2b-INF inhibits DNA biosynthesis and liver
mass restitution by affecting the expression of cell
cycle-associated genes, includin c-myc, p53 and c-erbB-2, in rat PH
models (86). Further experiments
have indicated that the suppression of hepatocyte proliferation
depends on the time at which α2b-IFN is administered (87), and the proliferation of hepatocytes
was found to be inhibited by granulocyte colony-stimulating factor
via upregulating the expression of IL-1β in a liver injury model
induced with dimethylnitrosamine (88). Mice with single deficiency of
myeloid differentiation factor 88 (MyD88), an adaptor protein for
the majority of Toll-like receptors, presented with enhanced
hepatocyte proliferation at 32 and 36 h post-PH (89). The decreased activation of STAT3
and induction of SOCS3 have also been detected in MyD88-null mice
following PH (89). These results
suggest that MyD88 has an antiproliferative effect in liver
regeneration by affecting IL-6 signaling pathways. A summary of the
cytokines involved in inhibiting liver regeneration is presented in
Table I.
 | Table I.Cytokines involved in inhibition of
liver regeneration. |
Table I.
Cytokines involved in inhibition of
liver regeneration.
| Cytokine | Source | Upstream | Downstream |
|---|
| TGF-β | NPCs (HSCs, KCs and
platelets), hepatocytes | TSP-1, CIMPR,
IL-1 | TGF-β receptors,
Smads (inhibit gene transcription) c-Jun, ROS (promote
apoptosis) |
| IL-1 | KCs | Heparin, PGE1,
PGE2, G-CSF | IL-1 receptor,
TGF-β, FGF19, β-Klotho, JNK, NF-κB, Erk1/2 |
| IFN-γ | iNKT cells | IL-4 | STAT1 and
downstream genes (RF-1, p21 and SOCS1) |
| α2b-IFN | Ectogenesis |
| Cell cycle-related
genes (c-myc, p53 and c-erbB-2) |
| G-CSF | Ectogenesis | IL-1 |
|
| MyD88 | Hepatocytes |
| STAT3 (inhibition),
SOCS3 (induction) |
Oncogene-associated pathways
P53
The role of p53, a negative regulator of growth and
tumor suppressor genes, is complex in the process of liver
regeneration and conclusions from different studies are
contradictory. The expression of p53 is upregulated, corresponding
to the G1/S transition, in the early stage of liver regeneration
following PH (9,90). It has been shown that the
overexpression of p53 by a transducing adenoviral vector encoding
wild-type p53, did not affect the process of liver regeneration
(91), however, other studies
showed that p53 may positively regulate hepatocyte proliferation in
rats. The suppression of the expression of p53 results in the
reduction of DNA replication in hepatocytes in vitro
(92). A study by Inoue et
al showed that the levels of p53 increased in cultured
hepatocytes induced with HGF, and inhibiting the expression of p53
by an antisense oligonucleotide resulted in a decrease in the
levels of TGF-α and of DNA synthesis (93). It was also found that upregulation
of the expression of p53 was synchronous with HGF followed by an
increase of TGF-α in a rat PH model, suggesting that p53 is
positively correlated with liver regeneration (93).
However, other findings have supported that p53
negatively controls liver regeneration. Previous studies have
provided indirect evidence of the anti-proliferation functions of
p53. Suppressing p53 with antisense oligonucleotides enabled
hepatocytes to recover from growth arrest induced by ultraviolet
(94). Increased expression of
p53, accompanied by a downregulation in the levels of proliferating
cell nuclear antigen (PCNA), was observed in rats treated with a
metallothionein antisense oligonucleotide, suggesting an inverse
correlation between p53 and liver regeneration (95). The detailed role of p53 in liver
regeneration was examined by Arora and Iversen (96). Increased weight gain of
remnant-regenerating liver, CYP isoform activities and expression
of PCNA were observed in rat PH models, in which the expression of
p53 was inhibited with an antisense oligonucleotide, whereas the
G1 cell populations and levels of P21 were also reduced
in the p53-inhibited rats, indicating that p53 controls hepatocyte
proliferation in a cell cycle checkpoint manner (96). Furthermore, loss of the expression
of p53 resulted in early entry into the cell cycle and prolonged
the proliferation of hepatocytes in p53 (−/−) mice following PH,
and the inhibitory mechanism of p53 was correlated with the
expression of mitotic genes, including Forkhead box M1, Aurka,
Large tumor suppressor kinase 2, Polo like kinase (Plk)2 and Plk4
(97). Abrogation of the p53
pathways also facilitates liver regeneration in Wild-type
p53-induced phosphatase 1-deficient mice following PH (98).
P53 can also suppress cell proliferation in
regeneration of damaged liver on a background of chronic disease or
drug treatment. An increased expression of p53 followed by the
induction of p21 was associated with the inhibition of hepatocyte
proliferation in PH models treated with 2-acetaminofluorene
(99). Impaired liver regeneration
and increased expression of p21 following partial PH was observed
in mice lacking c-Jun, a key regulator of hepatocyte proliferation,
whereas the activation of p53 eliminates the effect, indicating
that p53 is a proliferation inhibitor in liver regeneration
(100). The activation of a
p53-dependent checkpoint correlates with premitotic arrest and
abnormal cell death in transgenic mice overexpressing human
Aurora-A following PH (101). The
expression levels of p53 and p21 are also upregulated, coinciding
with cell cycle arrest and apoptosis in hepatocytes of liver injury
models induced by acetaminophen (102–104).
P21
P21, the expression product of the ras gene, also
termed wide-type 53-activated factor 1 or cyclin-dependent kinase
interacting protein 1, has been identified as a cyclin-dependent
kinase inhibitor and downstream gene of p53 (105–107). The negative control role of p21
has been confirmed in various studies. The expression of p21
increased maximally following peak hepatocyte DNA synthesis and
cell division, later than that of p53, in rat and mouse PH models,
indicating its association with the proliferation of liver cells
(90,108). The upregulation in levels of p21
have been examined not only hepatocytes, but also nonparenchymal
cells, in the process of liver regeneration (109). In addition to the data mentioned
above, overexpressed p21 also coincides with the arrestment of
liver regenerating in several disease and drug toxicity models
(65,110–115).
Although p21 is a classic downstream signal of p53,
the expression of p21 is induced by other factors in addition to
p53 in liver regeneration. Increased mRNA levels of p21 induced by
p53 were identified and involved in inhibiting hepatocyte
proliferation in 2-acetaminofluorene-treated rats (120). Upregulated levels of p53 and p21
were also correlated with impaired liver regeneration in acute
liver injury models induced by acetaminophen, and inhibition of the
p53/p21 pathway promoted hepatocyte proliferation (102,121). However, no difference in the gene
expression of p21 was found between p53-deficient and wild-type
mice following PH, although the post-transcriptional regulation may
be associated with p53 (116).
Furthermore, treatment with TGF-β and activin A upregulated the
expression of p21 in cultured primary hepatocytes (116). These data suggested that the
induction of p21 is controlled in a p53-dependent and independent
manner. Similar expression and localization of p21 was also found
in wild-type and p53-null mice treated with carbon tetrachloride,
and the increased levels of p21 might be correlated with
CCAAT/enhancer-binding protein (C/EBP) (117). Other experiments have shown that
levels of p21 were regulated by C/EBPα (122). Increased levels of p21 were
observed in c-Jun-knockout mice following PH, and inactivation of
the p53/p21 signaling pathway abrogated the suppression of liver
regeneration, suggesting that the expression of p21 may be
regulated by c-Jun (100).
B-cell lymphoma-2 (Bcl-2)
Important in the proliferation of tumor cells, Bcl-2
also inhibits hepatocyte proliferation in liver regeneration. Bcl-2
transgenic mice show delayed hepatocyte DNA synthesis following PH,
and a similar result is observed in cultured liver cells. In
addition, the overexpression of Bcl-2 is followed by decreased
expression levels of p107 and cyclin E (123). These results indicate that Bcl-2
may be a negative cell cycle modulator in liver regeneration.
P53 upregulated modulator of apoptosis (PUMA),
another member of the Bcl-2 family, is also negatively correlated
with liver regeneration. PUMA is downregulated, accompanied by
increased apoptosis and decreased proliferation, in the early phase
post-PH (124). Bcl-2-associated
X protein (Bax) inhibitor-1 (BI-1), which has a similar function to
that of Bcl-2 family proteins, also regulates liver cell
proliferation in vivo. Enhanced liver regeneration,
accompanied by the increased expression of cell cycle regulators,
including cyclin D1, cyclin D3, Cdk2 and Cdk4, have been observed
in BI-1-deficient mice following PH (125).
Other oncogene-associated
pathways
Several studies have shown that additional oncogenes
can inhibit liver regeneration. P27 is also a tumor suppressor gene
and cyclin-dependent kinase inhibitor. P27 was shown to be
minimally expressed in the mouse liver following PH in a
time-dependent manner, and p27 decreased the activity of CDK2 prior
to and following peak DNA synthesis, suggesting its negative role
in liver regeneration (108).
