Introduction
Lung cancer is the most common malignancy and the
leading cause of cancer-associated mortality worldwide, primarily
in the form of squamous cell carcinoma (1,2).
Many cases are diagnosed at an advanced stage, which may be
responsible for the poor prognosis in lung cancer patients. Thus,
determining novel and effective therapeutic targets is
indispensable for lung squamous cell carcinoma (LSCC) treatment,
and may provide novel insights into carcinogenesis.
Carcinogenesis is a multistep progress.
Tumor-suppressor genes or oncogenes serve critical roles in
regulating tumor-associated biological process, including
proliferation, apoptosis, migration and invasion (3–5).
Previous studies have identified many classic tumor suppressor
genes, including reversion-inducing-cysteine-rich protein with
kazal motifs (RECK), breast cancer 1 (BRCA1) and Ras
association domain family 1 isoform A (RASSF1A) (6–8).
These genes were hypermethylated in various types of tumors and
were involved in tumor-associated biological processes. For
example, promoter methylation of RECK contributes to
metastasis of osteosarcoma (9).
BRCA1 negatively mediates cell proliferation, and its mRNA
levels were downregulated by methylation (10). The epigenetically inactivated
RASSF1A gene was associated with poor prognosis and advanced
tumor stage (11). Therefore,
revealing the potential underlying mechanism and function of
deregulated tumor-associated genes may have great benefit for the
understanding of carcinogenesis. Protein tyrosine phosphatase
receptor-type O (PTPRO) is a candidate tumor suppressor belonging
to the protein tyrosine phosphatase (PTP) family, and is highly
conserved in different species (12). Previous studies have reported that
DNA methylation is involved in the regulation of PTPRO in
hepatocellular carcinomas (13),
lung cancer (14), chronic
lymphocytic leukemia (15),
esophageal squamous cell carcinoma (16) and colorectal cancer (17). Overexpression of PTPRO inhibits
cell proliferation and promotes apoptosis in hepatocellular
carcinoma and lymphoma (18,19),
while downregulation of PTPRO is associated with metastases
in breast cancer (20).
PTPRO regulates mammary epithelial transformation via
directly targeting the receptor tyrosine kinase ErbB2/human
epidermal growth factor receptor 2 (21). Although these studies suggested the
candidate tumor suppressor role of PTPRO, the expression and
biological function of PTPRO in LSCC remains to be fully
elucidated.
The present study assessed the methylation and
expression of PTPRO in LSCC cells and tissues, and the
effect of overexpression of PTPRO on tumor growth. The CpG
island of PTPRO exon 1 was hypermethylated in H520 and
SK-MES-1 cells. In LSCC patients, the significantly higher
methylation levels of PPTPRO was correlated with its
decreased mRNA levels. Furthermore, upregulation of PTPRO
significantly inhibited cell proliferation and colony formation
in vitro, and the tumorigenicity of H520 cells in
vivo. These data suggested that epigenetic regulation of
PTPRO expression is likely to be involved in the progression
of LSCC.
Materials and methods
Tissue samples
Primary tumors and corresponding adjacent healthy
tissues were obtained from 65 patients, including 40 men and 25
women, with a mean age 61.7 years, who were diagnosed in Department
of Thoracic Surgery, Hubei Cancer Hospital (Wuhan, China) between
March 2010 and July 2011. All the tumors used in this study were
squamous cell carcinoma, and tumor stages were confirmed by
pathologists according to the criterion of Union for International
Cancer Control. The clinical characteristics were obtained from
medical records. This study was approved by the ethical committees
of Hubei Cancer Hospital and written informed consent was obtained
prior to surgery. All tissue specimens were surgically resected and
immediately flash-frozen in liquid nitrogen, and stored at
−80°C.
Cell lines and cDNA transfection
The H520 and SK-MES-1 LSCC cell lines and the
BEAS-2B healthy human bronchial epithelial cell line were obtained
from the American Type Culture Collection (Manassas, VA, USA) and
cultured in the conditions as recommended (22). The cells were maintained in
RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum and 100
U/ml penicillin sodium at 37°C in an humidified atmosphere of 5%
CO2. To construct a vector stably expressing
PTPRO, a pcDNA 3.1-Hemagglutinin A (HA)-tagged vector
(Invitrogen; Thermo Fisher Scientific, Inc.) was purchased and used
in this study. The cDNA encoding the complete coding region of
human PTPRO cDNA was obtained from GeneBank (NM_030667.2).
The HA label was introduced to protein C in the vector, and the
E.coli strain of DH5a was also preserved in a laboratory at
Hubei Cancer Hospital. A pcDNA-PTPRO-HA expression vector was
established using a traditional method (23).
DNA extraction and methylation
analysis
Total amounts of DNA (2 µg) were extracted from
cells and tissues using a DNeasy Blood & Tissue kit (Qiagen
GmbH, Germany) according to manufacturer's protocol. The quantity
of DNA was tested by a NanoDrop spectrophotometer (Thermo Fisher
Scientific, Inc.), and stored at −80°C until use. All DNA samples
were treated using an EpiTect Bisulfite kit (Qiagen GmbH), and
converted-DNA was used as a template in next step analysis. For
bisulfite sequencing-polymerase chain reaction (BSP-PCR) analysis,
the PCR reaction was conducted in 50 µl solution containing
converted-DNA (200 ng), dNTP (200 nM for each), forward and reverse
primers (50 pM), and Taq DNA Polymerase (2.5 U; Thermo
Fisher Scientific, Inc.). The 4 µl PCR products (0.1 µg/µl) were
ligated into the PMD18T vector. Recombinant vectors were then
transformed to E. coli and the positive colonies were
selected for sequencing. As inactivation of tumor suppressor genes
may occur via hypermethylation of CpG islands upstream of the
transcription start site, the present study selected a target
region spanning from −405 to −74 (containing 23 CpG sites) in the
BSP analysis, and the primers for PTPRO (forward:
5′-GAGGTTGTTGTTATTTTATGGG-3′; reverse:
5′-TAAAACTACAACCTCAAACCCT-3′) were used. Methylation specific PCR
(MSP) assays were performed using a Techne-512 system (Techne,
Staffordshire, UK) and included an initial incubation at 95°C for 5
min, followed by 40 cycles of denaturation at 95°C for 30 sec,
annealing at 56°C for 20 sec, extension at 72°C for 20 sec and a
final extension step at 72°C for 10 min. One pair of primers for
methylated PTPRO (forward: 5′-TGTTGTTAGAGGATTACGGC-3′; reverse:
5′-CAAAAACGTACCAAACGCTA-3′) and unmethylated PTPRO (forward:
5′-TTTTGTTGTTAGAGGATTATGGT-3′; reverse:
5′-TCCAAAAACATACCAAACACTAC-3′) were used to amplify methylation and
unmethylation alleles of PTPRO, respectively. Quantitative (Q) MSP
was performed to detect the methylation levels of PTPRO in
tumors and matched healthy tissue, and the quantity of methylated
PTPRO was normalized to β-actin. Briefly, 10 ng
bisulfite-coverted DNA was used in the QMSP assay in 384-well
plates with a LightCycler480 system (Roche Diagnostics, Basel,
Switzerland). The PCR reaction included an initial incubation at
95°C for 5 min, followed by 45 cycles of denaturation at 95°C for
30 sec, 58°C for 10 sec, 72°C for 20 sec and 80°C for 1 sec. Each
plate consisted of clinical samples, water blanks and a positive
control. Serial dilutions of the H520 PTPRO
methylation-positive cell line were used for constructing the
calibration curve. QMSP analyses yield values are expressed as
ratios between two absolute measurements (PTPRO:β-actin
×100) (24). Each sample was
analyzed in duplicate.
