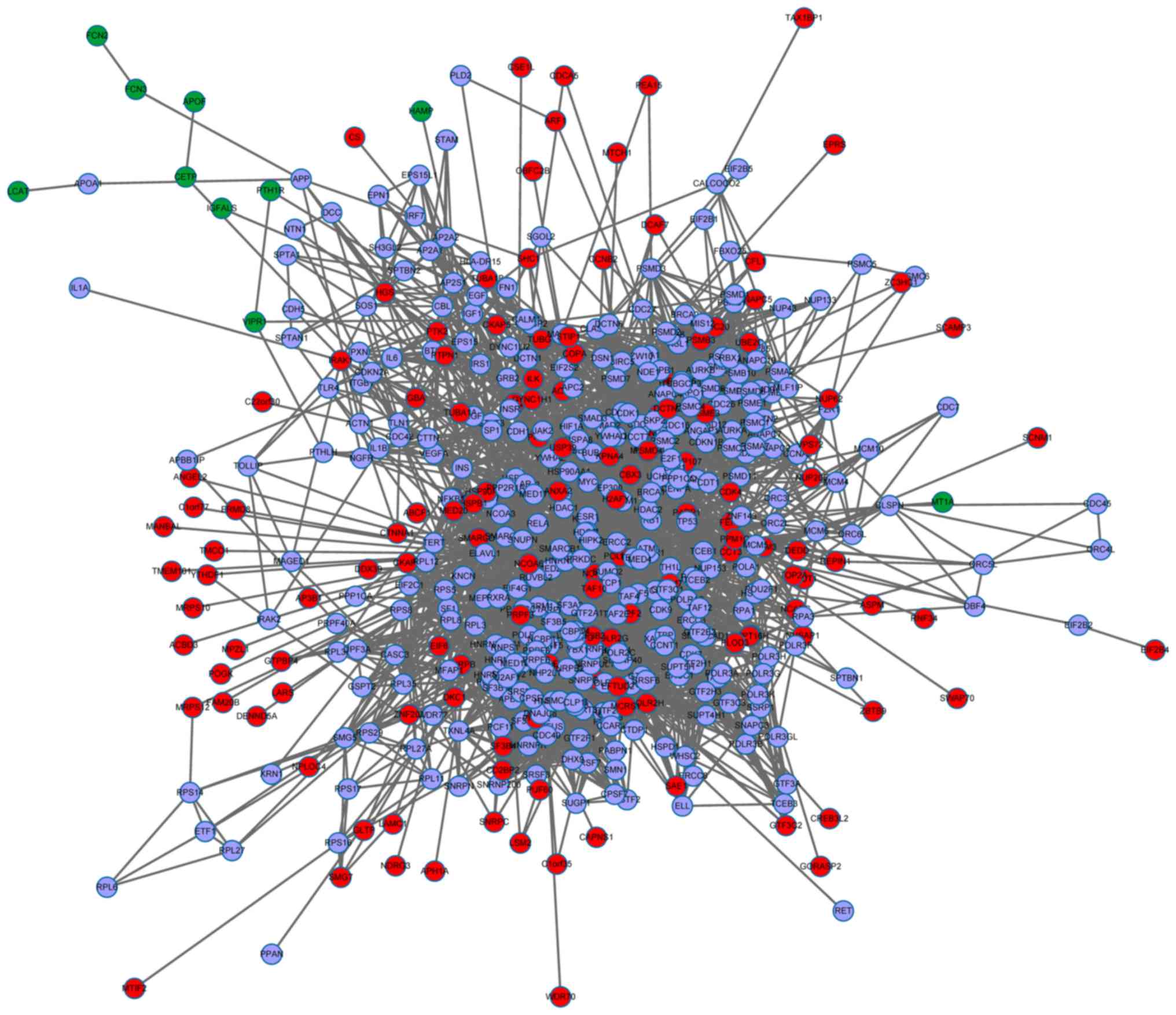
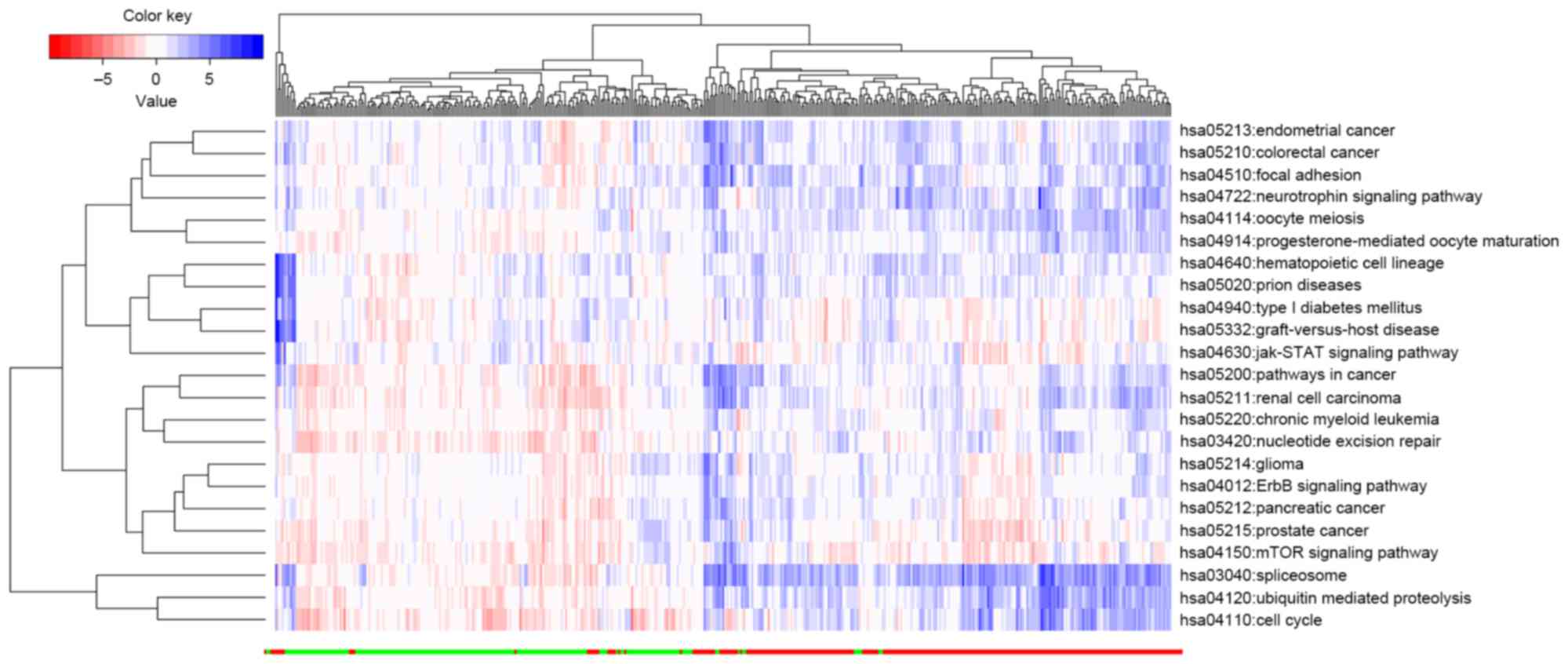
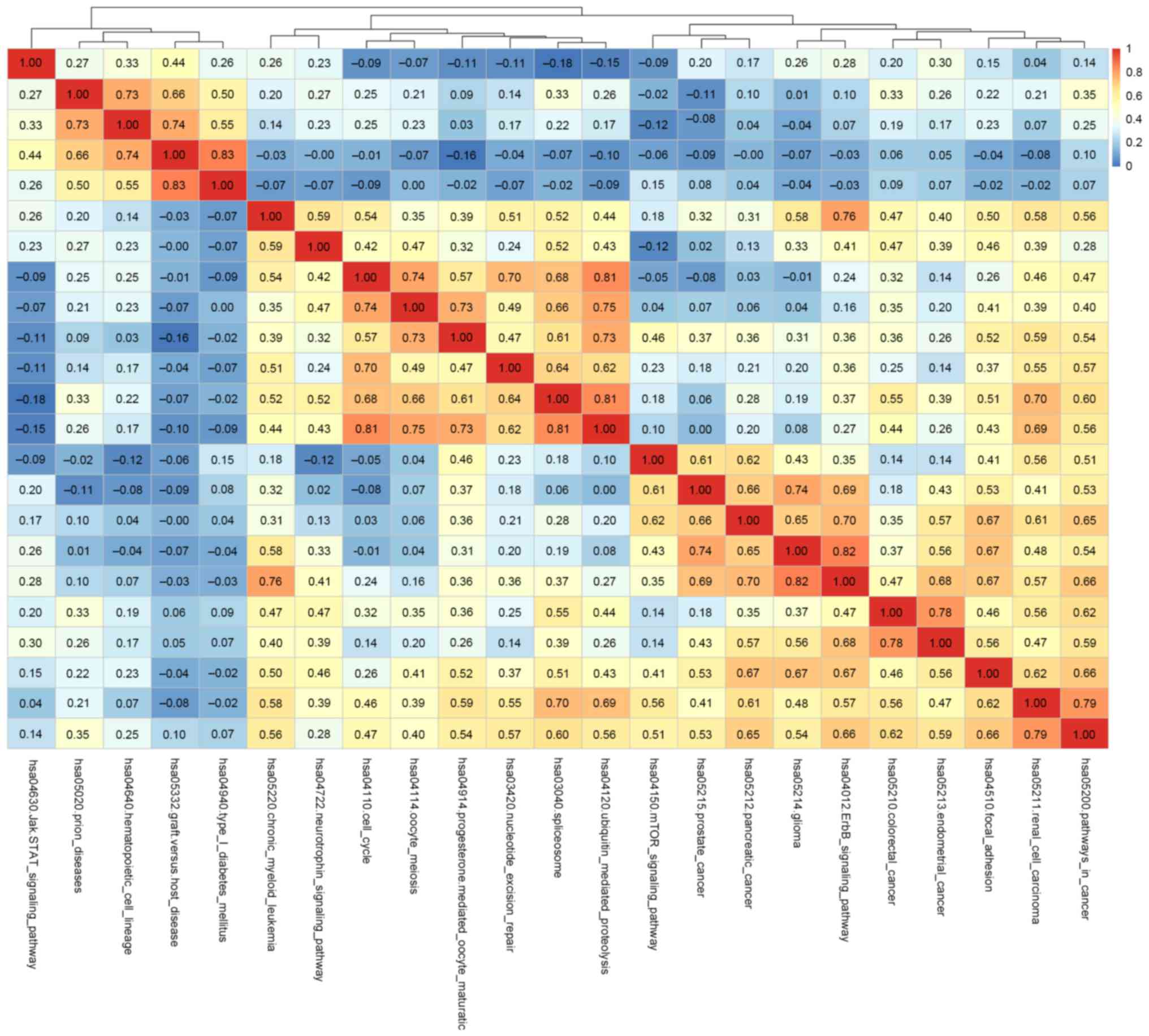