Fas, also known as APO-1 or CD95, and Fas ligand
(FasL) are essential in cell apoptosis and regulate the
proliferation of tumor cells (126). The expression of Fas decreased
significantly immediately in the rat liver and had recovered
gradually to a normal level of quiescent hepatocytes 14 days
following PH, suggesting that the Fas/FasL system was also
negatively correlated with liver regeneration (127).
N-Myc downstream-regulated gene 2 (NDRG2), is also a
known tumor suppressor gene and its expression is reduced or
undetectable in various types of tumor (128). The significant downregulation of
NDRG2 was examined in regenerating livers, and the overexpression
of NDRG2 resulted in a cell cycle arrest via upregulating the
p53/p21 pathway and inhibiting cyclin E-Cdk2 activity (129). These data indicate that NDRG2 has
a potential negative effect in liver regeneration. A summary of the
oncogenes involved in inhibiting liver regeneration is presented in
Table II.
 | Table II.Oncogenes involved in inhibition of
liver regeneration. |
Table II.
Oncogenes involved in inhibition of
liver regeneration.
| Oncogene | Upstream | Downstream | Function |
|---|
| P53 |
| P21
(upregulating) | Cell cycle
checkpoint |
| P21 | P53, TGF-β, activin
A, C/EBPα | Cell cycle-related
protein (downregulating) | Cell cycle
checkpoint |
| Bcl-2 |
| p107, cyclin E
(downregulating) | Negatively
modulates cell cycle |
| PUMA |
| Bax, macrophage
inflammatory protein-2 | Another member of
the Bcl-2 family |
| BI-1 |
| Cell cycle-related
protein (downregulating) | Negatively
modulates cell cycle |
| P27 |
| CDK2
(downregulating) | Negatively
modulates cell cycle |
| Fas |
| – | Induces
apoptosis |
| NDRG2 |
| P53/p21 | Negatively
modulates cell cycle |
Gene expression regulation-associated
pathways
Transcription factor-associated
pathways
Transcription factors important roles in liver
regeneration, however, the majority of them positively regulate
liver regeneration. Other transcription factors, which may have
inhibitory roles in liver regeneration, are less well
understood.
C/EBPα, a member of the CCAAT enhancer-binding
protein family, is expressed abundantly in the normal liver.
However, the expression of C/EBPα was found to decrease markedly in
proliferating cells following PH and in cultured hepatocytes
stimulated with EGF and insulin, suggesting an inverse correlation
between C/EBPα and liver regeneration (130). Positive correlation between p21
and C/EBPα has been observed in C/EBPα-knockout mice. C/EBPα did
not affect the mRNA levels of p21 but inhibited the proteolytic
degradation of p21, indicating that C/EBPα inhibited hepatocyte
proliferation via controlling the expression of p21 in a
post-transcriptional manner (122). In aged animal PH models, C/EBPα
also suppressed the expression of E2F via forming the
C/EBPα-Rb-E2F4 complex, which can bind to E2F-dependent promoters,
inhibiting the expression of E2F and inhibiting cell proliferation
(131).
Forkhead box (Fox)O1, a transcription factor
regulated by Akt, may also be an anti-proliferative factor in liver
regeneration. Deleting Akt1 and Akt2 was shown to result in
impairment of liver regeneration in mice following PH, whereas
liver-specific deletion of FoxO1 reversed this effect, indicating
that FoxO1 has a negative effect on liver regeneration (132).
MicroRNAs (miRNAs) and the inhibition of
liver regeneration
MicroRNAs (miRNAs) have an effect on the
post-transcriptional control of gene expression and their
association with liver regeneration has attracted increasing
interest. A number of miRNAs have been identified as liver
regeneration inhibitors, including miR-26a, miR-33, miR-34a,
miR-127, miR-150 and miR-378 (133).
Protein modification-associated
pathways
Previous studies have shown that certain
anti-proliferation inhibitors are involved in liver regeneration
via regulating the protein modification of downstream molecules,
including phosphorylation. Of note, SOCS, the natural terminators
of cytokine signaling, are involved via regulating the
phosphorylation of downstream molecules (76). Tyrosine phosphorylation-mediated
signaling, including the EGF and HGF pathways, is required in liver
regeneration, whereas protein tyrosine phosphatase 1B (PTP1B), a
key regulator of metabolism and cell growth, is involved in the
dephosphorylating process of the tyrosine kinase superfamily.
PTP1B-deficient mice present with enhanced cell proliferation and
increased phosphorylation of JNK1/2 and STAT3, the downstream
molecules of TNF-α and IL-6, which trigger liver regeneration
following PH (134). Similar
phosphorylation was observed in wild-type cells following the
silencing of PTP1B, whereas the overexpression of PTP1B led to the
opposite result. Accumulated tyrosine phosphorylation of EGFR and
hepatocyte growth factor receptor (HGFR), followed by increased
activation of Akt and ERK, were also examined in PTP1B-/− mice
following PH. These data indicate that PTP1B negatively regulates
liver regeneration via dephosphorylating key molecules of the
signaling pathways, which promotes cell proliferation.
The Hippo pathway is a novel pathway, which is
associated with organ size control and tumorigenesis (135). The Hippo pathway consists of
several core kinases, including the mammalian Ste20-like kinases
(Mst1/2) and Salvador 1, and activates large tumor suppressor
(Lats1/2), exerting effects via a series of phosphorylating
processes of these kinases. The final effect of the Hippo pathway
is to phosphorylate and inhibit the activity of Yes-associated
protein (YAP), a transcription co-activator, and the
transcriptional co-activator with PDZ-binding motif, which
regulates different proliferation-associated transcription factors.
Increased activation of YAP has been found in the early stage of
rat liver regeneration following PH, whereas the activation of
Mst1/2 and Lats1/2 were downregulated at this time (136). The levels of YAP and Mst1/2
kinase returned to their normal levels, as in quiescent liver
cells, when the liver size was almost restored, suggesting a
negative role of the Hippo pathway in liver regeneration.
Mg(2+)/Mn(2+)-dependent phosphatase 1A (PPM1A) is
another phosphatase identified in a previous study, which has
inhibitory potential in liver regeneration. The expression of PPM1A
was decreased, which was positively correlated with the expression
of Cyp3a11 in the early stage of liver regeneration in mice
(137). Furthermore, inhibition
of the expression of PPM1A leads to the enhanced proliferation of
HepG2 cells. The detailed mechanism of PPM1A in the control of
liver regeneration requires further investigation in the
future.
Mitogen-inducible gene-6 (mig-6), an adaptor protein
associated with the modulation of receptor tyrosine kinases,
including EGFR, is also known as receptor-associated late
transducer, gene33 and Errfi1 (138,139). In mig-6-knockout mice,
accelerated hepatocyte proliferation is observed in the early stage
of liver regeneration following PH. Downregulating the expression
of mig-6 also enhances EGFR signaling via the protein kinase B
pathway in mouse PH models (140), suggesting that mig-6 is a
negative regulator in liver regeneration. A summary of the protein
modification-associated pathways involved in the inhibiton of liver
regeneration is shown in Table
III.
 | Table III.Protein modification-associated
pathways in liver regeneration inhibition. |
Table III.
Protein modification-associated
pathways in liver regeneration inhibition.
| Name | Downstream | Function |
|---|
| SOCS | STAT3 | Downregulating
STAT3 phosphorylation |
| PTP1B | JNK1/2, STAT3,
EGFR, HGFR | Dephosphorylating
tyrosine kinases |
| Hippo pathway | YAP | Inhibiting
proliferation-associated transcription factors |
| PPM1A | Cyp3a11 |
|
| Mig-6 | Protein kinase B,
EGFR | Modulating
phosphorylation of EGFR |
Metabolism-associated pathways
Metabolism is an important function of the liver and
also affects the process of liver regeneration. Several metabolic
factors have been reported to be associated with the impairment of
liver regeneration. For example, very low-density lipoprotein has
been shown to suppress hepatocyte proliferation in in vitro
and in vivo experiments (141). The overadministration of glucose
has also been shown to arrest DNA synthesis and cell cycle in liver
injury models, indicating that glucose loading depresses liver
regeneration (142).
S-adenosylmethionine (SAMe), the primary methyl
donor in the liver, is important in the metabolism of cells and
regulates cell proliferation. Adenosine monophosphate-activated
protein kinase (AMPK), the principal energy sensor in cells, can be
inhibited by SAMe. A study by Vazquez-Chantada et al showed
that hepatocyte proliferation was inhibited by SAMe in mouse liver
regeneration (143). SAMe
inhibited the HGF-mediated phosphorylation of AMPK, which
upregulated cytoplasmic HuR and certain cell cycle proteins,
including cyclin D1 and A2 in liver cells. Furthermore,
phosphorylation of the LKB1/AMPK/endothelial nitric oxide synthase
cascade mediated by HGF was suppressed in mouse PH models
pretreated with SAMe. Evidence of the inhibitory function of SAMe
on AMPK and liver regeneration was also obtained in another study
(144).