5-Aza-2′-deoxycytidine (5-AZA)
treatment
For the demethylation assay, 1×105 H520
and SK-MES-1 cells were seeded into 6-well plates, cultured for 24
h and treated with 0, 2.5 or 5 µM 5-AZA (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). Fresh medium containing 5-AZA was changed
every 24 h for 3 days and the treated cells were harvested for
reverse transcription-quantitative (RT-q) PCR analysis.
RNA extraction and RT-qPCR
analysis
Total RNA was isolated from cells using
TRIzol® regent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Total RNA was
reverse transcribed into cDNA, and this procedure was performed
once using the PrimeScript RT reagent kit (Takara Bio, Inc., Otsu,
Japan), which was subsequently used for RT-qPCR analysis using an
ABI 7500 fast Sequence Detector (ABI, Carlsbad, CA, USA). The
reaction conditions were as follows: an initial predenaturation
step at 95°C for 10 min, followed by 40 cycles of denaturation at
95°C for 10 sec, annealing at 60°C for 20 sec and extension at 72°C
for 30 sec. β-actin served as the endogenous control for detection
of mRNA expression levels (25).
Relative quantification analysis was performed using the
2−ΔΔCq method (25).
The following primers for PTPRO (NM_030667.2, transcript
variant 1) were used: Forward, 5′-ACTGCCCCTTATCCACCTCA-3′ and
reverse, 5′-TGTTGCCCGAGGGAATTTCA-3′.
Cell proliferation and colony
formation assays
Cell proliferation was assessed by MTT assay. H520
and SK-MES-1 cells were seeded into 96-well culture plates at a
density of 1.5×103 per well. after 1–6 days, cells were
incubated with 20 µl MTT (5 mg/ml, Sigma-Aldrich; Merck KGaA) for 4
h at 37°C. The cell medium was removed and 150 µl dimethyl
sulfoxide was added to each well. The absorbance was measured at a
wavelength of 490 nm using a microtiter plate reader (Tecan Schweiz
AG, Männedorf, Switzerland). To investigate clonogenic ability,
cells were transfected with PTRPO or an empty vector, and
subsequently seeded into 6-well (200 cells per well) plates. The
culture medium was replaced every 3 days. After 2 weeks, the medium
was removed and the plates were washed twice using PBS. The
colonies were fixed in methanol at −20°C for 5 min, stained with
0.1% crystal violet (Sigma-Aldrich; Merck KGaA), and counted using
an inverted microscope (Nikon Corporation, Tokyo, Japan) in five
random fields.
Tumorigenicity analysis
Xenograft experiments were performed to evaluate the
tumorigenicity of H520 cells transfected with PTPRO or an
empty vector. Briefly, 1×107 H520 cells resuspended in
200 µl PBS were subcutaneously injected into the flanks of athymic
nude male mice (n=5; age, 4 weeks), which were obtained from the
Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). Animal experiments were conducted in accordance
with guidelines approved by the Institutional Animal Care and Use
Committee of Hubei Cancer Hospital. The mice were maintained at a
temperature of 18–22°C and humidity of 50–60% under 12:12 h
light-dark cycle with had free access food and water. Each mouse
was injected in left flank with the PTPRO vector, and in the
right flank with the empty vector. A total of 28 days after
injection, tumors were harvested, weighed and assayed for mRNA
expression.
Western blot analysis
Equal amount of protein extracts were lysed using
radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich; Merck
KGaA). Cells were centrifuged in a microcentrifuge at 12,000 × g
for 15 min at 4°C to collect the supernatant. Protein concentration
was determined using a Bicinchoninic Acid Protein Assay kit
(Pierce; Thermo Fisher Scientific, Inc.). A total of 30 µg protein
were separated by 10% SDS-PAGE and then transferred onto polyvinyl
fluoride membranes (Merck KGaA). The membranes were blocked with 5%
fat-free milk in Tris-buffered saline for 1 h, and incubated with
anti-PTPRO (1:1,000; catalog no. sc-365354; Santa Cruz
Biotechnology, Dallas, TX, USA) and anti-β-actin (1:2,000; catalog
no. 4970, Cell Signaling Technology, Inc., Danvers, MA, USA)
primary antibodies at 4°C overnight. The membranes were washed
three times with Tris-buffered saline containing 0.1% Tween and
incubated for 2 h at room temperature with a horseradish
peroxidase-conjugated goat anti-rabbit (catalog no. 7074; 1:1000;
Cell Signaling Technology, Inc.) or anti-mouse secondary antibody
(catalog no. sc-516102, 1:2000, Santa Cruz Biotechnology). Proteins
were visualized using a Bio-Rad ChemiDoc Imaging system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Statistical analysis was performed using Prism 5
software (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS
version 16.0 (SPSS, Inc., Chicago, IL, USA). Data are expressed as
the mean ± standard error of three independent experiments. Two-way
ANOVA followed by Bonferroni correction was used to determine the
statistical significance when the number of groups was more than
three. The methylation and expression levels of PTPRO in
tumors and healthy controls were compared using paired-samples
t-test. The overall survival of LSCC patients were analyzed using
the Log-rank test. All tests were two sided and P<0.05 was
considered to indicate a statistically significant difference.
Results
CpG island of PTPRO exon 1 is
hypermethylated in LSCC cells and tissues
As the methylation status of PTPRO in LSCC
cells is unclear, a BSP assay was performed in H520 and SK-MES-1
cells, and BEAS-2B cells served as a healthy control. As presented
in Fig. 1A and B, the CpG island
in the 1st exon was hypermethylated in H520 and SK-MES-1 cells,
while partially methylated in BEAS-2B cells. Following this, the
methylation status of PTPRO in LSCC tissues was assessed
using the MSP method. The intensity of methylated alleles was
noticeably increased compared with unmethylated alleles (Fig. 1C). To assess if a demethylation
agent could restore transcriptional activity, LSCC cells were
treated with 0, 2.5 or 5 µM 5-AZA for 72 h. The mRNA expression
levels of PTPRO were significantly increased following 5-AZA
treatment in all groups except the 2.5 µM treatment group in
SK-MES-1 cells (Fig. 1D). These
data demonstrated that the CpG island of PTPRO exon 1 was
hypermethylated in LSCC cells and tissues, suggesting that the
epigenetic regulation of PTPRO may serve a role in LSCC
tumorigenesis.