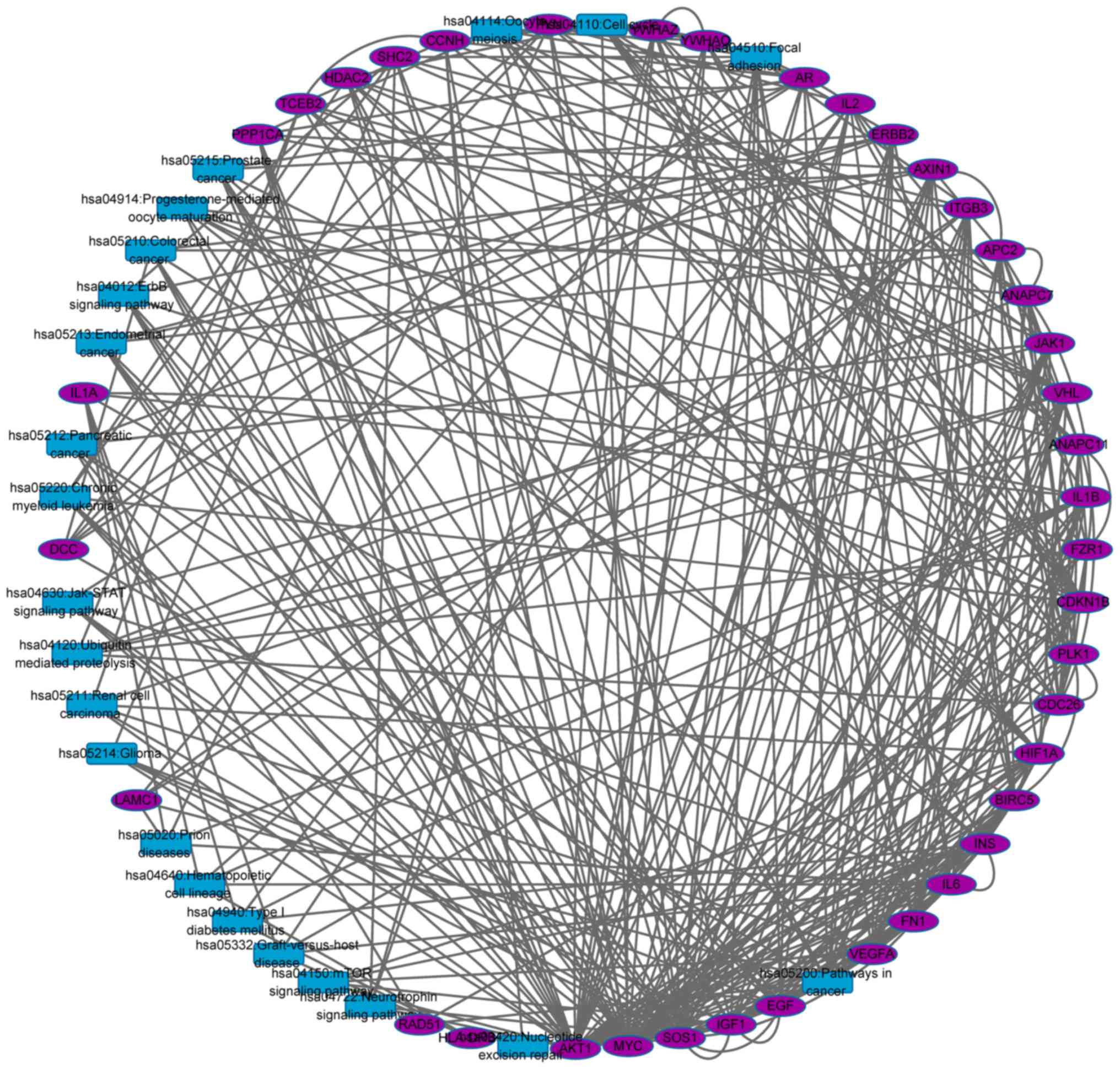
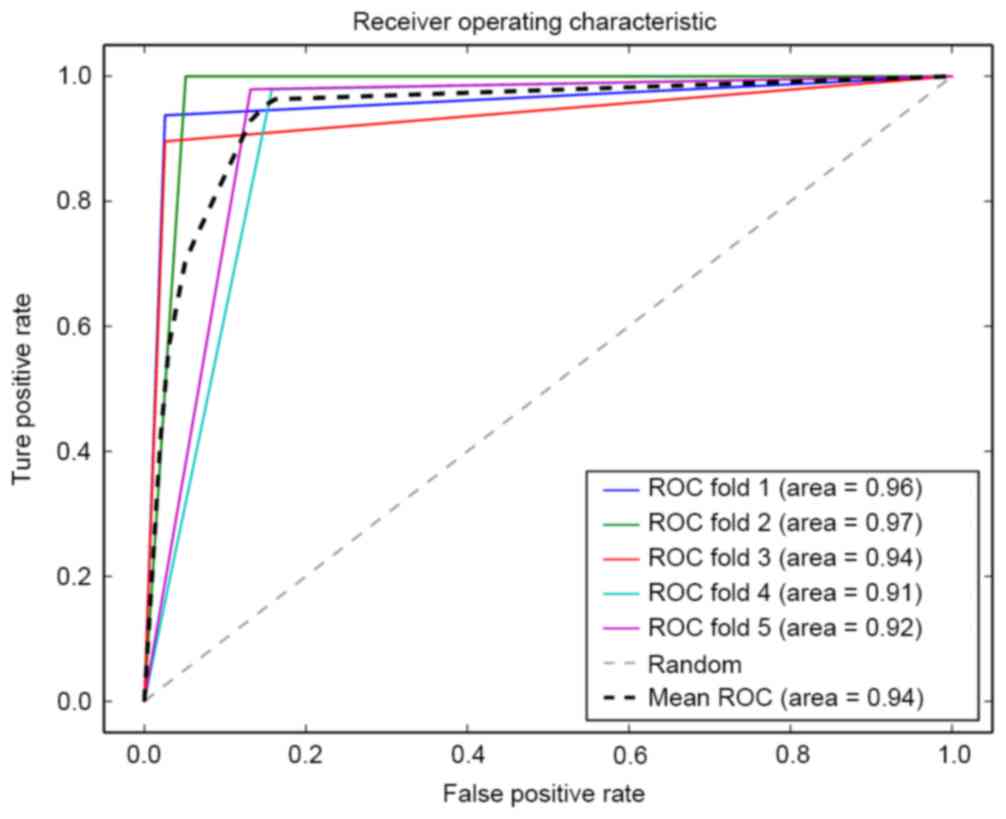
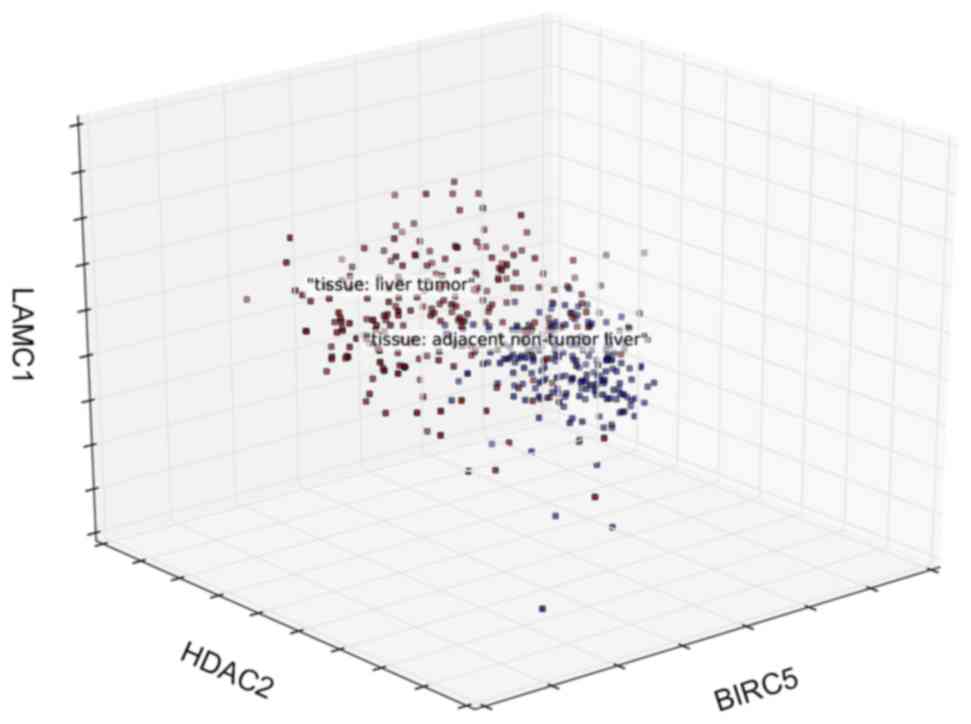