Sirtuin 1 (SIRT1) is an important metabolic
modulator of gluconeogenesis, fat, protein and bile acid (BA), and
is also associated with liver regeneration. Impaired hepatocyte
proliferation accompanied by BA accumulation and damaged farnesoid
X receptor (FXR) activity was observed in transgenic mice
overexpressing SIRT1 following PH (145). The overexpression of SIRT1
resulted in the persistent deacetylation of FXR, which was inversed
by 24-norursodeoxycholic acid, followed by decreased expression of
FXR-target genes, including small heterodimer partner and bile salt
export pump. These data indicate that SIRT1 may negatively regulate
liver regeneration by controlling the metabolism of BA.
Hormone-associated pathways
The association between hormones and liver
regeneration has been investigated in previous studies (146). Norepinephrine upregulates the
level of TGF-β via the α1-adrenergic receptor (α1-AR) in cultured
rat hepatocytes (147). The
levels of adrenal hormones are negatively correlated with DNA
synthesis and cell cycle arrest, in a circadian manner, during
liver regeneration in hepatectomized animals (148). The expression and function of
α1-AR are decreased, in contrast to the increased level of β2-AR,
in hepatocytes following PH. Of note, treatment with isoproterenol,
a β-AR agonist, inhibits 3H-thymidine incorporation of hepatocytes
isolated from PH models via upregulating the activation of
stress-activated protein kinase and inhibiting p42 MAP kinase. This
was inhibited by propranolol, a β-AR antagonist (149).
Hydrocortisone treatment results in the persistent
suppression of hepatocyte proliferation and increased expression of
P21 in PH models (150).
Pretreatment with dexamethasone also inhibits the proliferation of
hepatocytes, accompanied by reduced expression levels of TNF and
IL-6 (151). This suggests that
glucocorticoid hormone has an inhibitory effect on liver
regeneration. Short-term administration of ethinyl estradiol, a
potent promoter of hepatocarcinogenesis in female rats, leads to an
initial, transient increase in hepatocyte proliferation. However,
long-term treatment with ethinyl estradiol results in the
inhibition of liver regeneration, without affecting DNA synthesis,
following PH (152). Somatostatin
also inhibits DNA synthesis induced by HGF and EGF via a
cAMP-independent mechanism, indicating that it may be a potent
inhibitor of liver regeneration (153).
Pathological factors and liver
regeneration
Pathological alterations caused by various factors,
including ethanol, drugs and hepacivirus, on liver regeneration is
generally accepted. The abnormal expression of cytokines and
proliferative inhibitors, induced by acute or chronic pathological
changes, is associated with the impairment of liver regeneration.
Their association was discussed above.
Hepatitis, cirrhosis and hepatic fibrosis, the most
common pathological changes in the liver, negatively affect liver
regeneration. T cell-mediated hepatitis leads to reduced liver
regeneration via altered expression levels of certain cytokines and
associated signaling molecules, for example the upregulated
expression of p21, Smad2, TGF-β and IFN-γ, and downregulated
expression of cyclin D and activation of Stat3 (20). In addition to the involvement of
cytokines, other factors are associated with impaired regeneration
in liver disease models. For example, hypoxia in hepatocytes caused
by cirrhosis suppresses the hepatocyte proliferation induced by HGF
and c-Met in vivo (154).
The activation of HSCs, the primary cause of hepatic fibrosis, also
inhibits liver regeneration and can be attenuated by STI-571
(155).
The consumption of ethanol, which leads to
ethanol-induced liver injury, is another common harmful factor
affecting liver regeneration. Long-term ethanol intake inhibits
hepatic DNA synthesis following PH, which is associated with a
change in hepatocyte sensitivity to TNF (156). Acute and chronic ethanol
consumption can inhibit liver regeneration by different mechanisms.
The suppression of rat liver regeneration by acute ethanol is
accompanied by upregulated levels of p53, prohibitin and TGFβ-1
(157). Prolonging of the
activation of p42/44 MAPK and decreased expression of
γ-aminobutyric acid transport protein are also involved in the
damaged proliferation of hepatocytes (158,159). By contrast, prothymosin-α, p38
MAPK and p21 are associated with impaired liver regeneration
following chronic ethanol administration (158,160,161).
Fatty liver also exhibits impaired regenerative
capacity. The over-phosphorylation of Stat-3, inhibition of Jun
N-terminal kinase, and decreased expression of NF-κB and cyclinD1
were found to be associated with the inhibition of proliferation in
mouse nonalcoholic fatty liver disease models following PH
(162). Furthermore, enhanced
activation of STAT3 was found to be positively correlated with the
levels of p21 in the regenerating process of fatty liver models
(65).
The administration of drugs is another common factor
in liver injury and the inhibition of liver regeneration. For
example, amiloride, a weak diuretic and inhibitor of
Na+/H+ exchange, has the ability to suppress
DNA synthesis of proliferating hepatocytes in vitro and
in vivo (163,164). The amiloride analog, 5-(N,
N-hexamethylene)-amiloride, another inhibitor of
Na+/H+ exchange, not only inhibits DNA
synthesis, but also leads to apoptosis of hepatocytes in the rat
liver following PH. Induction of the p53/p21 pathway is associated
with the inhibitory function of 5-(N, N-hexamethylene)-amiloride
(164).
The toxicity caused by intake of certain
microelements can also damage liver regeneration. For example,
selenium, a non-metal element, was shown to arrest DNA synthesis
and cell cycle in rat PH models, which may be associated with the
reduction of glutathione (165).
Cadmium, a heavy metal element, is also harmful to liver
regeneration involving inhibition of the enzymatic activity of
thymidine kinase (166).
Other inhibitors of liver regeneration
There are certain proliferation inhibitors, which
cannot be classified into the categories mentioned above.
Regucalcin, a Ca2+-binding protein, was found to be
downregulated in the rat liver at 1–3 days post-PH. Inhibition of
nuclear DNA synthesis was found to be caused by regucalcin in
hepatocytes isolated from PH rats, and Ca2+ treatment
(1.0–25 µM), which led to a similar result, indicated the negative
effect for regucalcin in liver regeneration (167).
Peroxisome proliferator-activated receptor-γ
(PPAR-γ), a nuclear receptor, which induces cell-cycle arrest and
apoptosis in tumor cells, appears to have a negative correlation
with liver regeneration (168).
The expression of PPAR-γ was found to decrease in the rat liver at
24 h and increase at 48–120 h post-PH, whereas the consumption of
pioglitazone, a PPAR-γ agonist, suppressed the proliferation of
liver cells.
Vitamin D3 upregulated protein 1 (VDUP1), a
regulator of cellular metabolism and endoplasmic reticulum (ER)
stress, is also a suppressor of cell proliferation in various types
of tumor. A previous study showed that VDUP1 was negatively
correlated with liver regeneration (169). Enhanced proliferative responses
were observed in the process of liver regeneration in
VDUP1-knockout mice. The increased expression of cell-cycle
proteins and the activation of proliferative signals appeared
earlier in the VDUP1-knockout mice following PH.
Transmembrane and ubiquitin-like domain containing 1
(Tmub1), a novel gene, which is upregulated in the early stage of
liver regeneration, was shown to have a negative effect in
IL-6-induced hepatocyte proliferation via its interaction with
calcium modulating cyclophilin ligand in vitro (72). Another study showed that the
expression of Tmub1 may be regulated by IL-6 and C/EBPβ (170).
Conclusion and outlook
Proliferation-inhibiting signals are active in the
entire process of liver regeneration, a number of which are induced
in the early stage of liver regeneration when the cells are engaged
in proliferation. In addition, the expression of other inhibitors
are decreased at the same phase, but increased following peak
proliferation. They contribute to control of the degree of
regeneration and prevent oncogenesis through different mechanisms,
with the function of proliferation-inhibiting pathways being as
important as proliferation-promoting signals in liver
regeneration.
However, knowledge of the proliferation-inhibiting
pathways remains less than that of the proliferation-promoting
pathways in liver regeneration. The majority of the signaling
pathways described above remain to be fully elucidated. A focus is
required on the elucidation of proliferation-inhibiting pathways.
The identification of additional proliferation inhibitors, together
with their upstream and downstream molecules, is required, as is an
emphasis on the association between pro-proliferation and
anti-proliferation signals.
Acknowledgements
This study was supported by a grant from the
Natural Science Foundation of China (grant no. 81400651).