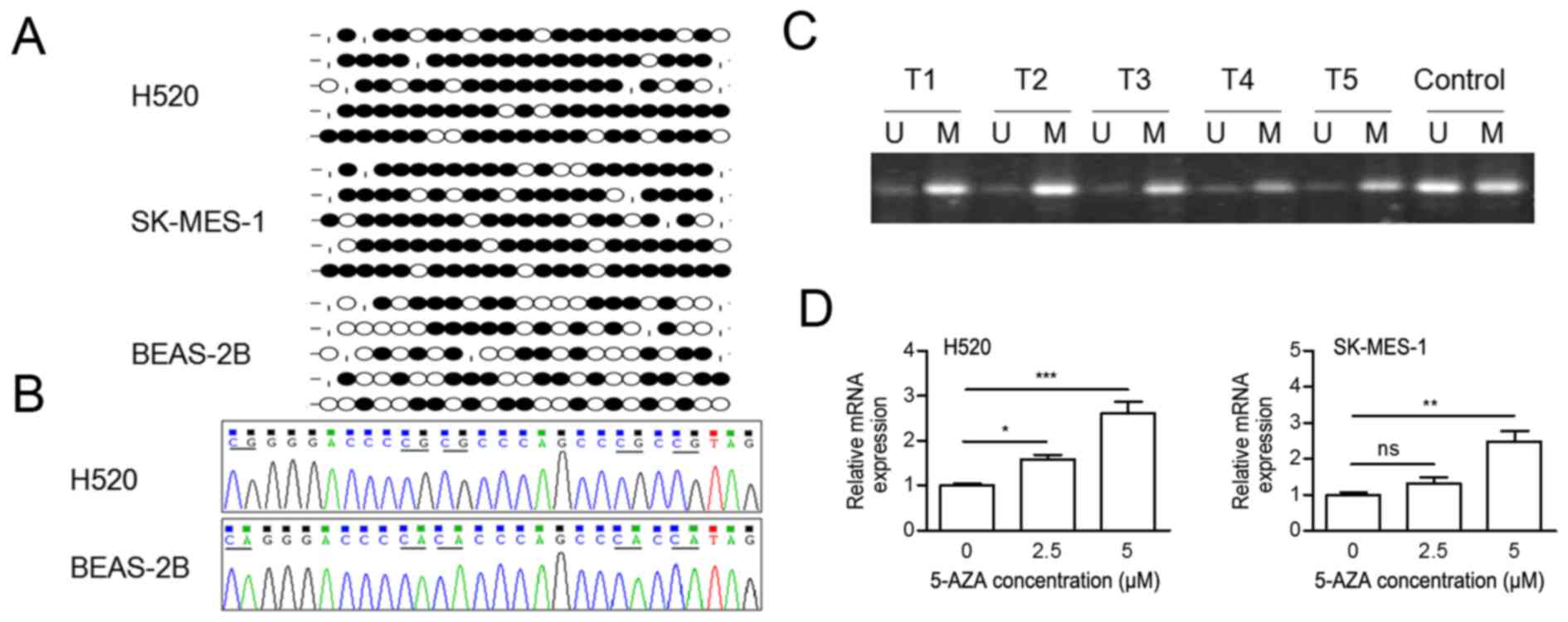 | Figure 1.CpG island of the PTPRO promoter
(from −405 to −74) is hypermethylated in LSCC cells. (A) Dot graph
of BSP data in H520 and SK-MES-1 LSCC cells and BEAS-2B healthy
control cells. The BSP-tested region contained 23 CpG sites. Black
dot, methylated; white dot, unmethylated; stub, not available. (B)
Representative sequences of BSP in H520 and BEAS-2B cells. (C)
Methylation-specific polymerase chain reaction of PTPRO in five
LSCC tissues. M, methylation alleles; U, unmethylation alleles. (D)
Reverse transcription-quantitative polymerase chain reaction
analysis of PTPRO mRNA expression levels after 0, 2.5 or 5 µM 5-AZA
treatment for 72 h. Data are presented as the mean ± standard
deviation. *P<0.05; **P<0.01; ***P<0.001. ns,
non-significant; 5AZA, 5-Aza-2′-deoxycytidine; BSP, bisulfite
sequencing; LSCC, lung squamous cell carcinoma, PTPRO, protein
tyrosine phosphatase receptor-type O. |
PTPRO is epigenetically downregulated
in LSCC tissues
To understand the methylation and mRNA levels of
PTPRO, QMSP and RT-qPCR analyses were performed in LSCC and
matched healthy tissues. The methylation levels of PTPRO
were significantly increased in LSCC tissues compared with healthy
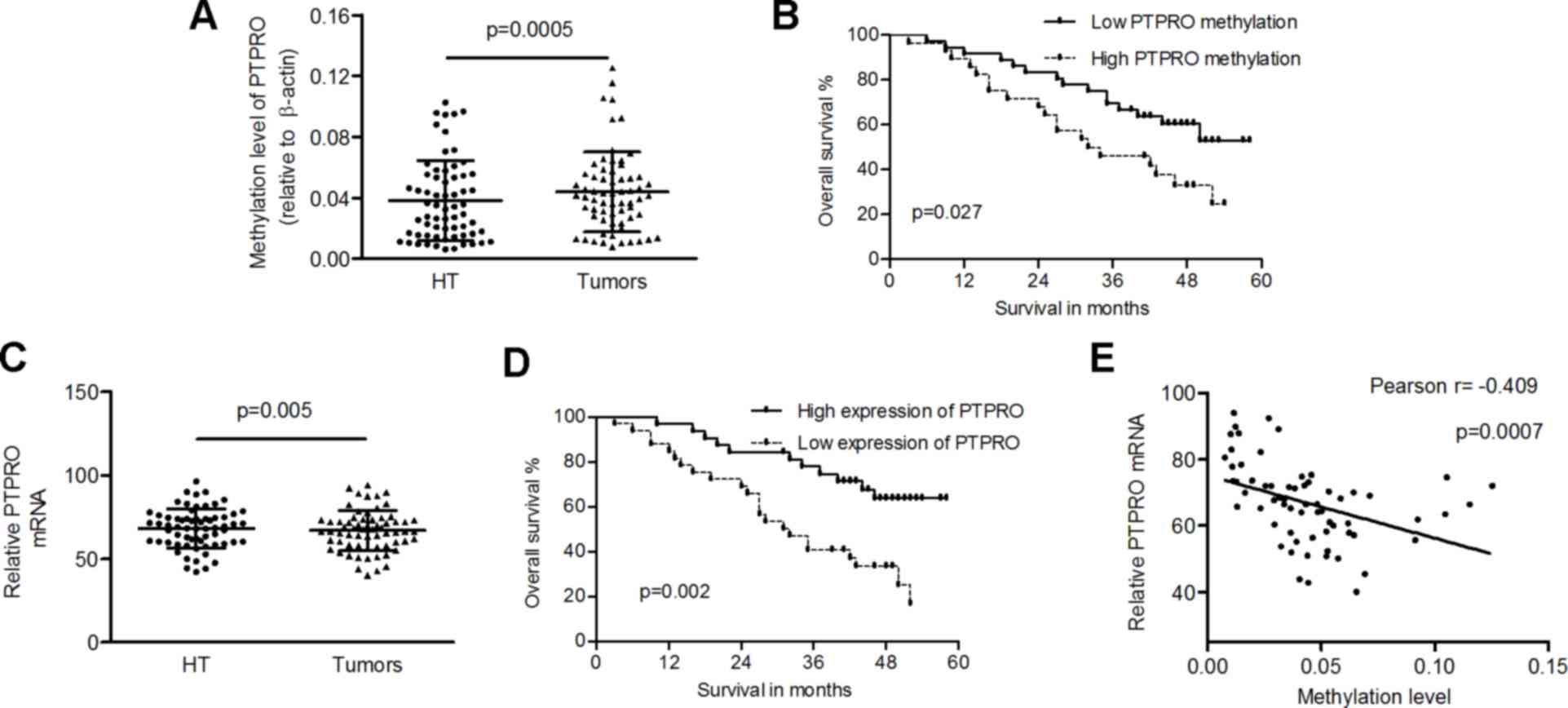
controls (0.0438±0.0263 vs. 0.0381±0.0264; P=0.0005; Fig. 2A). The mean methylation level of
PTPRO in tumors was used as cut-off to divide cases into two
groups (high- or low-methylation). A low level of PTPRO
methylation was significantly associated with high overall survival
probability in LSCC patients (P=0.027; Fig. 2B). Furthermore, the mRNA expression
levels of PTPRO were markedly reduced in tumors compared
with healthy tissues (66.87±12.11 vs. 68.24±11.81; P=0.005;
Fig. 2C), and low expression of
PTPRO (<the mean of mRNA levels of PTPRO in
tumors) was associated with poor prognosis of patients (P=0.002;
Fig. 2D). Pearson correlation
coefficient analysis identified an inverse correlation between
methylation and mRNA levels of PTPRO in tumors (Pearson
r=−0.409; P=0.0007; Fig. 2E).
Subsequently, the present study verified whether the
methylation or expression of PTPRO was associated with
clinicopathological features of patients. As presented Table I, mRNA expression levels of
PTPRO were significantly reduced in advanced tumors compared
with early-stage tumors (P=0.042). The methylation or mRNA levels
of PTPRO were not associated with other clinical parameters
(Table I). Univariate analysis
demonstrated that the high methylation of PTPRO, low
PTPRO mRNA, smoking, advanced tumor stage, higher T stage
