Introduction
Hepatocellular carcinoma (HCC) is one of the most
common types of malignancy worldwide, with ~630,000 new cases
reported each year (1). HCC
development is a complex process that is associated with numerous
risk factors, including several known environmental factors,
hepatitis, and alcohol and tobacco consumption (2,3).
There is increasing evidence supporting the role of genetic factors
in HCC risk (4). The lack of novel
therapeutic strategies for the treatment of HCC emphasizes the need
to determine the molecular mechanisms underlying liver cancer
development. MicroRNAs (miRNAs/miRs) are a class of endogenous,
non-coding RNAs 18–22 nucleotides in length, which are crucial for
gene expression regulation (5). In
the present study, differential miRNA expression between tumor
tissues and normal tissues was identified to be associated with
cancer progression via target gene regulation. More than half of
miRNAs are located in tumor-associated genomic regions or fragile
sites (6), and miRNAs may be
classified as tumor suppressors or oncogenes based on their
modulation of oncogenic and tumor suppressor pathways (7). Numerous miRNAs are involved in liver
cancer development, proliferation, apoptosis and differentiation
(8). For example, miR-150-5p
inhibits hepatoma cell migration and invasion by targeting matrix
metalloproteinase 14 (9) and
miR-486-5p suppresses tumor growth in HCC by targeting
phosphatidylinositol 3-kinase regulatory subunit α (10).
The present study demonstrated that miR-214
expression is lower in HCC tissues and liver cancer cell lines
compared with in matched normal tissues and the HL-7702 cell line,
indicating its role as a tumor suppressor in liver cancer. In
addition, Wnt3a was confirmed as a target gene of miR-214. Using
immunohistochemistry, the present study indicated that the
expression of Wnt3a was higher in HCC tissues compared with in
normal tissues. In addition, the function of miR-214 in liver
cancer cell proliferation was investigated, and overexpression of
miR-214 and Wnt3a silencing arrested the liver cancer cell cycle at
G1 phase. These results demonstrated that miR-214 may
suppress the growth of liver cancer cells by targeting Wnt3a.
Materials and methods
Human tissue samples and cell
lines
The SMMC-7721, HepG2 and Hep3B liver cancer cell
lines, and the HL-7702 normal liver cell line were provided by the
Key University, Ministry of Education (Xi'an, China). These cells
were cultured in Dulbecco's modified Eagle's medium (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Thermo Fisher Scientific, Inc.) at 37°C in a
humidified chamber containing 5% CO2. Tumor tissues and
matched normal tissues were obtained from 24 HCC patients (19
males, 5 females; age, 21–72 years). Patients were enrolled between
September 2011 and January 2013 at the First Affiliated Hospital of
Xi'an Jiaotong University (Xi'an, China). All patients provided
written informed consent. The present study was approved by the
Medical Ethical Committee of the College of Medicine, Xi'an
Jiaotong University (Xi'an, China).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.) from the cells or tissues according
to the manufacturer's protocol. cDNA synthesis was performed using
the Prime-Script RT reagent kit (Takara Biotechnology Co., Ltd.,
Dalian, China) according to the manufacturer's protocol. qPCR was
performed on cDNA using SYBR Premix Ex Taq (Takara Biotechnology
Co., Ltd.) according to the manufacturer's protocol. PCR
amplification was performed on a FTC-3000TM system (Funglyn Biotech
Inc., Toronto, ON, Canada). Thermocycling conditions were as
follows: Initial denaturation at 95°C for 30 sec, followed by 40
cycles at 95°C for 5 sec and at 60°C for 30 sec. U6 was used for
normalization in miRNA detection. The 2−ΔΔCq method was
used to quantify the relative expression levels of miR-214
(11). The primer sequences are
listed in Table I.
 | Table I.Sequences of primers, inhibitors and
UTRs used in the present study. |
Table I.
Sequences of primers, inhibitors and
UTRs used in the present study.
| Name | Sequence (5–3′) |
|---|
| U6 | RT:
CGCTTCACGAATTTGCGTGTCAT |
|
| Forward:
GCTTCGGCAGCACATATACTAAAAT |
|
| Reverse:
CGCTTCACGAATTTGCGTGTCAT |
| miR-214 | RT:
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACTGCCT |
|
| Forward:
ATCCAGTGCGTGTCGTG |
|
| Reverse:
TGCTACAGCAGGCACAGAC |
|
miR-214-inhibitor |
ACTGCCTGTCTGTGCCTGCTGT |
|
miR-214-inhibitor-ctrl |
CAGTACTTTTGTGTAGTACAA |
| si-ctrl-S |
UUCUCCGAACGUGUCACGUTT |
| si-ctrl-AS |
ACGUGACACGUUCGGAGAATT |
| siWnt3a-S |
CCCACUCGGAUACUUCUUATT |
| siWnt3a-AS |
UAAGAAGUAUCCGAGUGGGTT |
| Wnt3a |
CCCTGCCAGGGAACTGGCCTGCTGCC |
| 3′UTR-WS |
|
| Wnt3a |
TCGAGGCAGCAGGCCAGTTCCCTGG |
| 3′UTR-WA | CAGGGAGCT |
| Wnt3a |
CCCTGCCAGGGAACTGGCCACGTGCC |
| 3′UTR-MS |
|
| Wnt3a |
TCGAGGCACGTGGCCAGTTCCCTGG |
| 3′UTR-MA | CAGGGAGCT |
Plasmid vector constructs
EcoRI and HindIII sites were inserted
into the multiple cloning sites of the pcDNA6.2-GW/EmGFP vector
(Invitrogen; Thermo Fisher Scientific, Inc.). The primary miR-214
sequence was amplified by PCR from genomic DNA, as aforementioned,
and then cloned into the EcoRI and HindIII sites of
the pcDNA6.2-GW/EmGFP vector. The wild-type (wt) and mutant (mut)
Wnt3a 3′ untranslated region (UTR) sequences with SacI and
XhoI restriction enzymes were synthesized by Sangon Biotech.