References
|
1
|
Jia C: Advances in the regulation of liver
regeneration. Expert Rev Gastroenterol Hepatol. 5:105–121. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Elliott J: SOCS3 in liver regeneration and
hepatocarcinoma. Mol Interv. 8:19–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Willenbring H, Sharma AD, Vogel A, Lee AY,
Rothfuss A, Wang Z, Finegold M and Grompe M: Loss of p21 permits
carcinogenesis from chronically damaged liver and kidney epithelial
cells despite unchecked apoptosis. Cancer cell. 14:59–67. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakamura T, Tomita Y, Hirai R, Yamaoka K,
Kaji K and Ichihara A: Inhibitory effect of transforming growth
factor-beta on DNA synthesis of adult rat hepatocytes in primary
culture. Biochem Biophys Res Commun. 133:1042–1050. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Russell WE, Coffey RJ Jr, Ouellette AJ and
Moses HL: Type beta transforming growth factor reversibly inhibits
the early proliferative response to partial hepatectomy in the rat.
Proc Natl Acad Sci USA. 85:5126–5130. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liska V, Treska V, Mirka H, Kobr J, Sykora
R, Skalicky T, Sutnar A, Vycital O, Bruha J, Pitule P, et al:
Inhibition of transforming growth factor beta-1 augments liver
regeneration after partial portal vein ligation in a porcine
experimental model. Hepatogastroenterology. 59:235–240.
2012.PubMed/NCBI
|
|
7
|
Karkampouna S, Goumans MJ, Ten Dijke P,
Dooley S and Kruithof-de Julio M: Inhibition of TGFβ type I
receptor activity facilitates liver regeneration upon acute CCl
intoxication in mice. Arch Toxicol. 90:347–357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Braun L, Mead JE, Panzica M, Mikumo R,
Bell GI and Fausto N: Transforming growth factor beta mRNA
increases during liver regeneration: A possible paracrine mechanism
of growth regulation. Proc Natl Acad Sci USA. 85:1539–1543. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fausto N, Mead JE, Braun L, Thompson NL,
Panzica M, Goyette M, Bell GI and Shank PR: Proto-oncogene
expression and growth factors during liver regeneration. Symp
Fundam Cancer Res. 39:69–86. 1986.PubMed/NCBI
|
|
10
|
Bissell DM, Wang SS, Jarnagin WR and Roll
FJ: Cell-specific expression of transforming growth factor-beta in
rat liver. Evidence for autocrine regulation of hepatocyte
proliferation. J Clin Invest. 96:447–455. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nishikawa Y, Wang M and Carr BI: Changes
in TGF-beta receptors of rat hepatocytes during primary culture and
liver regeneration: Increased expression of TGF-beta receptors
associated with increased sensitivity to TGF-beta-mediated growth
inhibition. J Cell Physiol. 176:612–623. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jakowlew SB, Mead JE, Danielpour D, Wu J,
Roberts AB and Fausto N: Transforming growth factor-beta (TGF-beta)
isoforms in rat liver regeneration: Messenger RNA expression and
activation of latent TGF-beta. Cell Regul. 2:535–548.
1991.PubMed/NCBI
|
|
13
|
Bottinger EP, Factor VM, Tsang ML,
Weatherbee JA, Kopp JB, Qian SW, Wakefield LM, Roberts AB,
Thorgeirsson SS and Sporn MB: The recombinant proregion of
transforming growth factor beta1 (latency-associated peptide)
inhibits active transforming growth factor beta1 in transgenic
mice. Proc Natl Acad Sci USA. 93:5877–5882. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Strain AJ, Frazer A, Hill DJ and Milner
RD: Transforming growth factor beta inhibits DNA synthesis in
hepatocytes isolated from normal and regenerating rat liver.
Biochem Biophys Res Commun. 145:436–442. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Miyazono K, Kusanagi K and Inoue H:
Divergence and convergence of TGF-beta/BMP signaling. J Cell
Physiol. 187:265–276. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Attisano L and Wrana JL: Signal
transduction by the TGF-beta superfamily. Science. 296:1646–1647.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pañeda C, Gorospe I, Herrera B, Nakamura
T, Fabregat I and Varela-Nieto I: Liver cell proliferation requires
methionine adenosyltransferase 2A mRNA up-regulation. Hepatology.
35:1381–1391. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ten Dijke P, Goumans MJ, Itoh F and Itoh
S: Regulation of cell proliferation by Smad proteins. J Cell
Physiol. 191:1–16. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Macias-Silva M, Li W, Leu JI, Crissey MA
and Taub R: Up-regulated transcriptional repressors SnoN and Ski
bind Smad proteins to antagonize transforming growth factor-beta
signals during liver regeneration. J Biol Chem. 277:28483–28490.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hines IN, Kremer M, Isayama F, Perry AW,
Milton RJ, Black AL, Byrd CL and Wheeler MD: Impaired liver
regeneration and increased oval cell numbers following T
cell-mediated hepatitis. Hepatology. 46:229–241. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhong Z, Tsukada S, Rehman H, Parsons CJ,
Theruvath TP, Rippe RA, Brenner DA and Lemasters JJ: Inhibition of
transforming growth factor-beta/Smad signaling improves
regeneration of small-for-size rat liver grafts. Liver Transpl.
16:181–190. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schrum LW, Bird MA, Salcher O, Burchardt
ER, Grisham JW, Brenner DA, Rippe RA and Behrns KE: Autocrine
expression of activated transforming growth factor-beta(1) induces
apoptosis in normal rat liver. Am J Physiol Gastrointest Liver
Physiol. 280:G139–G148. 2001.PubMed/NCBI
|
|
23
|
Hayashi H, Sakai K, Baba H and Sakai T:
Thrombospondin-1 is a novel negative regulator of liver
regeneration after partial hepatectomy through transforming growth
factor-beta1 activation in mice. Hepatology. 55:1562–1573. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gohda E, Matsunaga T, Kataoka H and
Yamamoto I: TGF-beta is a potent inhibitor of hepatocyte growth
factor secretion by human fibroblasts. Cell Biol Int Rep.
16:917–926. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Satoh M and Yamazaki M: Tumor necrosis
factor stimulates DNA synthesis of mouse hepatocytes in primary
culture and is suppressed by transforming growth factor beta and
interleukin 6. J Cell Physiol. 150:134–139. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bouzahzah B, Fu M, Iavarone A, Factor VM,
Thorgeirsson SS and Pestell RG: Transforming growth factor-beta1
recruits histone deacetylase 1 to a p130 repressor complex in
transgenic mice in vivo. Cancer Res. 60:4531–4537. 2000.PubMed/NCBI
|
|
27
|
Moriuchi A, Hirono S, Ido A, Ochiai T,
Nakama T, Uto H, Hori T, Hayashi K and Tsubouchi H: Additive and
inhibitory effects of simultaneous treatment with growth factors on
DNA synthesis through MAPK pathway and G1 cyclins in rat
hepatocytes. Biochem Biophys Res Commun. 280:368–373. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takiya S, Tagaya T, Takahashi K, Kawashima
H, Kamiya M, Fukuzawa Y, Kobayashi S, Fukatsu A, Katoh K and Kakumu
S: Role of transforming growth factor beta 1 on hepatic
regeneration and apoptosis in liver diseases. J Clin Pathol.
48:1093–1097. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ueberham E, Löw R, Ueberham U, Schönig K,
Bujard H and Gebhardt R: Conditional tetracycline-regulated
expression of TGF-beta1 in liver of transgenic mice leads to
reversible intermediary fibrosis. Hepatology. 37:1067–1078. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leu JI, Crissey MA and Taub R: Massive
hepatic apoptosis associated with TGF-beta1 activation after Fas
ligand treatment of IGF binding protein-1-deficient mice. J Clin
Invest. 111:129–139. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Samson CM, Schrum LW, Bird MA, Lange PA,
Brenner DA, Rippe RA and Behrns KE: Transforming growth
factor-beta1 induces hepatocyte apoptosis by a c-Jun independent
mechanism. Surgery. 132:441–449. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Albright CD, Salganik RI, Craciunescu CN,
Mar MH and Zeisel SH: Mitochondrial and microsomal derived reactive
oxygen species mediate apoptosis induced by transforming growth
factor-beta1 in immortalized rat hepatocytes. J Cell Biochem.
89:254–261. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Villevalois-Cam L, Rescan C, Gilot D, Ezan
F, Loyer P, Desbuquois B, Guguen-Guillouzo C and Baffet G: The
hepatocyte is a direct target for transforming-growth factor beta
activation via the insulin-like growth factor II/mannose
6-phosphate receptor. J Hepatol. 38:156–163. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yasuda H, Mine T, Shibata H, Eto Y,
Hasegawa Y, Takeuchi T, Asano S and Kojima I: Activin A: An
autocrine inhibitor of initiation of DNA synthesis in rat
hepatocytes. J Clin Invest. 92:1491–1496. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schwall RH, Robbins K, Jardieu P, Chang L,
Lai C and Terrell TG: Activin induces cell death in hepatocytes in
vivo and in vitro. Hepatology. 18:347–356. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hillier SG and Miró F: Inhibin, activin,
and follistatin. Potential roles in ovarian physiology. Ann N Y
Acad Sci. 687:29–38. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kogure K, Zhang YQ, Kanzaki M, Omata W,
Mine T and Kojima I: Intravenous administration of follistatin:
Delivery to the liver and effect on liver regeneration after
partial hepatectomy. Hepatology. 24:361–366. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ichikawa T, Zhang YQ, Kogure K, Hasegawa
Y, Takagi H, Mori M and Kojima I: Transforming growth factor beta
and activin tonically inhibit DNA synthesis in the rat liver.