and lymph node metastasis were predictors of poor prognosis for
patients, whereas only mRNA expression levels of PTPRO
(P=0.005) and higher TNM stage (P=0.001) were identified as
significantly independent prognostic factors in multivariate
analysis, with relative risks of 2.826 and 3.714, respectively
(Table II). Taken together, these
data suggested that epigenetically downregulated PTPRO may be
involved in LSCC development and may be a potential prognostic
marker. Detailed clinical and molecular data of the patients are
presented in Table III.
 | Table I.Associations between
clinicopathological features and methylation or mRNA expression
levels of PTPRO in lung squamous cell carcinoma. |
Table I.
Associations between
clinicopathological features and methylation or mRNA expression
levels of PTPRO in lung squamous cell carcinoma.
| Variable | Total (n=65) | PTPRO
Methylation | P-value | PTPRO mRNA | P-value |
|---|
| Gender |
|
| 0.322 |
| 0.249 |
|
Female | 25 | 0.0471±0.0254 |
| 64.69±12.13 |
|
|
Male | 40 | 0.0417±0.0269 |
| 68.23±12.04 |
|
| Age (years) |
|
| 0.126 |
| 0.133 |
|
<60 | 28 | 0.0389±0.0264 |
| 69.11±13.26 |
|
|
≥60 | 37 | 0.0475±0.0259 |
| 65.17±11.04 |
|
| Smoking |
|
| 0.896 |
| 0.788 |
|
Never | 31 | 0.0411±0.0201 |
| 67.33±12.51 |
|
| Past,
current | 34 | 0.0462±0.0309 |
| 66.44±11.90 |
|
| TNM stage |
|
| 0.237 |
| 0.042 |
| I,
II | 34 | 0.0396±0.0233 |
| 70.55±12.49 |
|
| III,
IV | 31 | 0.0484±0.0289 |
| 62.82±10.42 |
|
| pT stage |
|
| 0.703 |
| 0.603 |
|
T1-2 | 43 | 0.0429±0.0261 |
| 67.56±13.13 |
|
|
T3-4 | 22 | 0.0455±0.0271 |
| 65.51±9.955 |
|
| pN stage |
|
| 0.263 |
| 0.286 |
| N0 | 17 | 0.0374±0.0222 |
| 70.64±3.001 |
|
|
N1-3 | 48 | 0.0461±0.0274 |
| 65.53±1.711 |
|
 | Table II.Clinical characteristics of lung
squamous cell carcinoma patients correlates with overall
survival. |
Table II.
Clinical characteristics of lung
squamous cell carcinoma patients correlates with overall
survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| PTPRO methylation
(low/high) | 2.108 | 1.068–4.163 | 0.032 |
|
|
|
| PTPRO mRNA
(high/low) | 2.971 | 1.442–6.121 | 0.003 | 2.826 | 1.364–5.853 | 0.005 |
| Gender
(female/male) | 1.15 | 0.569–2.326 | 0.696 |
|
|
|
| Age (<60 y/≥60
y) | 1.037 | 0.991–1.085 | 0.119 |
|
|
|
| Smoking
(never/past, current) | 2.113 | 1.042–4.286 | 0.038 |
|
|
|
| TNM stage (I,
II/III, IV) | 4.145 | 1.998–8.061 | 0.000 | 3.714 | 1.771–7.792 | 0.001 |
| T stage
(T1-2/T3-4) | 2.288 | 1.149–4.555 | 0.018 |
|
|
|
| N stage
(N0/N1-3) | 2.743 | 1.060–7.101 | 0.038 |
|
|
|
 | Table III.Detailed clinical and molecular data
of cases. |
Table III.
