Introduction
Preeclampsia (PE) is an increasingly problematic
pregnancy-related disorder around the world, especially in
developing countries, and it remains a major cause of maternal and
fetal mortality (1). This
life-threatening disease mainly manifests as new-onset hypertension
and proteinuria symptoms after 20 weeks of pregnancy and is
relieved after delivery, but it often has long-term effects on
maternal health. As the disease progresses, many patients suffer
from hypertensive disorders, cardiovascular dysfunction, and
chronic renal failure in addition to gestational diabetes mellitus
(GDM) (2–4). Researchers have revealed that PE can
increase the risk of asymptomatic cardiac abnormalities after birth
and is a specific risk factor for postpartum asymptomatic heart
failure (5). The exploration of
the molecular mechanisms underlying PE have become the focus of a
substantial amount of research.
Preeclampsia is a complex disease with systemic
involvement. Multiple factors have been found to be related to its
pathogenesis. These include hypoxia, the superficial nidation of
the placenta, damage to vascular endothelial cells, and immune
factors (6–8). The roles of many of these factors in
the pathogenesis of PE are in the early stage of exploration, and
the exact cause of PE therefore remains unknown. Scholars have come
to the consensus that in PE, an effective prognosis relies on an
early diagnosis. Traditional diagnostic methods include regular
blood pressure measurements, determining 24-h urine protein levels,
detecting placental thickness and homogeneity, obtaining a history
of preeclampsia and the patient's body-mass index. However, early
diagnostic efficiency is usually not unsatisfactory (9). In recent decades, to predict PE
early, many researchers have applied themselves to identifying the
mechanisms underlying PE. Serum markers, specific associated
proteins, long non-coding RNA (lncRNA), and even circular RNA
(circRNA) have been found to contribute to the onset of PE
(10–13). However, all of these discoveries
remain theoretical, and there is therefore a large distance between
laboratory findings and clinical applications.
Proteins are important organic factors that play key
roles in many physiological and pathological processes. In studying
proteins, scholars have identified a new type of polymer made of
amino acids that differs from protein. These are called peptides
and are usually less than 10 kDa. Peptides act like proteins but
are easier to acquire and transform. Performing deeper research
aimed at obtaining a comprehensive understanding of all peptides or
small proteins in a whole organism, collectively called the
peptidome, was suggested in 2000 (14,15).
In disease states, peptidome analyses have mainly been used to
select biomarkers, diagnose diseases, evaluate therapeutic
efficacy, and achieve early prevention. Single biomarkers are not
sensitive enough for early prevention, but peptidome analysis can
improve this deficiency and the rate of detection in diseases
(16,17). Peptidome analysis remains an
underdeveloped areas in PE patients. It therefore deserves to be
more deeply explored to identify differentially expressed peptides
that may be useful biomarkers for early screening in PE.
Amniotic fluid is a dynamic biological fluid that
forms the living environment of a developing fetus. An analysis of
the amniotic fluid can shed light on the functions of the placenta
and amniotic cavity. Abnormal components in the amniotic fluid
reflect the fetal and maternal state. The amniotic fluid has been
used to analyze disease-specific biomarkers associated with many
conditions, and the most common and most mature such application is
screening for Down's syndrome (18). It is easier to obtain amniotic
fluid than placental tissues during pregnancy. We therefore chose
amniotic fluid for research samples in our study. In this study, we
characterized the peptidome in PE using an ultracentrifugation
method to obtain a basic understanding of the ranges of peptide
expression. We then used liquid chromatography-tandem mass
spectrometry (LC-MS/MS) to quantify the expression of peptides that
were differentially up- and downregulated between normal and PE
pregnancies. An analysis of the functions of these peptides may
allow us to identify feasible biomarkers of PE.
Materials and methods
Sample collection
This study was permitted by the Human Research
Ethics Committee of Nanjing Medical University as well as the
Nanjing Maternal and Child Health Hospital and the Obstetrics and
Gynecology Hospital Affiliated to Nanjing Medical University.
Samples of amniotic fluid were obtained from women
with normal and PE pregnancies at 34–38 weeks of gestation at
Nanjing Maternal and Child Health Hospital, Obstetrics and
Gynecology Hospital Affiliated to Nanjing Medical University,
China, during January 14 and March 27. The pregnant participants
were fully informed regarding the aims and scope of the study and
freely signed informed consent forms. Preeclampsia patients were
diagnosed as first appearance of systolic blood pressure ≥140 mmHg
and (or) diastolic blood pressure ≥90 mmHg, companied with urine
protein quantification ≥300 mg per 24 h after 20 weeks of
gestation. Samples in our study were severe cases that blood
pressure more than 160/110 mmHg, proteinuria ≥5,000 mg/24 h,
headache or visual impairment, abnormal liver function with or
without right upper quadrant pain, and platelet less than
100×109/l. Women suffered from chronic hypertension,
acute or chronic heart disease, nephropathy, diabetes mellitus or
other maternal autoimmune diseases before pregnant were not
included in our research. All amniotic fluids were collected during
cesarean sections and placed on ice while en route to the
laboratory. The samples of amniotic fluid were centrifuged at 2,000
g for 20 min at 4°C to remove cell debris. The supernatant was then
mixed with protease inhibitors (Complete mini EDTA-free; Roche,
Mannheim, Germany) and stored at −80°C until used.