Hepatology. 34:918–925. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Takahashi H, Takeshita T and Yokomuro K:
Suppression of liver regeneration resulting from intravenous
injection of splenic glass adherent cells activated by poly I:C.
Immunobiology. 176:217–227. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nakamura T, Arakaki R and Ichihara A:
Interleukin-1 beta is a potent growth inhibitor of adult rat
hepatocytes in primary culture. Exp Cell Res. 179:488–497. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ichihara A: Growth and differentiation of
neonatal rat hepatocytes in primary culture. Nihon Sanka Fujinka
Gakkai Zasshi. 41:1021–1026. 1989.(In Japanese). PubMed/NCBI
|
|
42
|
Ichihara A: Mechanisms controlling growth
of hepatocytes in primary culture. Dig Dis Sci. 36:489–493. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Beyer HS and Theologides A: Tumor necrosis
factor-alpha is a direct hepatocyte mitogen in the rat. Biochem Mol
Biol Int. 29:1–4. 1993.PubMed/NCBI
|
|
44
|
Okamura N, Tsukada K, Sakaguchi T, Ohtake
M, Yoshida K and Muto T: Enhanced liver regeneration by FK 506 can
be blocked by interleukin-1 alpha and interleukin-2. Transplant
Proc. 24:413–415. 1992.PubMed/NCBI
|
|
45
|
Nagata Y, Tanaka N and Orita K:
Endotoxin-induced liver injury after extended hepatectomy and the
role of Kupffer cells in the rat. Surg Today. 24:441–448. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Boulton R, Woodman A, Calnan D, Selden C,
Tam F and Hodgson H: Nonparenchymal cells from regenerating rat
liver generate interleukin-1alpha and -1beta: A mechanism of
negative regulation of hepatocyte proliferation. Hepatology.
26:49–58. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Scotte M, Masson S, Lyoumi S, Hiron M,
Ténière P, Lebreton JP and Daveau M: Cytokine gene expression in
liver following minor or major hepatectomy in rat. Cytokine.
9:859–867. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Uemura T, Miyazaki M, Hirai R, Matsumoto
H, Ota T, Ohashi R, Shimizu N, Tsuji T, Inoue Y and Namba M:
Different expression of positive and negative regulators of
hepatocyte growth in growing and shrinking hepatic lobes after
portal vein branch ligation in rats. Int J Mol Med. 5:173–179.
2000.PubMed/NCBI
|
|
49
|
Stärkel P, Lambotte L, Sempoux C, De
Saeger C, Saliez A, Maiter D and Horsmans Y: After portal branch
ligation in the rat, cellular proliferation is associated with
selective induction of c-Ha-ras, p53, cyclin E, and Cdk2. Gut.
49:119–130. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Anderson SP, Yoon L, Richard EB, Dunn CS,
Cattley RC and Corton JC: Delayed liver regeneration in peroxisome
proliferator-activated receptor-alpha-null mice. Hepatology.
36:544–554. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tsutsumi R, Kamohara Y, Eguchi S, Azuma T,
Fujioka H, Okudaira S, Yanaga K and Kanematsu T: Selective
suppression of initial cytokine response facilitates liver
regeneration after extensive hepatectomy in rats.
Hepatogastroenterology. 51:701–704. 2004.PubMed/NCBI
|
|
52
|
Hou Z, Yanaga K, Kamohara Y, Eguchi S,
Tsutsumi R, Furui J and Kanematsu T: A new suppressive agent
against interleukin-1beta and tumor necrosis factor-alpha enhances
liver regeneration after partial hepatectomy in rats. Hepatol Res.
26:40–46. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhu RZ, Xiang D, Xie C, Li JJ, Hu JJ, He
HL, Yuan YS, Gao J, Han W and Yu Y: Protective effect of
recombinant human IL-1Ra on CCl4-induced acute liver injury in
mice. World J Gastroenterol. 16:2771–2779. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Goss JA, Mangino MJ and Flye MW: Kupffer
cell autoregulation of IL-1 production by PGE2 during hepatic
regeneration. J Surg Res. 52:422–428. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Goss JA, Mangino MJ, Callery MP and Flye
MW: Prostaglandin E2 downregulates Kupffer cell production of IL-1
and IL-6 during hepatic regeneration. Am J Physiol. 264:G601–G608.
1993.PubMed/NCBI
|
|
56
|
Higashitsuji H, Arii S, Furutani M, Mise
M, Monden K, Fujita S, Ishiguro S, Kitao T, Nakamura T, Nakayama H,
et al: Expression of cytokine genes during liver regeneration after
partial hepatectomy in rats. J Surg Res. 58:267–274. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sgroi A, Gonelle-Gispert C, Morel P,
Baertschiger RM, Niclauss N, Mentha G, Majno P, Serre-Beinier V and
Buhler L: Interleukin-1 receptor antagonist modulates the early
phase of liver regeneration after partial hepatectomy in mice. PLoS
One. 6:e254422011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zheng YB, Zhang XH, Huang ZL, Lin CS, Lai
J, Gu YR, Lin BL, Xie DY, Xie SB, Peng L and Gao ZL:
Amniotic-fluid-derived mesenchymal stem cells overexpressing
interleukin-1 receptor antagonist improve fulminant hepatic
failure. PLoS One. 7:e413922012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Franco-Gou R, Roselló-Catafau J,
Casillas-Ramirez A, Massip-Salcedo M, Rimola A, Calvo N, Bartrons R
and Peralta C: How ischaemic preconditioning protects small liver
grafts. J Pathol. 208:62–73. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhao Y, Meng C, Wang Y, Huang H, Liu W,
Zhang JF, Zhao H, Feng B, Leung PS and Xia Y: IL-1β inhibits
β-Klotho expression and FGF19 signaling in hepatocytes. Am J
Physiol Endocrinol Metab. 310:E289–E300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Streetz KL, Luedde T, Manns MP and
Trautwein C: Interleukin 6 and liver regeneration. Gut. 47:309–312.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sun R, Jaruga B, Kulkarni S, Sun H and Gao
B: IL-6 modulates hepatocyte proliferation via induction of
HGF/p21cip1: Regulation by SOCS3. Biochem Biophys Res Commun.
338:1943–1949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kamohara Y, Sugiyama N, Mizuguchi T,
Inderbitzin D, Lilja H, Middleton Y, Neuman T, Demetriou AA and
Rozga J: Inhibition of signal transducer and activator
transcription factor 3 in rats with acute hepatic failure. Biochem
Biophys Res Commun. 273:129–135. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wustefeld T, Rakemann T, Kubicka S, Manns
MP and Trautwein C: Hyperstimulation with interleukin 6 inhibits
cell cycle progression after hepatectomy in mice. Hepatology.
32:514–522. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Torbenson M, Yang SQ, Liu HZ, Huang J,
Gage W and Diehl AM: STAT-3 overexpression and p21 up-regulation
accompany impaired regeneration of fatty livers. Am J Pathol.
161:155–161. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Quétier I, Brezillon N, Duriez M, Massinet
H, Giang E, Ahodantin J, Lamant C, Brunelle MN, Soussan P and
Kremsdorf D: Hepatitis B virus HBx protein impairs liver
regeneration through enhanced expression of IL-6 in transgenic
mice. J Hepatol. 59:285–291. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kakumu S, Fukatsu A, Shinagawa T, Kurokawa
S and Kusakabe A: Localisation of intrahepatic interleukin 6 in
patients with acute and chronic liver disease. J Clin Pathol.
45:408–411. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Napoli J, Bishop GA and McCaughan GW:
Increased intrahepatic messenger RNA expression of interleukins 2,
6, and 8 in human cirrhosis. Gastroenterology. 107:789–798. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Malaguarnera M, Di Fazio I, Romeo MA,
Restuccia S, Laurino A and Trovato BA: Elevation of interleukin 6
levels in patients with chronic hepatitis due to hepatitis C virus.