Co., Ltd. (Shanghai, China) and cloned between the SacI and
XhoI sites of pmirGLO Dual-Luciferase miRNA Target
Expression Vector (Promega Corporation, Madison, WI, USA). The
3′UTR sequences are listed in Table
I. Wnt3a was cloned into a GV230 vector (Shanghai GeneChem Co.,
Ltd., Shanghai, China).
Luciferase activity assay
Potential target genes for miR-214 were predicted
using RegRNA online software version 2.0 (http://regrna2.mbc.nctu.edu.tw). A potential
miR-214-binding site in the 3′-UTR of Wnt3a mRNA was identified. To
determine whether Wnt3a was a direct target of miR-214, HepG2 cells
were co-transfected with miR-214 and wt Wnt3a 3′UTR or mut Wnt3a
3′UTR pmirGLO plasmid, using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) as the transfection reagent, according to
manufacturer's protocol. HepG2 cells co-transfected with miR-214
and pmirGLO vector were used as the control. After 48 h, luciferase
activity was detected using the Dual-Luciferase Reporter Assay
system (Promega Corporation) according to the manufacturer's
protocol.
Cell proliferation assay
Cells were seeded into 96-well plates at a density
of 5,000 cells/well. Cells were transfected with miR-214, miR-214
inhibitor, small interfering RNA(si) Wnt3a, or their respective
controls using Lipofectamine 2000 as the transfection reagent,
according to manufacturer's protocol. The miR-214 inhibitor was
synthesized by Sangon Biotech Co., Ltd. The siWnt3a and the
corresponding control (ctrl) siRNA were purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). Cell Counting kit-8 (CCK8;
7Sea Biotech, Shanghai, China) assay was performed according to the
manufacturer's protocol. Optical density at 450 nm was measured 24,
48 and 72 h post-transfection using the FLUOstar OPTIMA microplate
reader (BMG Labtech GmbH, Ortenberg, Germany), in order to measure
cell proliferation.
Cell cycle analysis
HepG2 and Hep3B cells were transfected with miR-214,
miR-control (ctrl), miR-214 inhibitor, inhibitor-ctrl, siWnt3a or
si-ctrl (Shanghai GenePharma Co., Ltd.) using Lipofectamine 2000
according to the manufacturer's instructions. At 48 h
post-transfection, 1×106 cells were harvested and washed
in PBS, then fixed with 70% ice-cold ethanol at 4°C overnight. The
cells were washed in PBS and then incubated at 4°C with 0.1 mg/ml
RNase A and 0.05 mg/ml propidium iodide (PI; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) for 30 min. Fluorescence-activated cell
sorting was performed using a BD FACSort flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA). ModFit LT version 4.0
(Verity Software House, Inc., Topsham, ME, USA) was used to analyze
cell cycle distributions.
Immunohistochemistry
Formaldehyde-fixed, paraffin-embedded tissue samples
were sectioned at 5 µm, after which the samples were deparaffinized
in xylene and hydrated using graded alcohol. Antigen retrieval was
performed with citrate buffer 0.01 M (pH 6.0), followed by blocking
endogenous peroxidases with 3% H2O2 for 20
min, and blocking of non-specific binding with normal goat serum
(Beijing Biosynthesis Biotechnology Co., Ltd., Beijing, China) at
room temperature for 20 min. The slides were then incubated with
anti-Wnt3a primary antibodies [FT20][a21](1:100; Beijing
Biosynthesis Biotechnology Co., Ltd., Beijing, China, bs-1700R) at
4°C overnight, followed by incubation with the HRP-conjugated
secondary antibody (Beijing ZhongShan-Golden Bridge Biological
Technology Co., Ltd., Beijing, China) [FT22][a23] for 30 min at
37°C. Detection was performed using 3,3′-diaminobenzidine
[FT25][a26] for 3 min at room temperature (Beijing ZhongShan-Golden
Bridge Biological Technology Co., Ltd., Beijing, China) and Harris
hematoxylin [FT27][a28] for 30 sec at room temperature. Finally, a
Leica Photo Microscope (Leica Microsystems GmbH, Wetzlar, Germany)
was used to obtain digital images.