Detailed clinical and molecular data
of cases.
| IID | Methy (H) | Methy (T) | mRNA (H) | mRNA (T) | Gender | Age | Smoking | TNM | Survival | Months |
|---|
| 1 | 0.0416 | 0.0445 | 78.02 | 73.09 | M | 57 | current | T3N2M0 | No | 16 |
| 2 | 0.0253 | 0.0384 | 71.13 | 71.283 | M | 66 | current | T1N2M0 | No | 32 |
| 3 | 0.0553 | 0.0623 | 56.214 | 57.89 | M | 45 | past | T3N0M0 | No | 52 |
| 4 | 0.0363 | 0.0487 | 67.083 | 64.097 | F | 61 | never | T2N0M0 | Yes | 52 |
| 5 | 0.00652 | 0.0149 | 79.032 | 78.37 | M | 55 | current | T1N1M0 | Yes | 50 |
| 6 | 0.0272 | 0.0368 | 57.734 | 51.98 | M | 59 | never | T2N1M0 | No | 50 |
| 7 | 0.0966 | 0.1252 | 76.04 | 72.01 | M | 58 | current | T1N2M0 | Yes | 54 |
| 8 | 0.0357 | 0.0427 | 74.32 | 72.094 | M | 72 | never | T3N0M0 | Yes | 41 |
| 9 | 0.0623 | 0.0619 | 52.77 | 60.67 | F | 64 | never | T3N0M0 | Yes | 41 |
| 10 | 0.0422 | 0.0534 | 74.04 | 70.32 | M | 62 | never | T2N1M0 | Yes | 43 |
| 11 | 0.0342 | 0.0415 | 68.32 | 74.89 | M | 59 | never | T2N1M0 | No | 44 |
| 12 | 0.0164 | 0.0268 | 96.43 | 92.384 | M | 60 | current | T1N1M0 | Yes | 51 |
| 13 | 0.1026 | 0.1154 | 69.05 | 66.391 | M | 66 | current | T4N1M0 | No | 19 |
| 14 | 0.0949 | 0.1045 | 60.21 | 63.42 | F | 68 | current | T1N1M0 | No | 42 |
| 15 | 0.0713 | 0.0643 | 70.14 | 70.06 | M | 72 | past | T2N2M0 | No | 34 |
| 16 | 0.0532 | 0.0503 | 60.69 | 64.382 | M | 68 | current | T3N2M0 | No | 13 |
| 17 | 0.0106 | 0.0483 | 66.985 | 66.45 | F | 55 | never | T2N2M0 | Yes | 52 |
| 18 | 0.0214 | 0.0312 | 84.376 | 89.103 | F | 53 | never | T1N0M0 | Yes | 47 |
| 19 | 0.0123 | 0.0164 | 62.165 | 70.01 | F | 62 | never | T2N2M0 | Yes | 57 |
| 20 | 0.0226 | 0.0324 | 58.54 | 53.75 | M | 64 | current | T1N1M0 | No | 12 |
| 21 | 0.0351 | 0.0456 | 81.23 | 75.376 | F | 59 | never | T2N1M0 | Yes | 49 |
| 22 | 0.0556 | 0.0575 | 44.01 | 50.09 | F | 77 | never | T3N1M1 | No | 16 |
| 23 | 0.0434 | 0.0537 | 62.93 | 61.067 | M | 62 | past | T1N2M0 | No | 24 |
| 24 | 0.0153 | 0.0292 | 72.71 | 67.651 | M | 69 | never | T3N1M0 | Yes | 49 |
| 25 | 0.0441 | 0.0439 | 59.92 | 51.06 | M | 68 | current | T3N1M0 | No | 25 |
| 26 | 0.0532 | 0.0528 | 54.04 | 52.39 | F | 72 | never | T2N0M0 | Yes | 9 |
| 27 | 0.0503 | 0.0645 | 60.87 | 57.09 | F | 75 | never | T1N1M0 | No | 48 |
| 28 | 0.0413 | 0.0392 | 58.36 | 55.09 | F | 55 | never | T1N0M0 | Yes | 46 |
| 29 | 0.0096 | 0.0126 | 74.09 | 73.213 | M | 67 | current | T2N0M0 | Yes | 47 |
| 30 | 0.0634 | 0.0714 | 73.98 | 69.08 | M | 63 | past | T2N1M0 | No | 46 |
| 31 | 0.00921 | 0.0105 | 85.362 | 82.99 | M | 62 | current | T2N1M0 | Yes | 42 |
| 32 | 0.04632 | 0.0443 | 50.093 | 42.78 | F | 63 | never | T1N2M0 | No | 27 |
| 33 | 0.02752 | 0.03648 | 59.986 | 57.853 | M | 67 | current | T3N2M0 | Yes | 39 |
| 34 | 0.0264 | 0.0336 | 70.84 | 67.895 | F | 77 | never | T1N1M0 | Yes | 52 |
| 35 | 0.0144 | 0.0293 | 63.468 | 60.432 | M | 57 | past | T2N2M0 | Yes | 49 |
| 36 | 0.01123 | 0.01293 | 64.35 | 65.783 | M | 65 | current | T2N3M0 | No | 9 |