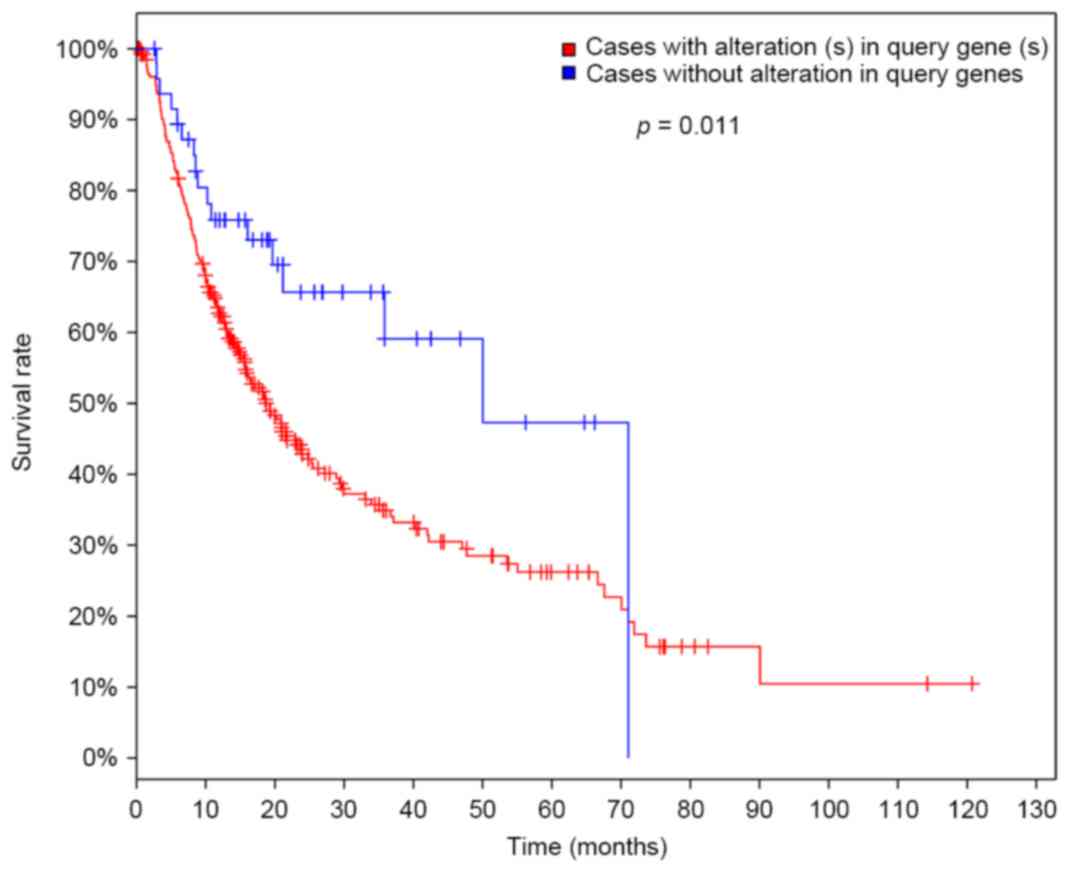
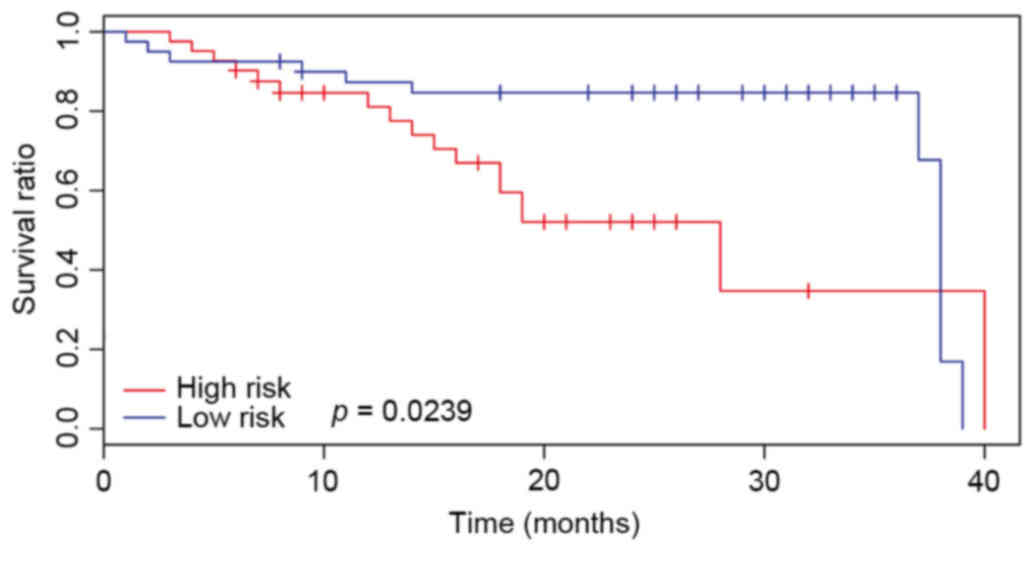
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perz JF, Armstrong GL, Farrington LA,
Hutin YJ and Bell BP: The contributions of hepatitis B virus and
hepatitis C virus infections to cirrhosis and primary liver cancer
worldwide. J Hepatol. 45:529–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marrero JA, Kudo M and Bronowicki JP: The
challenge of prognosis and staging for hepatocellular carcinoma.