Western blot analysis
Total proteins were extracted using
radioimmunoprecipitation assay lysis buffer (Xi'an Wolsen
Biotechnology Co., Ltd., Xi'an, China). Protein concentration was
determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop;
Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Equal amounts
(30 µg) of extracted protein samples were separated by 10% SDS-PAGE
and were then transferred onto a polyvinylidene fluoride membrane,
which was blocked with 5% non-fat milk in TBS containing 0.05%
Tween-20 (TBST) for 1 h at room temperature. Membranes were then
incubated with rabbit anti-human Wnt3a (cat no. bs-1700R; 1:100;
Beijing Biosynthesis Biotechnology Co., Ltd.) and rabbit anti-human
GAPDH (cat no. 10494-1-AP; 1:2,000; ProteinTech Group, Inc.,
Chicago, IL, USA) at 4°C overnight. Subsequently, the membrane was
washed 3 times with TBST and incubated with secondary goat
anti-rabbit antibody (cat no. 111-035-144; 1:1,000; Jackson
ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 2 h at
room temperature. Protein bands were visualized using Immobilon
Western Chemiluminescent HRP substrate (EMD Millipore, Billerica,
MA, USA). Blots were semi-quantified by densitometry using Quantity
One imaging software (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Statistical analysis
Statistical analysis was performed with SPSS
software version 13.0 (SPSS, Inc., Chicago, IL, USA). Student's
t-test or one-way analysis of variance followed by a post hoc Tukey
test were used to analyze the data from three independent
experiments. P<0.05 was considered to indicate a statistically
significant difference.
Results
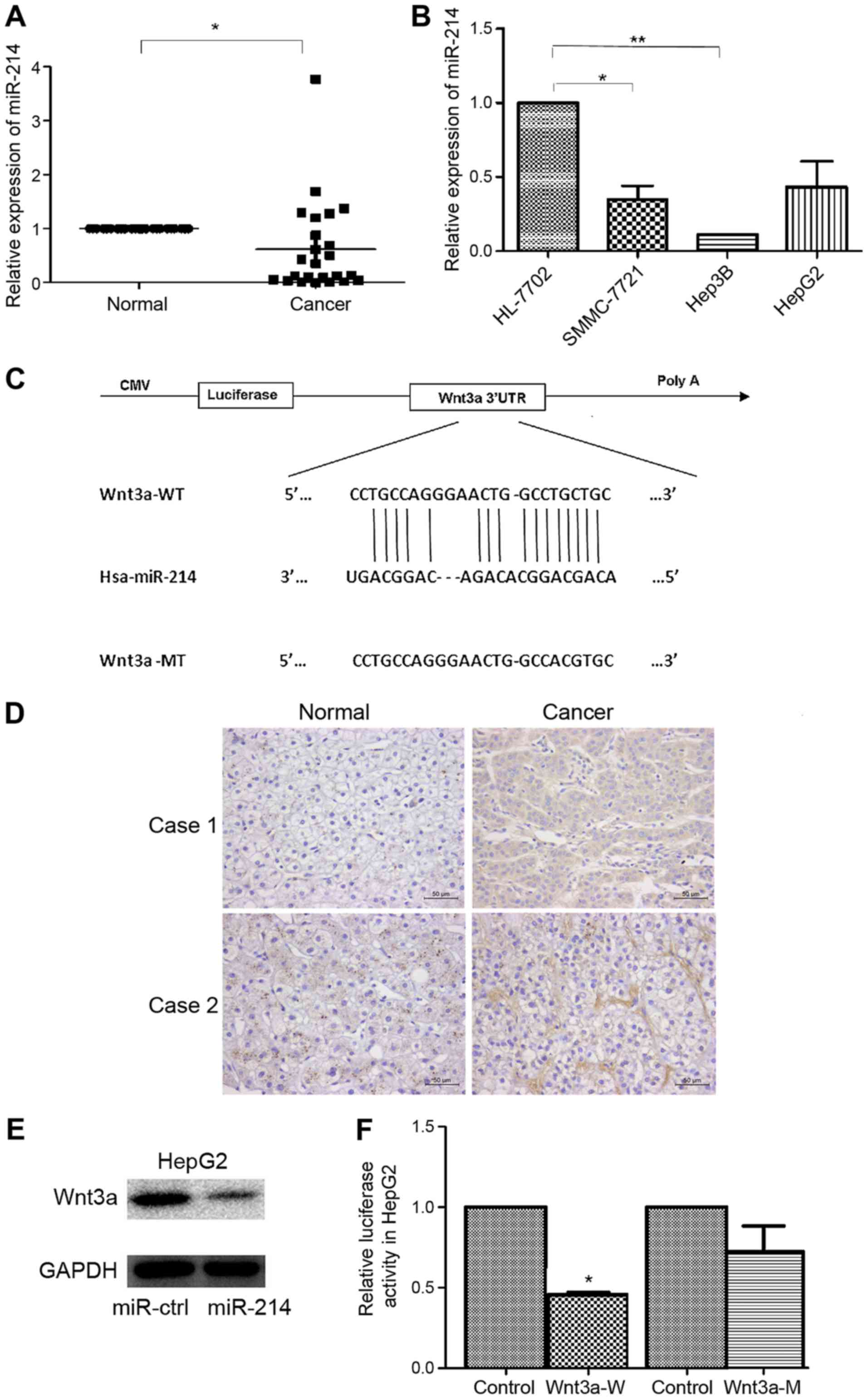
miR-214 is downregulated in liver
cancer and targets Wnt3a
To explore the expression of miR-214 in liver cancer
development, the expression levels of miR-214 were detected in 24
pairs of HCC and matched normal tissue samples using RT-qPCR.