| 37 | 0.0261 | 0.0359 | 67.94 | 71.67 | M | 53 | current | T1N0M0 | No | 40 |
| 38 | 0.0292 | 0.0366 | 68.17 | 65.332 | F | 73 | never | T3N1M0 | No | 35 |
| 39 | 0.0061 | 0.0077 | 81.653 | 80.56 | M | 58 | past | T2N0M0 | Yes | 48 |
| 40 | 0.04493 | 0.0523 | 60.41 | 58.24 | F | 70 | never | T3N1M1 | No | 3 |
| 41 | 0.0236 | 0.0413 | 68.56 | 64.02 | F | 52 | never | T2N1M0 | Yes | 46 |
| 42 | 0.0958 | 0.0914 | 59.054 | 55.67 | F | 64 | current | T1N2M0 | No | 14 |
| 43 | 0.0142 | 0.0317 | 71.23 | 68.541 | M | 66 | current | T2N2M0 | No | 22 |
| 44 | 0.01116 | 0.01157 | 90.01 | 94.03 | M | 55 | past | T1N0M0 | Yes | 58 |
| 45 | 0.08357 | 0.01094 | 78.55 | 77.83 | F | 59 | never | T3N1M0 | Yes | 48 |
| 46 | 0.0607 | 0.0555 | 61.64 | 60.098 | M | 51 | never | T3N1M0 | No | 32 |
| 47 | 0.0578 | 0.0586 | 71.62 | 68.154 | M | 53 | current | T1N1M0 | Yes | 31 |
| 48 | 0.0545 | 0.0523 | 57.43 | 50.82 | M | 64 | current | T1N1M0 | Yes | 43 |
| 49 | 0.0945 | 0.1054 | 76.841 | 74.54 | F | 69 | never | T3N2M0 | Yes | 10 |
| 50 | 0.0582 | 0.0693 | 48.68 | 45.469 | M | 60 | never | T3N2M0 | No | 9 |
| 51 | 0.00843 | 0.01128 | 74.71 | 73.52 | M | 55 | past | T3N1M0 | Yes | 45 |
| 52 | 0.0446 | 0.0431 | 73.12 | 66.376 | F | 68 | never | T1N0M0 | No | 35 |
| 53 | 0.0882 | 0.0925 | 64.89 | 61.87 | M | 56 | current | T2N0M0 | Yes | 46 |
| 54 | 0.00948 | 0.01237 | 88.56 | 89.82 | F | 59 | never | T1N1M0 | Yes | 46 |
| 55 | 0.01512 | 0.02538 | 73.02 | 72.02 | M | 78 | past | T3N2M0 | No | 18 |
| 56 | 0.0197 | 0.0336 | 69.332 | 66.365 | M | 63 | never | T3N0M0 | No | 27 |
| 57 | 0.0703 | 0.0655 | 42.24 | 40.08 | F | 58 | current | T1N2M0 | No | 31 |
| 58 | 0.01692 | 0.02335 | 83.01 | 82.32 | M | 64 | never | T1N0M0 | Yes | 52 |
| 59 | 0.01808 | 0.04049 | 44.33 | 43.9 | M | 47 | current | T2N1M1 | No | 6 |
| 60 | 0.0156 | 0.01964 | 74.841 | 73.546 | F | 57 | never | T3N2M0 | Yes | 37 |
| 61 | 0.00938 | 0.01015 | 90.091 | 87.569 | M | 50 | never | T2N1M0 | Yes | 53 |
| 62 | 0.02085 | 0.02322 | 71.841 | 65.323 | F | 52 | past | T3N0M0 | No | 28 |
| 63 | 0.0203 | 0.02813 | 73.841 | 72.04 | F | 59 | current | T2N2M0 | No | 20 |
| 64 | 0.03256 | 0.0463 | 47.65 | 56.33 | M | 52 | current | T2N3M0 | No | 27 |
| 65 | 0.01014 | 0.01379 | 83.01 | 87.83 | M | 62 | never | T3N0M0 | Yes | 49 |
PTPRO inhibits cell viability in
vitro
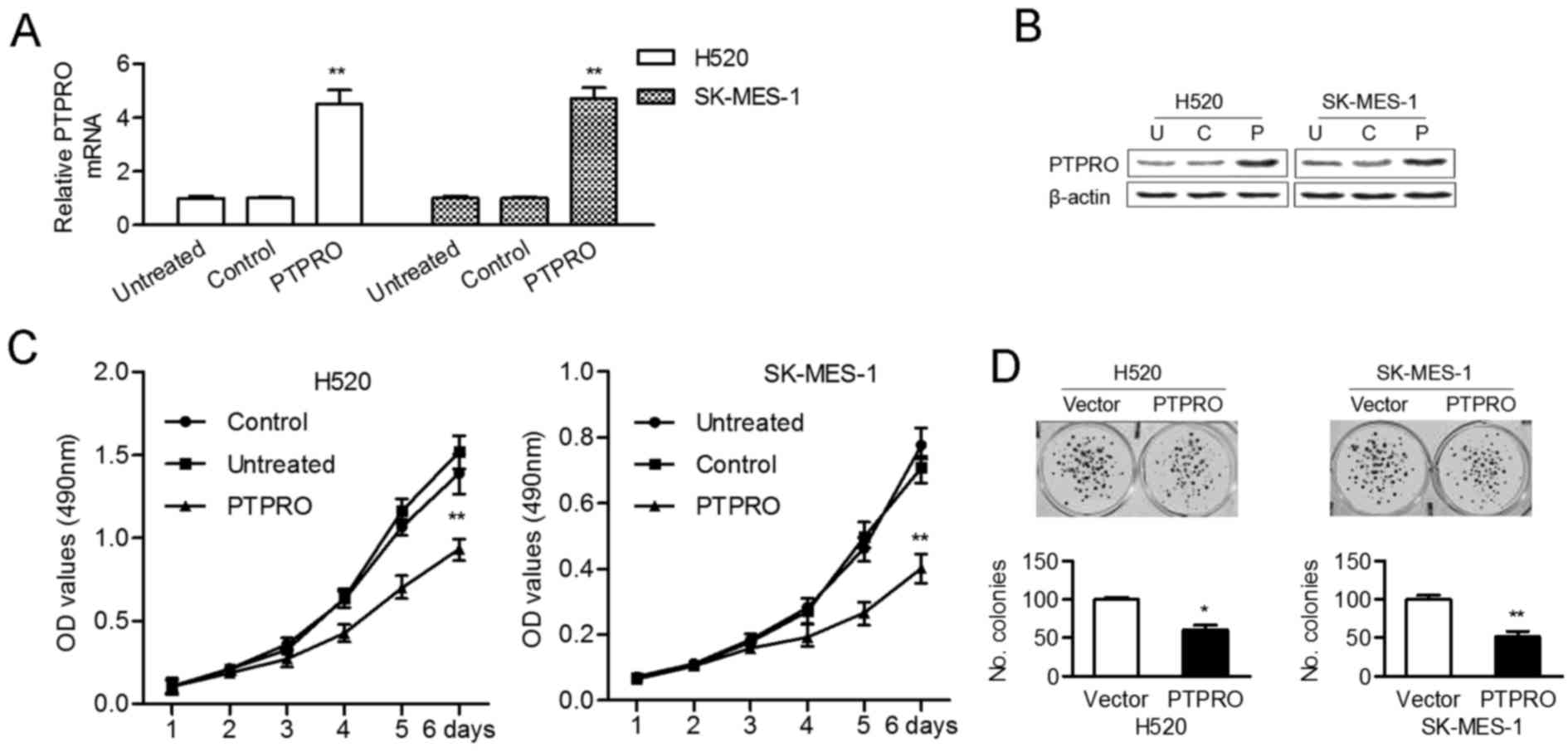
The expression levels of PTPRO in transfected
cells and control cells were detected. The mRNA (Fig. 3A) and protein (Fig. 3B) expression levels of PTPRO
were upregulated in H520 and SK-MES-1 cells transfected with stably
expressing PTPRO vectors in comparison with control cells
and non-transfected cells. The results of MTT demonstrated that
when compared with the empty vector group (control) and
non-transfected group (untreated), the proliferation of LSCC cells
was significantly inhibited in overexpressing PTPRO cells
(Fig. 3C). Colony formation assay
was performed to evaluate the effect of PTPRO on LSCC cells.