Oncologist. 15:(Suppl 4). S23–S33. 2010. View Article : Google Scholar
|
|
4
|
Farazi PA and DePinho RA: Hepatocellular
carcinoma pathogenesis: From genes to environment. Nat Rev Cancer.
6:674–687. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li D and Satomura S: Biomarkers for
hepatocellular carcinoma (HCC): An update. Adv Exp Med Biol.
867:179–193. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reuter JA, Spacek DV and Snyder MP:
High-throughput sequencing technologies. Mol Cell. 58:586–597.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schmidt D, Wilson MD, Spyrou C, Brown GD,
Hadfield J and Odom DT: ChIP-seq: Using high-throughput sequencing
to discover protein-DNA interactions. Methods. 48:240–248. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mathew R, Karantza-Wadsworth V and White
E: Role of autophagy in cancer. Nat Rev Cancer. 7:961–967. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Astolfi A, Landuzzi L, Nicoletti G, De
Giovanni C, Croci S, Palladini A, Ferrini S, Iezzi M, Musiani P,
Cavallo F, et al: Gene expression analysis of immune-mediated
arrest of tumorigenesis in a transgenic mouse model of
HER-2/neu-positive basal-like mammary carcinoma. Am J Pathol.
166:1205–1216. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kamb A, Gruis NA, Weaver-Feldhaus J, Liu
Q, Harshman K, Tavtigian SV, Stockert E, Day RS III, Johnson BE and
Skolnick MH: A cell cycle regulator potentially involved in genesis
of many tumor types. Science. 264:436–440. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bergers G and Benjamin LE: Tumorigenesis
and the angiogenic switch. Nat Rev Cancer. 3:401–410. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Arwert EN, Hoste E and Watt FM: Epithelial
stem cells, wound healing and cancer. Nat Rev Cancer. 12:170–180.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Berkeley C: Linear models and empirical
Bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:Article32004.PubMed/NCBI
|
|
14
|
Aoki-Kinoshita K, Kanehisa M and Bergman
N: Comparative genomics. Journal. 2007.
|
|
15
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Köhn HF and Hubert LJ: Hierarchical
cluster analysis. Wiley StatsRef: Statistics Reference Online.
2006.
|
|
17
|
Liaw A and Wiener M: Classification and
regression by randomForest. R News. 2:18–22. 2002.
|
|
18
|
Vazquez A, Flammini A, Maritan A and
Vespignani A: Global protein function prediction from
protein-protein interaction networks. Nat Biotechnol. 21:697–700.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Colcombet J and Hirt H: Arabidopsis MAPKs:
A complex signalling network involved in multiple biological
processes. Biochem J. 413:217–226. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stuart JM, Segal E, Koller D and Kim SK: A
gene-coexpression network for global discovery of conserved genetic
modules. Science. 302:249–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zinkin NT, Grall F, Bhaskar K, Otu HH,
Spentzos D, Kalmowitz B, Wells M, Guerrero M, Asara JM, Libermann
TA and Afdhal NH: Serum proteomics and biomarkers in hepatocellular
carcinoma and chronic liver disease. Clin Cancer Res. 14:470–477.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lodish H, Berk A, Kaiser CA, Krieger M,
Bretscher A, Ploegh H and Amon A: Molecular Cell Biology. 7th. W H
Freeman and Company; New York: 2012
|
|
23
|
Przulj N, Wigle DA and Jurisica I:
Functional topology in a network of protein interactions.