miR-214 was significantly downregulated in HCC tissue samples
compared with in matched non-tumor tissues, as presented in
Fig. 1A. In addition, miR-214 was
downregulated in the examined liver cancer cells (SMMC-7721, Hep3B,
HepG2) compared with in normal HL-7702 hepatocytes (Fig. 1B). These findings suggested that
miR-214 was downregulated in HCC tissues and liver cancer cell
lines; therefore, miR-214 may act as a potential anti-oncogenic
miRNA in liver cancer.
The prediction of miR-214 targets was acquired using
RegRNA. Wnt3a was identified as a potential target gene of miR-214.
A miR-214 binding site was identified in the 3′UTR of Wnt3a
(Fig. 1C). Immunohistochemistry
demonstrated that Wnt3a protein expression was increased in HCC
samples compared with in normal tissues (Fig. 1D). The protein expression levels of
Wnt3a were also measured by western blotting in HepG2 cells
transfected with miR-214 or miR-ctrl, and the results demonstrated
that miR-214 overexpression was able to reduce Wnt3a protein
expression (Fig. 1E).
Dual-luciferase reporter assay was employed to determine whether
Wnt3a was a direct target of miR-214. Luciferase activity was
significantly reduced following co-transfection with miR-214 and wt
Wnt3a 3′UTR compared with the control; however, luciferase activity
was not altered following co-transfection with miR-214 and mut
Wnt3a 3′UTR (Fig. 1F). These
results provided direct evidence that Wnt3a was a target of
miR-214.
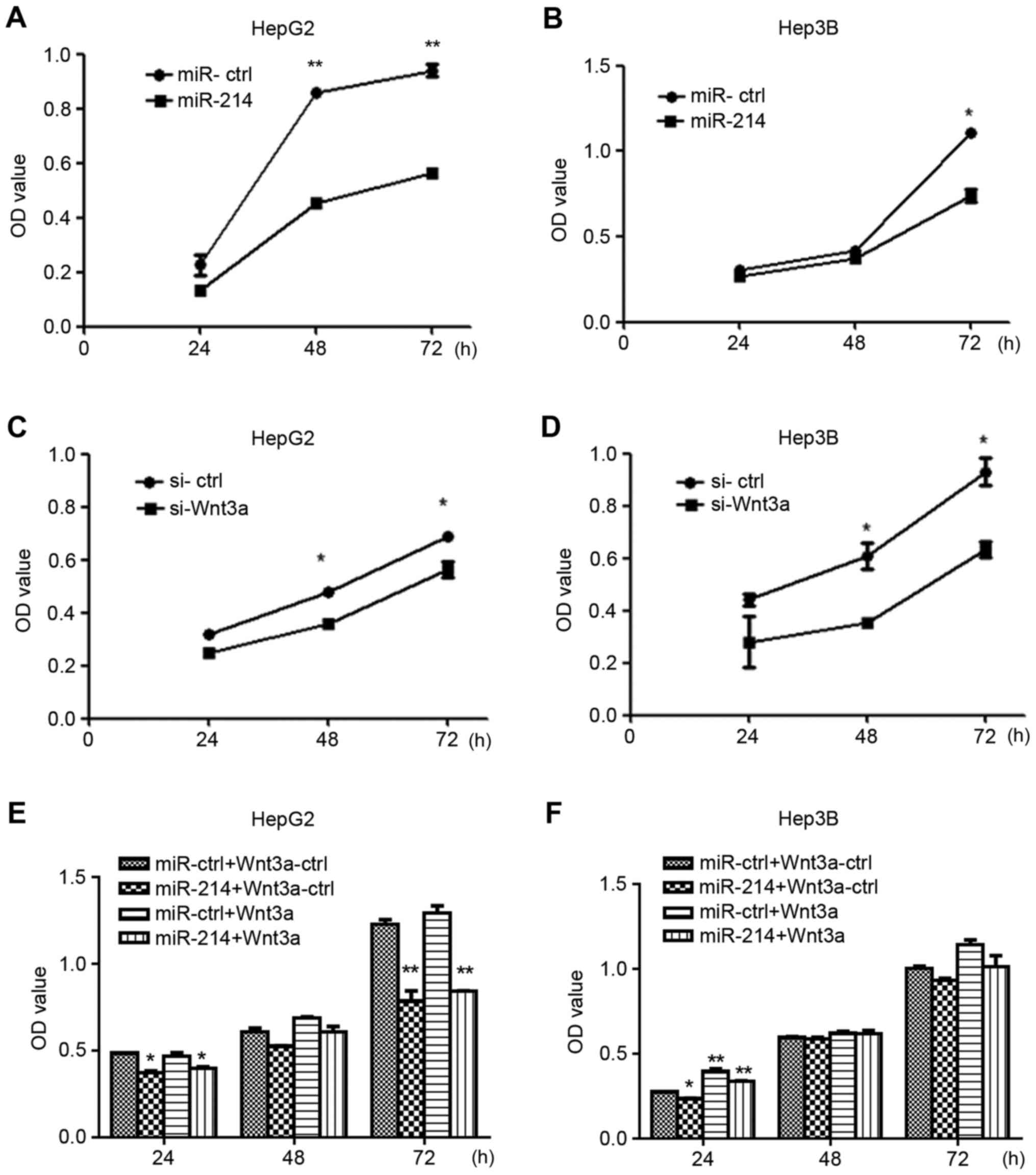
miR-214 suppresses liver cancer cell
growth via Wnt3a targeting
To explore the role of miR-214 in liver cancer cell
growth, HepG2 and Hep3B cells were transfected with miR-214,
miR-ctrl, siWnt3a or si-ctrl. Overexpression of miR-214 suppressed
the growth of HepG2 and Hep3B cells following transfection for 24,
48 and 72 h compared with control vector-transfected cells
(Fig. 2A and B). The results of
Wnt3a silencing were consistent with those of miR-214
overexpression. Based on CCK8 assays (Fig. 2C and D), siWnt3A inhibited the
growth of HepG2 and Hep3B cells. To further demonstrate that
miR-214 suppressed the growth of HepG2 and Hep3B by targeting
Wnt3a, Wnt3a overexpression vector and miR-ctrl or miR-214 were
co-transfected into HepG2 and Hep3B cells. The results demonstrated
that overexpression of Wnt3a can mitigate the growth inhibition