As a result, the number of colonies were significantly reduced in
H520 and SK-MES-1 cells transfected with PTPRO expressing
vectors, compared with control and untreated cells (Fig. 3D). These in vitro analyses
demonstrated the inhibitory effect of PTPRO on cell
viability.
PTPRO impairs the tumorigenicity of
H520 cells in vivo
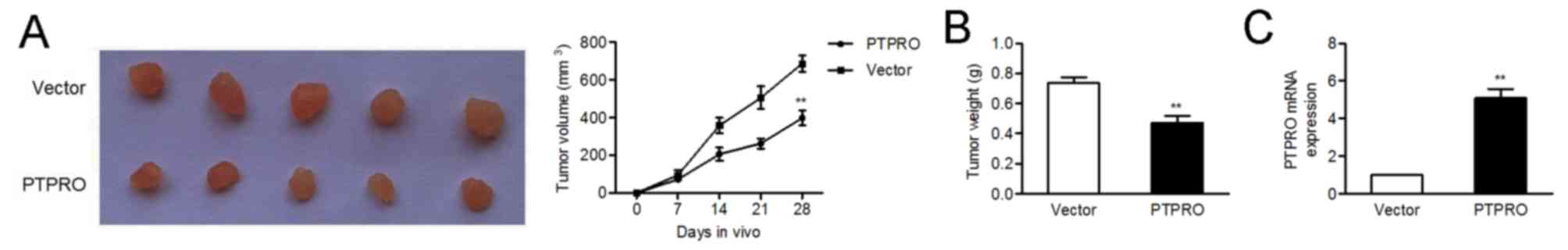
The inhibitory role of PTPRO was further
confirmed using a xenograft model of H520 cells in nude mice. As
expected, there was a significant reduction in tumor volume and
weight in the PTPRO overexpression group compared with the
empty vector group (Fig. 4A and
B). In addition, mRNA expression levels of PTPRO were
upregulated in tumors injected with PTPRO vectors (Fig. 4C).
Discussion
Many LSCC patients are already in the advanced
stages when diagnosed, rendering treatment difficult. The
initiation and progression of squamous cell carcinoma is a complex
process involving the abnormalities of a variety of oncogenes and
tumor suppressors (26,27). The present study focused on the
novel tumor suppressor PTPRO.
It is well known that the cell tyrosine
phosphorylation levels are co-regulated by PTP and protein tyrosine
kinase (PTK); dysfunction of tyrosine phosphatase is closely
associated with the occurrence of a variety of human tumors
(28). Previous studies have
demonstrated that overexpression of PTP in cancer cells may reverse
the malignant transformation induced by PTK (29). PTPRO is a member of the PTP
family that has been reported to be frequently methylated, and is
characterized as a tumor suppressor gene in the occurrence and
development of many malignancies (13–16).
The present study examined H520 and SK-MES-1 LSCC cells and LSCC
cases for analysis. The CpG island of PTPRO exon1 was
revealed to be hypermethylated, which was consistent with previous
studies (14). Additionally, in
the present study, high methylation or low expression of
PTPRO were associated with poor prognosis. Similar results
have been reported in breast (21,30)
and colorectal (31) cancer. Li
et al (30) reported that
methylation of PTPRO was an independent predictor for
survival. However, mRNA expression of PTPTO, rather than
methylation, was an independent factor in the present study. All
these data strongly suggested that PTPRO is involved in
tumorigenesis and may serve as a valuable prognostic marker in
cancers.
Although the function and underlying mechanism of
PTPRO has been documented in former studies (18–20),
the biological effect of PTPRO in LSCC remains unclear. The
present study demonstrated that ectopic PTPRO expression
significantly inhibited the proliferation rate and colony formation
ability of cells. Previous investigations observed a similar effect
in lung adenocarcinoma and lymphoma (14,19).
PTPRO was also reported to be involved in other critical
biological processes including angiogenesis, metastasis and
apoptosis (18,20,21).
Therefore, PTPRO may serve as a multi-functional regulator
in tumorigenesis. In the present study, tumorigenicity analysis
confirmed the tumor suppressive effect of PTPRO in vivo.
Taken together, these findings expanded current knowledge of
PTPRO in LSCC, suggesting the potential value of
PTPRO as a therapeutic target.
In conclusion, the present study demonstrated that
PTPRO inhibits tumor growth in vitro and in
vivo, indicating the tumor suppressive function of PTPRO
in LSCC. This study highlights PTPRO as an epigenetically silenced
gene, and a candidate tumor-suppressor of LSCC.
References
|
1
|
Derman BA, Mileham KF, Bonomi PD, Batus M
and Fidler MJ: Treatment of advanced squamous cell carcinoma of the
lung: A review. Transl Lung Cancer Res. 4:524–532. 2015.PubMed/NCBI
|
|
2
|
Minami H, Isomoto H, Inoue H, Akazawa Y,
Yamaguchi N, Ohnita K, Takeshima F, Hayashi T, Nakayama T and Nakao
K: Significance of background coloration in endoscopic detection of
early esophageal squamous cell carcinoma. Digestion. 89:6–11. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weißenborn C, Ignatov T, Ochel HJ, Costa
SD, Zenclussen AC, Ignatova Z and Ignatov A: GPER functions as a
tumor suppressor in triple-negative breast cancer cells. J Cancer
Res Clin Oncol. 140:713–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Calì G, Insabato L, Conza D, Bifulco G,
Parrillo L, Mirra P, Fiory F, Miele C, Raciti GA, Di Jeso B, et al:
GRP78 mediates cell growth and invasiveness in endometrial cancer.
J Cell Physiol. 229:1417–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tan B, Anaka M, Deb S, Freyer C, Ebert LM,
Chueh AC, Al-Obaidi S, Behren A, Jayachandran A, Cebon J, et al:
FOXP3 over-expression inhibits melanoma tumorigenesis via effects
on proliferation and apoptosis. Oncotarget. 5:264–276.
2014.PubMed/NCBI
|
|
6
|
Correa TC, Brohem CA, Winnischofer SM, da
Silva Cardeal LB, Sasahara RM, Taboga SR, Sogayar MC and
Maria-Engler SS: Downregulation of the RECK-tumor and metastasis
suppressor gene in glioma invasiveness. J Cell Biochem. 99:156–167.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Romagnolo AP, Romagnolo DF and Selmin OI:
BRCA1 as target for breast cancer prevention and therapy.