Bioinformatics. 20:340–348. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fabregat I: Dysregulation of apoptosis in
hepatocellular carcinoma cells. World J Gastroenterol. 15:513–520.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakanishi K, Sakamoto M, Yamasaki S, Todo
S and Hirohashi S: Akt phosphorylation is a risk factor for early
disease recurrence and poor prognosis in hepatocellular carcinoma.
Cancer. 103:307–312. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Borlak J, Meier T, Halter R, Spanel R and
Spanel-Borowski K: Epidermal growth factor-induced hepatocellular
carcinoma: Gene expression profiles in precursor lesions, early
stage and solitary tumours. Oncogene. 24:1809–1819. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Höpfner M, Sutter AP, Huether A, Schuppan
D, Zeitz M and Scherübl H: Targeting the epidermal growth factor
receptor by gefitinib for treatment of hepatocellular carcinoma. J
Hepatol. 41:1008–1016. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shachaf CM, Kopelman AM, Arvanitis C,
Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B,
Cardiff RD, et al: MYC inactivation uncovers pluripotent
differentiation and tumour dormancy in hepatocellular cancer.
Nature. 431:1112–1117. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marrero JA and Lok AS: Newer markers for
hepatocellular carcinoma. Gastroenterology. 127:(5 Suppl 1).
S113–S119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tovar V, Alsinet C, Villanueva A, Hoshida
Y, Chiang DY, Solé M, Thung S, Moyano S, Toffanin S, Mínguez B, et
al: IGF activation in a molecular subclass of hepatocellular
carcinoma and pre-clinical efficacy of IGF-1R blockage. J Hepatol.
52:550–559. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bekaii-Saab T, Williams N, Plass C, Calero
MV and Eng C: A novel mutation in the tyrosine kinase domain of
ERBB2 in hepatocellular carcinoma. BMC Cancer. 6:2782006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fornari F, Gramantieri L, Ferracin M,
Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM,
Bolondi L and Negrini M: MiR-221 controls CDKN1C/p57 and CDKN1B/p27
expression in human hepatocellular carcinoma. Oncogene.
27:5651–5661. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Whittaker S, Marais R and Zhu AX: The role
of signaling pathways in the development and treatment of
hepatocellular carcinoma. Oncogene. 29:4989–5005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kerbel RS: Tumor angiogenesis. N Engl J
Med. 358:2039–2049. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kaseb AO, Hanbali A, Cotant M, Hassan MM,
Wollner I and Philip PA: Vascular endothelial growth factor in the
management of hepatocellular carcinoma: A review of literature.
Cancer. 115:4895–4906. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nischalke HD, Coenen M, Berger C,
Aldenhoff K, Müller T, Berg T, Krämer B, Körner C, Odenthal M,
Schulze F, et al: The toll-like receptor 2 (TLR2)-196 to −174
del/ins polymorphism affects viral loads and susceptibility to
hepatocellular carcinoma in chronic hepatitis C. Int J Cancer.
130:1470–1475. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Timofeeva OA, Zhang X, Ressom HW, Varghese
RS, Kallakury BV, Wang K, Ji Y, Cheema A, Jung M, Brown ML, et al:
Enhanced expression of SOS1 is detected in prostate cancer
epithelial cells from African-American men. Int J Oncol.
35:751–760. 2009.PubMed/NCBI
|
|
38
|
De S, Dermawan JK and Stark GR: EGF
receptor uses SOS1 to drive constitutive activation of NFκB in
cancer cells. Proc Natl Acad Sci USA. 111:11721–11726. 2014.
View Article : Google Scholar : PubMed/NCBI
|