caused by overexpression of miR-214 (Fig. 2E and F).
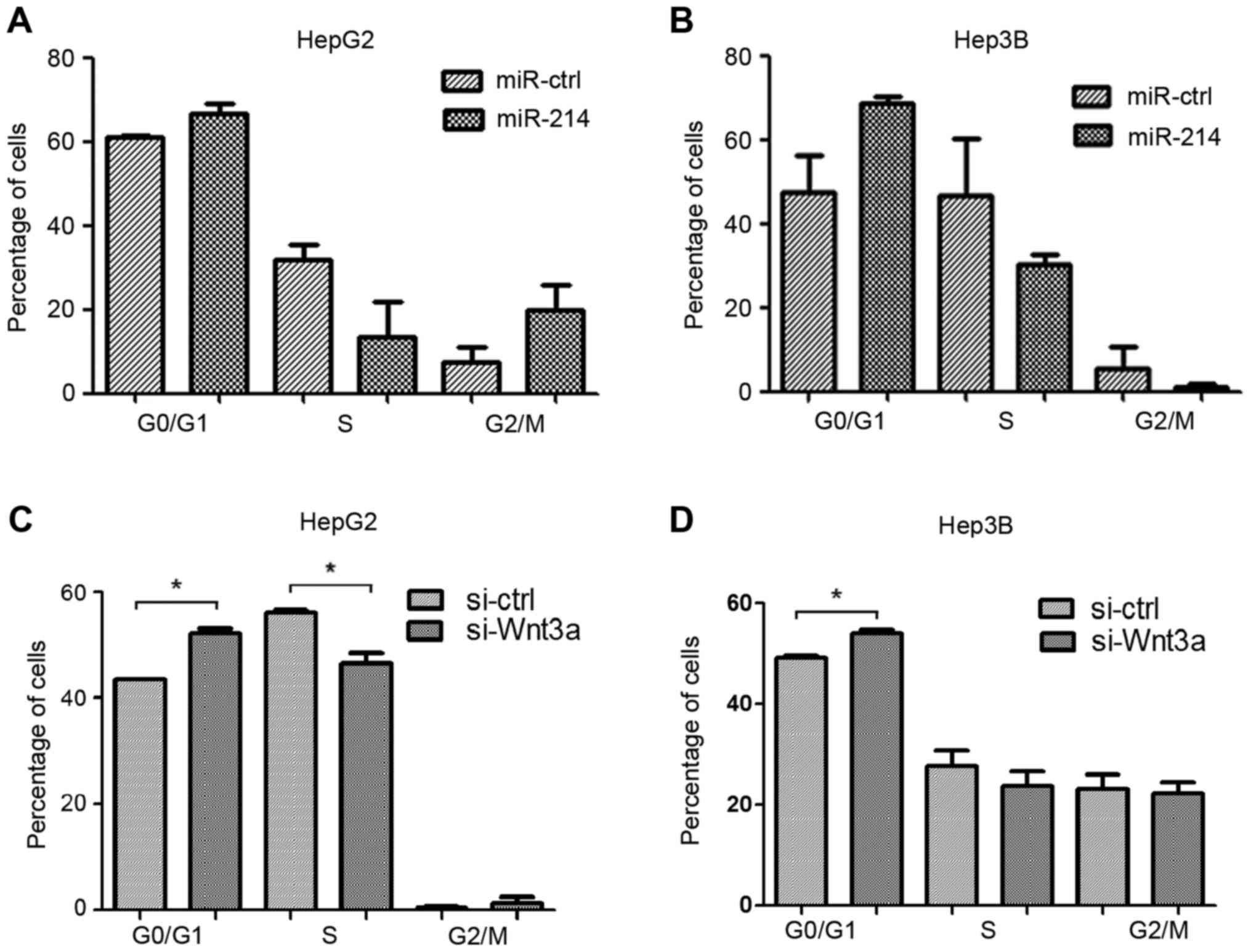
Overexpression of miR-214 and
silencing of Wnt3a inhibits liver cancer cell proliferation via
cell cycle regulation
HepG2 and Hep3B cells were transfected with miR-214
or miR-ctrl, and siWnt3a or si-ctrl. Cell cycle distribution, as
detected by flow cytometry, revealed an accumulation of HepG2 and
Hep3B cells in G1 phase in the miR-214 overexpression
groups compared with the miR-ctrl groups (Fig. 3A and B). In addition, the number of
cells in G1 phase was increased following transfection
with siWnt3a compared with the control groups (Fig. 3C and D). These results suggested
that overexpression of miR-214 or Wnt3a silencing may induce
G1 cell cycle arrest.
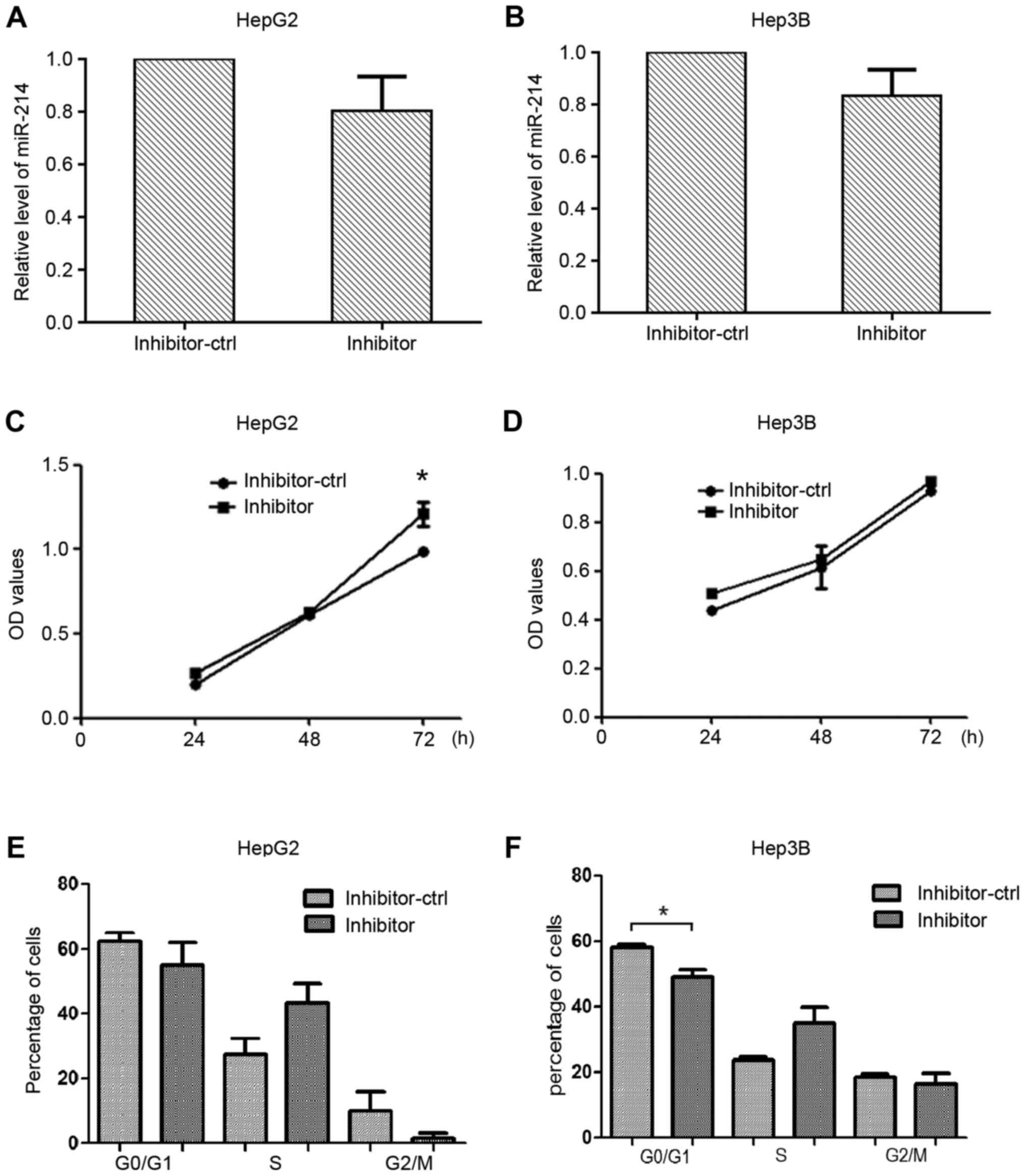
Knockdown of miR-214 contributes to
liver cancer cell proliferation
miR-214 inhibitor was used to further investigate
the effects of miR-214 silencing on liver cancer cell
proliferation. HepG2 and Hep3B cells were transfected with miR-214
inhibitor or inhibitor-ctrl, and the expression levels of miR-214
were detected (Fig. 4A and B).