J Gastroenterol. 32:211–215. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Tangkijvanich P, Vimolket T, Theamboonlers
A, Kullavanijaya P, Suwangool P and Poovorawan Y: Serum
interleukin-6 and interferon-gamma levels in patients with
hepatitis B-associated chronic liver disease. Asian Pac J Allergy
Immunol. 18:109–114. 2000.PubMed/NCBI
|
|
71
|
Suh HN, Lee SH, Lee MY, Lee YJ, Lee JH and
Han HJ: Role of Interleukin-6 in the control of DNA synthesis of
hepatocytes: Involvement of PKC, p44/42 MAPKs, and PPARdelta. Cell
Physiol Biochem. 22:673–684. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu M, Liu H, Wang X, Chen P and Chen H:
IL-6 induction of hepatocyte proliferation through the
Tmub1-regulated gene pathway. Int J Mol Med. 29:1106–1112.
2012.PubMed/NCBI
|
|
73
|
Campbell JS, Prichard L, Schaper F,
Schmitz J, Stephenson-Famy A, Rosenfeld ME, Argast GM, Heinrich PC
and Fausto N: Expression of suppressors of cytokine signaling
during liver regeneration. J Clin Invest. 107:1285–1292. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sakuda S, Tamura S, Yamada A, Miyagawa Ji,
Yamamoto K, Kiso S, Ito N, Imanaka K, Wada A, Naka T, et al:
Activation of signal transducer and activator transcription 3 and
expression of suppressor of cytokine signal 1 during liver
regeneration in rats. J Hepatol. 36:378–384. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Luedde T, Wuestefeld T and Trautwein C: A
new player in the team: SOCS-3 socks it to cytokine signaling in
the regenerating liver. Hepatology. 34:1254–1256. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Heinrich PC, Behrmann I, Haan S, Hermanns
HM, Müller-Newen G and Schaper F: Principles of interleukin
(IL)-6-type cytokine signalling and its regulation. Biochem J.
374:1–20. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Riehle KJ, Campbell JS, McMahan RS,
Johnson MM, Beyer RP, Bammler TK and Fausto N: Regulation of liver
regeneration and hepatocarcinogenesis by suppressor of cytokine
signaling 3. J Exp Med. 205:91–103. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Seki E, Kondo Y, Iimuro Y, Naka T, Son G,
Kishimoto T, Fujimoto J, Tsutsui H and Nakanishi K: Demonstration
of cooperative contribution of MET- and EGFR-mediated STAT3
phosphorylation to liver regeneration by exogenous suppressor of
cytokine signalings. J Hepatol. 48:237–245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Brand S, Dambacher J, Beigel F, Zitzmann
K, Heeg MH, Weiss TS, Prüfer T, Olszak T, Steib CJ, Storr M, et al:
IL-22-mediated liver cell regeneration is abrogated by SOCS-1/3
overexpression in vitro. Am J Physiol Gastrointest Liver Physiol.
292:G1019–G1028. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yang XP, Schaper F, Teubner A, Lammert F,
Heinrich PC, Matern S and Siewert E: Interleukin-6 plays a crucial
role in the hepatic expression of SOCS3 during acute inflammatory
processes in vivo. J Hepatol. 43:704–710. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Aydemir TB, Chang SM, Guthrie GJ, Maki AB,
Ryu MS, Karabiyik A and Cousins RJ: Zinc transporter ZIP14
functions in hepatic zinc, iron and glucose homeostasis during the
innate immune response (endotoxemia). PLoS One. 7:e486792012.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Gui Y, Yeganeh M, Ramanathan S, Leblanc C,
Pomerleau V, Ferbeyre G, Saucier C and Ilangumaran S: SOCS1
controls liver regeneration by regulating HGF signaling in
hepatocytes. J Hepatol. 55:1300–1308. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Sun R, Park O, Horiguchi N, Kulkarni S,
Jeong WI, Sun HY, Radaeva S and Gao B: STAT1 contributes to dsRNA
inhibition of liver regeneration after partial hepatectomy in mice.
Hepatology. 44:955–966. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yin S, Wang H, Bertola A, Feng D, Xu MJ,
Wang Y and Gao B: Activation of invariant natural killer T cells
impedes liver regeneration by way of both IFN-γ- and IL-4-dependent
mechanisms. Hepatology. 60:1356–1366. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Cosgrove BD, Cheng C, Pritchard JR, Stolz
DB, Lauffenburger DA and Griffith LG: An inducible autocrine
cascade regulates rat hepatocyte proliferation and apoptosis
responses to tumor necrosis factor-alpha. Hepatology. 48:276–288.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Theocharis SE, Margeli AP and Tsokos MG:
Alpha 2b-interferon inhibits rat liver regeneration after partial
hepatectomy without affecting thymidine kinase activity. J Lab Clin
Med. 125:588–596. 1995.PubMed/NCBI
|
|
87
|
Theocharis SE, Margeli AP, Skaltsas SD,
Skopelitou AS, Mykoniatis MG and Kittas CN: Effect of
interferon-alpha2b administration on rat liver regeneration after
partial hepatectomy. Dig Dis Sci. 42:1981–1986. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ogiso T, Nagaki M, Takai S, Tsukada Y,
Mukai T, Kimura K and Moriwaki H: Granulocyte colony-stimulating
factor impairs liver regeneration in mice through the up-regulation
of interleukin-1beta. J Hepatol. 47:816–825. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Vaquero J, Campbell JS, Haque J, McMahan
RS, Riehle KJ, Bauer RL and Fausto N: Toll-Like Receptor 4 and
myeloid differentiation factor 88 provide mechanistic insights into
the cause and effects of interleukin-6 activation in mouse liver
regeneration. Hepatology. 54:597–608. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Thompson NL, Mead JE, Braun L, Goyette M,
Shank PR and Fausto N: Sequential protooncogene expression during
rat liver regeneration. Cancer Res. 46:3111–3117. 1986.PubMed/NCBI
|
|
91
|
Drazan KE, Shen XD, Csete ME, Zhang WW,
Roth JA, Busuttil RW and Shaked A: In vivo adenoviral-mediated
human p53 tumor suppressor gene transfer and expression in rat
liver after resection. Surgery. 116:197–204. 1994.PubMed/NCBI
|
|
92
|
Lennartsson P, Stenius U and Högberg J:
p53 expression and TGF-alpha-induced replication of hepatocytes
isolated from rats exposed to the carcinogen diethylnitrosamine.
Cell Biol Toxicol. 15:31–39. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Inoue Y, Tomiya T, Yanase M, Arai M, Ikeda
H, Tejima K, Ogata I, Kimura S, Omata M and Fujiwara K: p53 May
positively regulate hepatocyte proliferation in rats. Hepatology.
36:336–344. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Tsuji K and Ogawa K: Recovery from
ultraviolet-induced growth arrest of primary rat hepatocytes by p53
antisense oligonucleotide treatment. Mol Carcinog. 9:167–174. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Arora V, Iversen PL and Ebadi M:
Manipulation of metallothionein expression in the regenerating rat
liver using antisense oligonucleotides. Biochem Biophys Res Commun.
246:711–718. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Arora V and Iversen PL: Antisense
oligonucleotides targeted to the p53 gene modulate liver
regeneration in vivo. Drug Metab Dispos. 28:131–138.
2000.PubMed/NCBI
|
|
97
|
Kurinna S, Stratton SA, Coban Z,
Schumacher JM, Grompe M, Duncan AW and Barton MC: p53 regulates a
mitotic transcription program and determines ploidy in normal mouse
liver. Hepatology. 57:2004–2013. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zhang L, Liu L, He Z, Li G, Liu J, Song Z,
Jin H, Rudolph KL, Yang H, Mao Y, et al: Inhibition of wild-type
p53-induced phosphatase 1 promotes liver regeneration in mice by
direct activation of mammalian target of rapamycin. Hepatology.
61:2030–2041. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Ohlson LC, Koroxenidou L and Hällström IP:
Inhibition of in vivo rat liver regeneration by
2-acetylaminofluorene affects the regulation of cell cycle-related
proteins. Hepatology. 27:691–696. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Stepniak E, Ricci R, Eferl R, Sumara G,
Sumara I, Rath M, Hui L and Wagner EF: c-Jun/AP-1 controls liver
regeneration by repressing p53/p21 and p38 MAPK activity. Genes
Dev. 20:2306–2314. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Li CC, Chu HY, Yang CW, Chou CK and Tsai
TF: Aurora-A overexpression in mouse liver causes p53-dependent
premitotic arrest during liver regeneration. Mol Cancer Res.
7:678–688. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Fan X, Jiang Y, Wang Y, Tan H, Zeng H,
Wang Y, Chen P, Qu A, Gonzalez FJ, Huang M and Bi H: Wuzhi tablet
(Schisandra Sphenanthera extract) protects against
acetaminophen-induced hepatotoxicity by inhibition of CYP-mediated
bioactivation and regulation of NRF2-ARE and p53/p21 pathways. Drug
Metab Dispos. 42:1982–1990. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wang Y, Jiang Y, Fan X, Tan H, Zeng H,
Wang Y, Chen P, Huang M and Bi H: Hepato-protective effect of
resveratrol against acetaminophen-induced liver injury is
associated with inhibition of CYP-mediated bioactivation and
regulation of SIRT1-p53 signaling pathways. Toxicol Lett.