Anticancer Agents Med Chem. 15:4–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thaler S, Hähnel PS, Schad A, Dammann R
and Schuler M: RASSF1A mediates p21Cip1/Waf1-dependent cell cycle
arrest and senescence through modulation of the Raf-MEK-ERK pathway
and inhibition of Akt. Cancer Res. 69:1748–1757. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang L, Ge J, Ma T, Zheng Y, Lv S, Li Y
and Liu S: Promoter hypermethylation of the cysteine protease RECK
may cause metastasis of osteosarcoma. Tumour Biol. 36:9511–9516.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Magdinier F, Ribieras S, Lenoir GM,
Frappart L and Dante R: Down-regulation of BRCA1 in human sporadic
breast cancer; analysis of DNA methylation patterns of the putative
promoter region. Oncogene. 17:3169–3176. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dammann R, Schagdarsurengin U, Strunnikova
M, Rastetter M, Seidel C, Liu L, Tommasi S and Pfeifer GP:
Epigenetic inactivation of the Ras-association domain family 1
(RASSF1A) gene and its function in human carcinogenesis. Histol
Histopathol. 18:665–677. 2003.PubMed/NCBI
|
|
12
|
Wiggins RC, Wiggins JE, Goyal M, Wharram
BL and Thomas PE: Molecular cloning of cDNAs encoding human GLEPP1,
a membrane protein tyrosine phosphatase: Characterization of the
GLEPP1 protein distribution in human kidney and assignment of the
GLEPP1 gene to human chromosome 12p12-p13. Genomics. 27:174–181.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Motiwala T, Ghoshal K, Das A, Majumder S,
Weichenhan D, Wu YZ, Holman K, James SJ, Jacob ST and Plass C:
Suppression of the protein tyrosine phosphatase receptor type O
gene (PTPRO) by methylation in hepatocellular carcinomas. Oncogene.
22:6319–6331. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Motiwala T, Kutay H, Ghoshal K, Bai S,
Seimiya H, Tsuruo T, Suster S, Morrison C and Jacob ST: Protein
tyrosine phosphatase receptor-type O (PTPRO) exhibits
characteristics of a candidate tumor suppressor in human lung
cancer. Proc Natl Acad Sci USA. 101:13844–13849. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Motiwala T, Majumder S, Kutay H, Smith DS,
Neuberg DS, Lucas DM, Byrd JC, Grever M and Jacob ST: Methylation
and silencing of protein tyrosine phosphatase receptor type O in
chronic lymphocytic leukemia. Clin Cancer Res. 13:3174–3181. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
You YJ, Chen YP, Zheng XX, Meltzer SJ and
Zhang H: Aberrant methylation of the PTPRO gene in peripheral blood
as a potential biomarker in esophageal squamous cell carcinoma
patients. Cancer Lett. 315:138–144. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Laczmanska I and Sasiadek MM: Tyrosine
phosphatases as a superfamily of tumor suppressors in colorectal
cancer. Acta Biochim Pol. 58:467–470. 2011.PubMed/NCBI
|
|
18
|
Hou J, Xu J, Jiang R, Wang Y, Chen C, Deng
L, Huang X, Wang X and Sun B: Estrogen-sensitive PTPRO expression
represses hepatocellular carcinoma progression by control of STAT3.
Hepatology. 57:678–688. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen L, Juszczynski P, Takeyama K, Aguiar
RC and Shipp MA: Protein tyrosine phosphatase receptor-type O
truncated (PTPROt) regulates SYK phosphorylation, proximal
B-cell-receptor signaling and cellular proliferation. Blood.
108:3428–3433. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Z, Hou J, Ren L, He J, Sun B, Sun LZ
and Wang S: Protein tyrosine phosphatase receptor type O expression
in the tumor niche correlates with reduced tumor growth,
angiogenesis, circulating tumor cells and metastasis of breast
cancer. Oncol Rep. 33:1908–1914. 2015.PubMed/NCBI
|
|
21
|
Yu M, Lin G, Arshadi N, Kalatskaya I, Xue
B, Haider S, Nguyen F, Boutros PC, Elson A, Muthuswamy LB, et al:
Expression profiling during mammary epithelial cell
three-dimensional morphogenesis identifies PTPRO as a novel
regulator of morphogenesis and ErbB2-mediated transformation. Mol
Cell Biol. 32:3913–3924. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chien W, Yin D, Gui D, Mori A, Frank JM,
Said J, Kusuanco D, Marchevsky A, McKenna R and Koeffler HP:
Suppression of cell proliferation and signaling transduction by
connective tissue growth factor in non-small cell lung cancer
cells. Mol Cancer Res. 4:591–598. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Werstuck GH, Lentz SR, Dayal S, Hossain
GS, Sood SK, Shi YY, Zhou J, Maeda N, Krisans SK, Malinow MR and
Austin RC: Homocysteine-induced endoplasmic reticulum stress causes
dysregulation of the cholesterol and triglyceride biosynthetic
pathways. J Clin Invest. 107:1263–1273. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jerónimo C, Henrique R, Hoque MO, Ribeiro
FR, Oliveira J, Fonseca D, Teixeira MR, Lopes C and Sidransky D:
Quantitative RARbeta2 hypermethylation: A promising prostate cancer
marker. Clin Cancer Res. 10:4010–4014. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Coutinho-Camillo CM, Lourenço SV, de
Araújo Lima L, Kowalski LP and Soares FA: Expression of
apoptosis-regulating miRNAs and target mRNAs in oral squamous cell
carcinoma. Cancer Genet. 208:382–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang X, Xu Y, He C, Guo X, Zhang J, He C,
Zhang L, Kong M, Chen B and Zhu C: Elevated expression of CCAT2 is
associated with poor prognosis in esophageal squamous cell
carcinoma. J Surg Oncol. 111:834–839. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jacob ST and Motiwala T: Epigenetic
regulation of protein tyrosine phosphatases: Potential molecular
targets for cancer therapy. Cancer Gene Ther. 12:665–672. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matozaki T and Kasuga M: Roles of
protein-tyrosine phosphatases in growth factor signalling. Cell
Signal. 8:13–19. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li SY, Li R, Chen YL, Xiong LK, Wang HL,
Rong L and Luo RC: Aberrant PTPRO methylation in tumor tissues as a
potential biomarker that predicts clinical outcomes in breast
cancer patients. BMC Genet. 15:672014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Asbagh LA, Vazquez I, Vecchione L,
Budinska E, De Vriendt V, Baietti MF, Steklov M, Jacobs B, Hoe N,
Singh S, et al: The tyrosine phosphatase PTPRO sensitizes colon
cancer cells to anti-EGFR therapy through activation of
SRC-mediated EGFR signaling. Oncotarget. 5:10070–10083. 2014.
View Article : Google Scholar : PubMed/NCBI
|