Cell viability was measured using the CCK8 assay; transfection with
the miR-214 inhibitor increased the proliferation of HepG2 cells
compared with the control group (Fig.
4C and D). The number of cells in G1 phase was reduced in the
Hep3B cells transfected with miR-214 inhibitor compared with the
control group (Fig. 4E and F).
Discussion
Previous studies have supported the role of miRNAs
in tumorigenesis, including human liver cancer, and their functions
as tumor suppressors or oncogenes (12,13).
These small non-protein-coding RNAs are able to target numerous
genes, including oncogenes, leading to the degradation of target
mRNAs and inhibition of translation (14). miRNA dysregulation is associated
with human cancer; specifically, miR-214 is a tumor suppressor of
HCC (15). The present study
revealed that miR-214 is downregulated in HCC cancer tissues and
liver cancer cell lines, and may act as a tumor suppresser via
Wnt3a signaling.
The Wnt/β-catenin signaling pathway is highly
conserved and serves an essential role in regulating a series of
genes associated with processes including embryonic development,
stem cell maintenance and tissue homeostasis, and its disruption is
a common cause of numerous types of cancer (16–18).
In addition, the pathway is important during the development of HCC
(19). Previous studies have
demonstrated that β-catenin-activating mutations may associate with
HCC, thus establishing an association between Wnt signaling and HCC
(20,21) and providing novel insight into the
complex network underlying HCC. Wnts act through distinct canonical
and noncanonical pathways (22).
The canonical Wnt/β-catenin signaling pathway is involved in the
transition from cell proliferation to myogenic differentiation
(23). β-catenin is a central
regulator in the Wnt/β-catenin pathway (24,25)
and is present in the plasma membrane, cytoplasm and nucleus. The
interplay between the cell cycle and Wnt signaling is complex and
requires further study (26,27).
Wnt3a is a member of the Wnt family located on human chromosome 17
(17q21) and is an important molecule in the Wnt/β-catenin pathway
that can active the canonical Wnt signaling pathway (28). The overexpression of Wnt3a
expression serves an important role in hepatocarcinogenesis
(29).
In conclusion, the present study demonstrated that
Wnt3a is a target gene of miR-214. In addition, the results
indicated that miR-214 may inhibit cell proliferation in liver
cancer cells via Wnt3a targeting. These data provide experimental
evidence to suggest that miR-214 acts as a tumor suppressor by
suppressing the Wnt/β-catenin signaling pathway. Therefore, miR-214
may be considered a potential molecular therapeutic target for the
treatment of liver cancer.
Acknowledgements
The present study was supported by the Scientific
Research and Sharing Platform Construction Project of Shaanxi
Province (grant no. 2015FWPT-14) and Research Support Project of
New Teacher of Xi'an Jiaotong University (grant no. YX1K078).
References
|
1
|
Kew MC: Epidemiology of chronic hepatitis
B virus infection, hepatocellular carcinoma, and hepatitis B
virus-induced hepatocellular carcinoma. Pathol Biol (Paris).
58:273–277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sherman M: Hepatocellular carcinoma: New
and emerging risks. Dig Liver Dis. 42 Suppl 3:S215–S222. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dragani TA: Risk of HCC: Genetic
heterogeneity and complex genetics. J Hepatol. 52:252–257. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Feo F, De Miglio MR, Simile MM, Muroni MR,
Calvisi DF, Frau M and Pascale RM: Hepatocellular carcinoma as a
complex polygenic disease. Interpretive analysis of recent
developments on genetic predisposition. Biochim Biophys Acta.
1765:126–147. 2006.PubMed/NCBI
|
|
5
|
Rajendiran S, Parwani AV, Hare RJ,
Dasgupta S, Roby RK and Vishwanatha JK: MicroRNA-940 suppresses
prostate cancer migration and invasion by regulating MIEN1. Mol
Cancer. 13:2502014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA, Liu CG, Sevignani C, Ferracin M,
Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al:
MicroRNA profiling reveals distinct signatures in B cell chronic
lymphocytic leukemias. Proc Natl Acad Sci USA. 101:11755–11760.