236:82–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zeng H, Li D, Qin X, Chen P, Tan H, Zeng
X, Li X, Fan X, Jiang Y, Zhou Y, et al: Hepatoprotective effects of
schisandra sphenanthera extract against lithocholic acid-induced
cholestasis in male mice are associated with activation of the
pregnane X receptor pathway and promotion of liver regeneration.
Drug Metab Dispos. 44:337–342. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Shih TY and Weeks MO: Oncogenes and
cancer: The p21 ras genes. Cancer Invest. 2:109–123. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
el-Deiry WS, Tokino T, Velculescu VE, Levy
DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW and
Vogelstein B: WAF1, a potential mediator of p53 tumor suppression.
Cell. 75:817–825. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Harper JW, Adami GR, Wei N, Keyomarsi K
and Elledge SJ: The p21 Cdk-interacting protein Cip1 is a potent
inhibitor of G1 cyclin-dependent kinases. Cell. 75:805–816. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Albrecht JH, Poon RY, Ahonen CL, Rieland
BM, Deng C and Crary GS: Involvement of p21 and p27 in the
regulation of CDK activity and cell cycle progression in the
regenerating liver. Oncogene. 16:2141–2150. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Sasaki Y, Hayashi N, Morita Y, Ito T,
Kasahara A, Fusamoto H, Sato N, Tohyama M and Kamada T: Cellular
analysis of c-Ha-ras gene expression in rat liver after CCl4
administration. Hepatology. 10:494–500. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Hui TT, Mizuguchi T, Sugiyama N, Avital I,
Rozga J and Demetriou AA: Immediate early genes and p21 regulation
in liver of rats with acute hepatic failure. Am J Surg.
183:457–463. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Nango R, Terada C and Tsukamoto I: Jun
N-terminal kinase activation and upregulation of p53 and
p21(WAF1/CIP1) in selenite-induced apoptosis of regenerating liver.
Eur J Pharmacol. 471:1–8. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Chang ML, Chen TH, Chang MY and Yeh CT:
Cell cycle perturbation in the hepatocytes of HCV core transgenic
mice following common bile duct ligation is associated with
enhanced p21 expression. J Med Virol. 81:467–472. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Weymann A, Hartman E, Gazit V, Wang C,
Glauber M, Turmelle Y and Rudnick DA: p21 is required for
dextrose-mediated inhibition of mouse liver regeneration.
Hepatology. 50:207–215. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Lehmann K, Tschuor C, Rickenbacher A, Jang
JH, Oberkofler CE, Tschopp O, Schultze SM, Raptis DA, Weber A, Graf
R, et al: Liver failure after extended hepatectomy in mice is
mediated by a p21-dependent barrier to liver regeneration.
Gastroenterology. 143:1609–1619.e4. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Buitrago-Molina LE, Marhenke S, Longerich
T, Sharma AD, Boukouris AE, Geffers R, Guigas B, Manns MP and Vogel
A: The degree of liver injury determines the role of p21 in liver
regeneration and hepatocarcinogenesis in mice. Hepatology.
58:1143–1152. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Albrecht JH, Meyer AH and Hu MY:
Regulation of cyclin-dependent kinase inhibitor p21(WAF1/Cip1/Sdi1)
gene expression in hepatic regeneration. Hepatology. 25:557–563.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Serfas MS, Goufman E, Feuerman MH, Gartel
AL and Tyner AL: p53-independent induction of p21WAF1/CIP1
expression in pericentral hepatocytes following carbon
tetrachloride intoxication. Cell Growth Differ. 8:951–961.
1997.PubMed/NCBI
|
|
118
|
Wu H, Wade M, Krall L, Grisham J, Xiong Y
and Van Dyke T: Targeted in vivo expression of the cyclin-dependent
kinase inhibitor p21 halts hepatocyte cell-cycle progression,
postnatal liver development and regeneration. Genes Dev.
10:245–260. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Jaime M, Pujol MJ, Serratosa J, Pantoja C,
Canela N, Casanovas O, Serrano M, Agell N and Bachs O: The
p21(Cip1) protein, a cyclin inhibitor, regulates the levels and the
intracellular localization of CDC25A in mice regenerating livers.
Hepatology. 35:1063–1071. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Trautwein C, Will M, Kubicka S, Rakemann
T, Flemming P and Manns MP: 2-acetaminofluorene blocks cell cycle
progression after hepatectomy by p21 induction and lack of cyclin E
expression. Oncogene. 18:6443–6453. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Fan X, Chen P, Tan H, Zeng H, Jiang Y,
Wang Y, Wang Y, Hou X, Bi H and Huang M: Dynamic and coordinated
regulation of KEAP1-NRF2-ARE and p53/p21 signaling pathways is
associated with acetaminophen injury responsive liver regeneration.
Drug Metab Dispos. 42:1532–1539. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Timchenko NA, Harris TE, Wilde M, Bilyeu
TA, Burgess-Beusse BL, Finegold MJ and Darlington GJ:
CCAAT/enhancer binding protein alpha regulates p21 protein and
hepatocyte proliferation in newborn mice. Mol Cell Biol.
17:7353–7361. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Vail ME, Chaisson ML, Thompson J and
Fausto N: Bcl-2 expression delays hepatocyte cell cycle progression
during liver regeneration. Oncogene. 21:1548–1555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Chen S, Zheng J, Hao Q, Yang S, Wang J,
Chen H, Chen L, Zhou Y, Yu C, Jiao B and Cai Z: p53-insensitive
PUMA down-regulation is essential in the early phase of liver
regeneration after partial hepatectomy in mice. J Hepatol.
52:864–871. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Bailly-Maitre B, Bard-Chapeau E, Luciano
F, Droin N, Bruey JM, Faustin B, Kress C, Zapata JM and Reed JC:
Mice lacking bi-1 gene show accelerated liver regeneration. Cancer
Res. 67:1442–1450. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Wyllie AH: Apoptosis and carcinogenesis.
Eur J Cell Biol. 73:189–197. 1997.PubMed/NCBI
|
|
127
|
Kiba T, Saito S, Numata K and Sekihara H:
Fas (APO-1/CD95) mRNA is down-regulated in liver regeneration after
hepatectomy in rats. J Gastroenterol. 35:34–38. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Hu W, Fan C, Jiang P, Ma Z, Yan X, Di S,
Jiang S, Li T, Cheng Y and Yang Y: Emerging role of N-myc
downstream-regulated gene 2 (NDRG2) in cancer. Oncotarget.
7:209–223. 2016.PubMed/NCBI
|
|
129
|
Yang JD, Li Y, Wu L, Zhang Z, Han T, Guo
H, Jiang N, Tao K, Ti Z, Liu X, et al: NDRG2 in rat liver
regeneration: Role in proliferation and apoptosis. Wound Repair
Regen. 18:524–531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Mischoulon D, Rana B, Bucher NL and Farmer
SR: Growth-dependent inhibition of CCAAT enhancer-binding protein
(C/EBP alpha) gene expression during hepatocyte proliferation in
the regenerating liver and in culture. Mol Cell Biol. 12:2553–2560.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Iakova P, Awad SS and Timchenko NA: Aging
reduces proliferative capacities of liver by switching pathways of
C/EBPalpha growth arrest. Cell. 113:495–506. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Pauta M, Rotllan N, Fernández-Hernando A,
Langhi C, Ribera J, Lu M, Boix L, Bruix J, Jimenez W, Suárez Y, et
al: Akt-mediated foxo1 inhibition is required for liver
regeneration. Hepatology. 63:1660–1674. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Chen X, Zhao Y, Wang F, Bei Y, Xiao J and
Yang C: MicroRNAs in liver regeneration. Cell Physiol Biochem.
37:615–628. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Revuelta-Cervantes J, Mayoral R, Miranda
S, González-Rodríguez A, Fernández M, Martín-Sanz P and Valverde
AM: Protein tyrosine phosphatase 1B (PTP1B) deficiency accelerates
hepatic regeneration in mice. Am J Pathol. 178:1591–1604. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Hong L, Cai Y, Jiang M, Zhou D and Chen L:
The Hippo signaling pathway in liver regeneration and
tumorigenesis. Acta Biochim Biophys Sin (Shanghai). 47:46–52. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Grijalva JL, Huizenga M, Mueller K,
Rodriguez S, Brazzo J, Camargo F, Sadri-Vakili G and Vakili K:
Dynamic alterations in Hippo signaling pathway and YAP activation
during liver regeneration. Am J Physiol Gastrointest Liver Physiol.