2004; View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wiemer EA: The role of microRNAs in
cancer: No small matter. Eur J Cancer. 43:1529–1544. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z,
Zhan Q, Deng X, Chen H, Shen B, et al: miR-150-5p Inhibits Hepatoma
cell migration and invasion by targeting MMP14. PLoS One.
9:e1155772014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang XP, Hou J, Shen XY, Huang CY, Zhang
XH, Xie YA and Luo XL: MicroRNA-486-5p, which is downregulated in
hepatocellular carcinoma, suppresses tumor growth by targeting
PIK3R1. FEBS J. 282:579–594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kerr TA, Korenblat KM and Davidson NO:
MicroRNAs and liver disease. Transl Res. 157:241–252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang S and He X: The role of microRNAs in
liver cancer progression. Br J Cancer. 104:235–240. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Di Leva G and Croce CM: miRNA profiling of
cancer. Curr Opin Genet Dev. 23:3–11. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shih TC, Tien YJ, Wen CJ, Yeh TS, Yu MC,
Huang CH, Lee YS, Yen TC and Hsieh SY: MicroRNA-214 downregulation
contributes to tumor angiogenesis by inducing secretion of the
hepatoma-derived growth factor in human hepatoma. J Hepatol.
57:584–591. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zeng XC, Liu FQ, Yan R, Yi HM, Zhang T,
Wang GY, Li Y and Jiang N: Downregulation of miR-610 promotes
proliferation and tumorigenicity and activates Wnt/β-catenin
signaling in human hepatocellular carcinoma. Mol Cancer.
13:2612014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reya T and Clevers H: Wnt signalling in
stem cells and cancer. Nature. 434:843–850. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gheinani A Hashemi, Burkhard FC, Rehrauer
H, Fournier C Aquino and Monastyrskaya K: MicroRNA miR-199a-5p
regulates smooth muscle cell proliferation and morphology by
targeting Wnt2 signaling pathway. J Biol Chem. 13:7067–7086. 2015.
View Article : Google Scholar
|
|
19
|
Qu B, Liu BR, Du YJ, Chen J, Cheng YQ, Xu
W and Wang XH: Wnt/β-catenin signaling pathway may regulate the
expression of angiogenic growth factors in hepatocellular
carcinoma. Oncol Lett. 7:1175–1178. 2014.PubMed/NCBI
|
|
20
|
de La Coste A, Romagnolo B, Billuart P,
Renard CA, Buendia MA, Soubrane O, Fabre M, Chelly J, Beldjord C,
Kahn A and Perret C: Somatic mutations of the beta-catenin gene are
frequent in mouse and human hepatocellular carcinomas. Proc Natl
Acad Sci USA. 95:8847–8851. 1998; View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Miyoshi Y, Iwao K, Nagasawa Y, Aihara T,
Sasaki Y, Imaoka S, Murata M, Shimano T and Nakamura Y: Activation
of the beta-catenin gene in primary hepatocellular carcinomas by
somatic alterations involving exon 3. Cancer Res. 58:2524–2527.
1998.PubMed/NCBI
|
|
22
|
Shi X and Garry DJ: Muscle stem cells in
development, regeneration, and disease. Genes Dev. 20:1692–1708.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tanaka S, Terada K and Nohno T: Canonical
Wnt signaling is involved in switching from cell proliferation to
myogenic differentiation of mouse myoblast cells. J Mol Signal.
6:122011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Anastas JN and Moon RT: WNT signalling
pathways as therapeutic targets in cancer. Nat Rev Cancer.
13:11–26. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li D, Liu W, Wang X, Wu J, Quan W, Yao Y,
Bals R, Ji S, Wu K, Guo J and Wan H: Cathelicidin, an antimicrobial
peptide produced by macrophages, promotes colon cancer by
activating the Wnt/β-catenin pathway. Oncotarget. 6:2939–2950.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gougelet A and Colnot S: A complex
interplay between Wnt/β-catenin signalling and the cell cycle in
the adult liver. Int J Hepatol. 2012:8161252012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He TC, Sparks AB, Rago C, Hermeking H,
Zawel L, da Costa LT, Morin PJ, Vogelstein B and Kinzler KW:
Identification of c-MYC as a target of the APC pathway. Science.
281:1509–1512. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Constantinou T, Baumann F, Lacher MD,
Saurer S, Friis R and Dharmarajan A: SFRP-4 abrogates
Wnt-3a-induced beta-catenin and Akt/PKB signalling and reverses a
Wnt-3a-imposed inhibition of in vitro mammary differentiation. J
Mol Signal. 3:102008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pan LH, Yao M, Cai Y, Gu JJ, Yang XL, Wang
L and Yao DF: Oncogenic Wnt3a expression as an estimable prognostic
marker for hepatocellular carcinoma. World J Gastroenterol.
22:3829–3836. 2016. View Article : Google Scholar : PubMed/NCBI
|