307:G196–G204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Pondugula SR, Flannery PC, Apte U, Babu
JR, Geetha T, Rege SD, Chen T and Abbott KL: Mg2+/Mn2+-dependent
phosphatase 1A is involved in regulating pregnane X
receptor-mediated cytochrome p450 3A4 gene expression. Drug Metab
Dispos. 43:385–391. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Keeton AB, Xu J, Franklin JL and Messina
JL: Regulation of Gene33 expression by insulin requires MEK-ERK
activation. Biochim Biophys Acta. 1679:248–255. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Ballaro C, Ceccarelli S, Tiveron C,
Tatangelo L, Salvatore AM, Segatto O and Alemà S: Targeted
expression of RALT in mouse skin inhibits epidermal growth factor
receptor signalling and generates a Waved-like phenotype. EMBO Rep.
6:755–761. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Reschke M, Ferby I, Stepniak E, Seitzer N,
Horst D, Wagner EF and Ullrich A: Mitogen-inducible gene-6 is a
negative regulator of epidermal growth factor receptor signaling in
hepatocytes and human hepatocellular carcinoma. Hepatology.
51:1383–1390. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Leffert HL and Weinstein DB: Growth
control of differentiated fetal rat hepatocytes in primary
monolayer culture. IX. Specific inhibition of DNA synthesis
initiation by very low density lipoprotein and possible
significance to the problem of liver regeneration. J Cell Biol.
70:20–32. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Chanda S and Mehendale HM: Nutritional
impact on the final outcome of liver injury inflicted by model
hepatotoxicants: Effect of glucose loading. FASEB J. 9:240–245.
1995.PubMed/NCBI
|
|
143
|
Vazquez-Chantada M, Ariz U, Varela-Rey M,
Embade N, Martínez-Lopez N, Fernández-Ramos D, Gómez-Santos L,
Lamas S, Lu SC, Martínez-Chantar ML and Mato JM: Evidence for
LKB1/AMP-activated protein kinase/endothelial nitric oxide synthase
cascade regulated by hepatocyte growth factor,
S-adenosylmethionine, and nitric oxide in hepatocyte proliferation.
Hepatology. 49:608–617. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Varela-Rey M, Beraza N, Lu SC, Mato JM and
Martinez-Chantar ML: Role of AMP-activated protein kinase in the
control of hepatocyte priming and proliferation during liver
regeneration. Exp Biol Med (Maywood). 236:402–408. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Garcia-Rodriguez JL, Barbier-Torres L,
Fernández-Álvarez S, Gutiérrez-de Juan V, Monte MJ, Halilbasic E,
Herranz D, Álvarez L, Aspichueta P, Marín JJ, et al: SIRT1 controls
liver regeneration by regulating bile acid metabolism through
farnesoid X receptor and mammalian target of rapamycin signaling.
Hepatology. 59:1972–1983. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Leffert HL: Growth control of
differentiated fetal rat hepatocytes in primary monolayer culture.
VII. Hormonal control of DNA synthesis and its possible
significance to the problem of liver regeneration. J Cell Biol.
62:792–801. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Houck KA, Cruise JL and Michalopoulos G:
Norepinephrine modulates the growth-inhibitory effect of
transforming growth factor-beta in primary rat hepatocyte cultures.
J Cell Physiol. 135:551–555. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Barbason H, Bouzahzah B, Herens C,
Marchandise J, Sulon J and Van Cantfort J: Circadian
synchronization of liver regeneration in adult rats: The role
played by adrenal hormones. Cell Tissue Kinet. 22:451–460.
1989.PubMed/NCBI
|
|
149
|
Spector MS, Auer KL, Jarvis WD, Ishac EJ,
Gao B, Kunos G and Dent P: Differential regulation of the
mitogen-activated protein and stress-activated protein kinase
cascades by adrenergic agonists in quiescent and regenerating adult
rat hepatocytes. Mol Cell Biol. 17:3556–3565. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Krasil'nikov MA, Bogdanova NN and Adler
VV: Inhibition of proliferation of normal and neoplastic liver
cells under conditions of prolonged hormonal stimulation.
Biokhimiia. 54:656–661. 1989.(In Russian). PubMed/NCBI
|
|
151
|
Nagy P, Kiss A, Schnur J and Thorgeirsson
SS: Dexamethasone inhibits the proliferation of hepatocytes and
oval cells but not bile duct cells in rat liver. Hepatology.
28:423–429. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Yager JD, Zurlo J, Sewall CH, Lucier GW
and He H: Growth stimulation followed by growth inhibition in
livers of female rats treated with ethinyl estradiol.
Carcinogenesis. 15:2117–2123. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Kokudo N, Kothary PC, Eckhauser FE,
Nakamura T and Raper SE: Inhibition of DNA synthesis by
somatostatin in rat hepatocytes stimulated by hepatocyte growth
factor or epidermal growth factor. Am J Surg. 163:169–173. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Corpechot C, Barbu V, Wendum D, Chignard
N, Housset C, Poupon R and Rosmorduc O: Hepatocyte growth factor
and c-Met inhibition by hepatic cell hypoxia: A potential mechanism
for liver regeneration failure in experimental cirrhosis. Am J
Pathol. 160:613–620. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Kuo WL, Yu MC, Lee JF, Tsai CN, Chen TC
and Chen MF: Imatinib mesylate improves liver regeneration and
attenuates liver fibrogenesis in CCL4-treated mice. J Gastrointest
Surg. 16:361–369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Akerman PA, Cote PM, Yang SQ, McClain C,
Nelson S, Bagby G and Diehl AM: Long-term ethanol consumption
alters the hepatic response to the regenerative effects of tumor
necrosis factor-alpha. Hepatology. 17:1066–1073. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Lumpkin CK Jr, Moore TL, Tarpley MD,
Taylor JM, Badger TM and McClung JK: Acute ethanol and selected
growth suppressor transcripts in regenerating rat liver. Alcohol.
12:357–362. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Chen J, Ishac EJ, Dent P, Kunos G and Gao
B: Effects of ethanol on mitogen-activated protein kinase and
stress-activated protein kinase cascades in normal and regenerating
liver. Biochem J. 334:669–676. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Zhang MN, Gong Y and Minuk GY: The effects
of acute ethanol exposure on inhibitors of hepatic regenerative
activity in the rat. Mol Cell Biochem. 207:109–114. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Silverman AL, Smith MR, Sasaki D,
Mutchnick MG and Diehl AM: Altered levels of prothymosin
immunoreactive peptide, a growth-related gene product, during liver
regeneration after chronic ethanol feeding. Alcohol Clin Exp Res.
18:616–619. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Koteish A, Yang S, Lin H, Huang J and
Diehl AM: Ethanol induces redox-sensitive cell-cycle inhibitors and
inhibits liver regeneration after partial hepatectomy. Alcohol Clin
Exp Res. 26:1710–1718. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Yang SQ, Lin HZ, Mandal AK, Huang J and
Diehl AM: Disrupted signaling and inhibited regeneration in obese
mice with fatty livers: Implications for nonalcoholic fatty liver
disease pathophysiology. Hepatology. 34:694–706. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Koch KS and Leffert HL: Increased sodium
ion influx is necessary to initiate rat hepatocyte proliferation.
Cell. 18:153–163. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Ramalho FS, Ramalho LN, Castro-E-Silva
Júnior O, Zucoloto S and Corrêa FM: Angiotensin-converting enzyme
inhibition by lisinopril enhances liver regeneration in rats. Braz
J Med Biol Res. 34:125–127. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
LeBoeuf RA, Laishes BA and Hoekstra WG:
Effects of selenium on cell proliferation in rat liver and
mammalian cells as indicated by cytokinetic and biochemical
analysis. Cancer Res. 45:5496–5504. 1985.PubMed/NCBI
|
|
166
|
Theocharis SE, Margeli AP, Spiliopoulou C,
Skaltsas S, Kittas C and Koutselinis A: Hepatic stimulator
substance administration enhances regenerative capacity of
hepatocytes in cadmium-pretreated partially hepatectomized rats.
Dig Dis Sci. 41:1475–1480. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Yamaguchi M and Kanayama Y:
Calcium-binding protein regucalcin inhibits deoxyribonucleic acid
synthesis in the nuclei of regenerating rat liver. Mol Cell
Biochem. 162:121–126. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Yamamoto Y, Ono T, Dhar DK, Yamanoi A,
Tachibana M, Tanaka T and Nagasue N: Role of peroxisome
proliferator-activated receptor-gamma (PPARgamma) during liver
regeneration in rats. J Gastroenterol Hepatol. 23:930–937. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Kwon HJ, Won YS, Yoon YD, Yoon WK, Nam KH,
Choi IP, Kim DY and Kim HC: Vitamin D3 up-regulated protein 1
deficiency accelerates liver regeneration after partial hepatectomy
in mice. J Hepatol. 54:1168–1176. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Liu M, Yuan T, Liu H and Chen P:
CCAAT/enhancer-binding protein β regulates interleukin-6-induced
transmembrane and ubiquitin-like domain containing 1 gene
expression in hepatocytes. Mol Med Rep. 10:2177–2183.
2014.PubMed/NCBI
|