Ultrafiltration and LC-MS/MS
The amniotic fluid samples were centrifuged (12,000
g, 4°C, 20 min), and the supernatant was transferred into a new
centrifuge tube and mixed with acetonitrile (20% v/v). The mixture
was then vortexed and incubated at room temperature for
approximately 25 min (19).
Molecular weight cut-off (MWCO) filters (10,000 Da; Millipore,
Billerica, MA, USA) were washed with H2O (0.5 ml) before
they were used, and the processed samples were centrifuged through
the filters according to the manufacturer's recommendations. The
concentrations of the peptides in each of the samples were
determined using the bicinchonic acid method (BCA) (Pierce
Biotechnology, Inc., Rockford, IL, USA). The filtrates were then
concentrated and lyophilized.
LC-MS/MS is commonly used as a method for
identifying peptides or low molecular weight proteins according to
their detection sensitivity and throughput. We labeled the peptides
derived from each sample with isotopomeric dimethyl labels, which
was based on the inter-reaction of primary amines with deuterated
and 13C-labeled formaldehyde to generate a Schiff base
rapidly reduced by the addition of cyanoborohydride (20,21).
The labeled samples were simultaneously detected using LC-MS/MS
based on their abundance, which was reflected by mass differences
in the dimethyl labels.
The lyophilized samples were dissolved in 0.1%
formic acid, filtered through a 0.45-µm membrane and then set
aside. We chose an LC Packings C18 trap column (Acclaim PepMap100,
75 µm × 20 mm) to perform reverse-phase chromatography and
separated the peptides using an Ultimate 3000 nano-LC system
(Dionex) with a linear gradient in liquid phase containing (A) 0.1%
formic acid dissolved in water and (B) 0.1% formic acid dissolved
in acetonitrile (22,23). After the samples were transferred
into the MALDI TOF/TOF (Ultraflextreme; Bruker Daltonics, Bremen,
Germany) instrument under the positive ion mode, the mass spectral
analysis strategy went as follows: full scan analysis was operated
over the m/z range 600–5,000 at 2 spectra/s, with the capillary
voltage and cone voltage were maintained at 3.9 kV and 40 V.
For MS analysis, each position (10 random positions
on each sample) was irradiated with 200 laser pulses, and 2000
single-shot spectra were collected. Mass Lynx™ software (v.4.1,
Waters Corp., Milford, MA, USA) was used to analyze the protein
databases using the MS/MS spectral data for the protein ID. The
human taxonomy was searched for protein IDs in the NCBI and Matrix
Science databases (http://www.matrixscience.com/, ©2016). The MS/MS data
were searched using the Mascot database (http://www.matrixscience.com, ©2016).
Bioinformatics
Each peptide isoelectric point (pI) was calculated
using an online pI/Mw tool (http://web.expasy.org/compute_pi/, ©2017). A GO
analysis was then performed to investigate whether the identified
peptide precursors favored any compartments or protein classes. The
peptides were classified according to both their cellular
components and their molecular functions using annotations obtained
from the UniProt Database (http://www.uniprot.org/, ©2002–2017, last modified
January 10, 2017).
Statistical analysis
Data from quantitative experiments were analyzed
using unpaired Student's t-tests where appropriate in Statistical
Program for Social Sciences (SPSS) v.22 and GraphPad Prism 5.0. The
statistical significance of the results was determined using
multiple comparisons followed by Student's t-tests. Significance
was set at P<0.05. All data are shown as the means ± standard
deviations (SDs). The heat map was generated by software HemI
Version 1.0.0.
Results
Characteristics of the study
population
The basic information relating to the subjects
involved in our study are summarized in Table I. There was no difference in the
ages, number of pregnancies, BMI and gestational week between the
groups (P>0.05). Every subject delivered by caesarean section at
the appropriate time, and the newborn babies were all healthy. No
cases of asphyxia neonatorum were observed. There were differences
between the two groups in systolic pressure, diastolic pressure,
proteinuria level and neonatal weight (P<0.05).
 | Table I.Comparison of characteristics between
PE and normal pregnancies. |
Table I.
Comparison of characteristics between
PE and normal pregnancies.
| Characteristic | PE (n=3) | Normal pregnancy
(n=3) |
|---|
| Age (years) | 28.67±1.24 | 29.67±1.70 |
| Number of
pregnancies | 2.67±1.26 | 2.33±0.47 |
| BMI
(kg/m2) | 31.06±0.96 | 30.76±1.32 |
| Gestation week | 35.87±1.25 | 36.23±0.98 |
| Systolic pressure
(mmHg) |
166.33±1.25a | 118.67±2.49 |
| Diastolic pressure
(mmHg) |
84.67±2.05a | 71.33±3.40 |
| Proteinuria level
(mg/24 h) |
4806.60±1518.06a | N/A |
| Mode of devliviery CS
(%) | 100 | 100 |
| Birth weight
(g) |
3073.33±92.86a | 3133.33±62.36 |
| Apgar score | 8.67±0.47 | 9.33±0.47 |
Peptide identification and
quantitative analysis
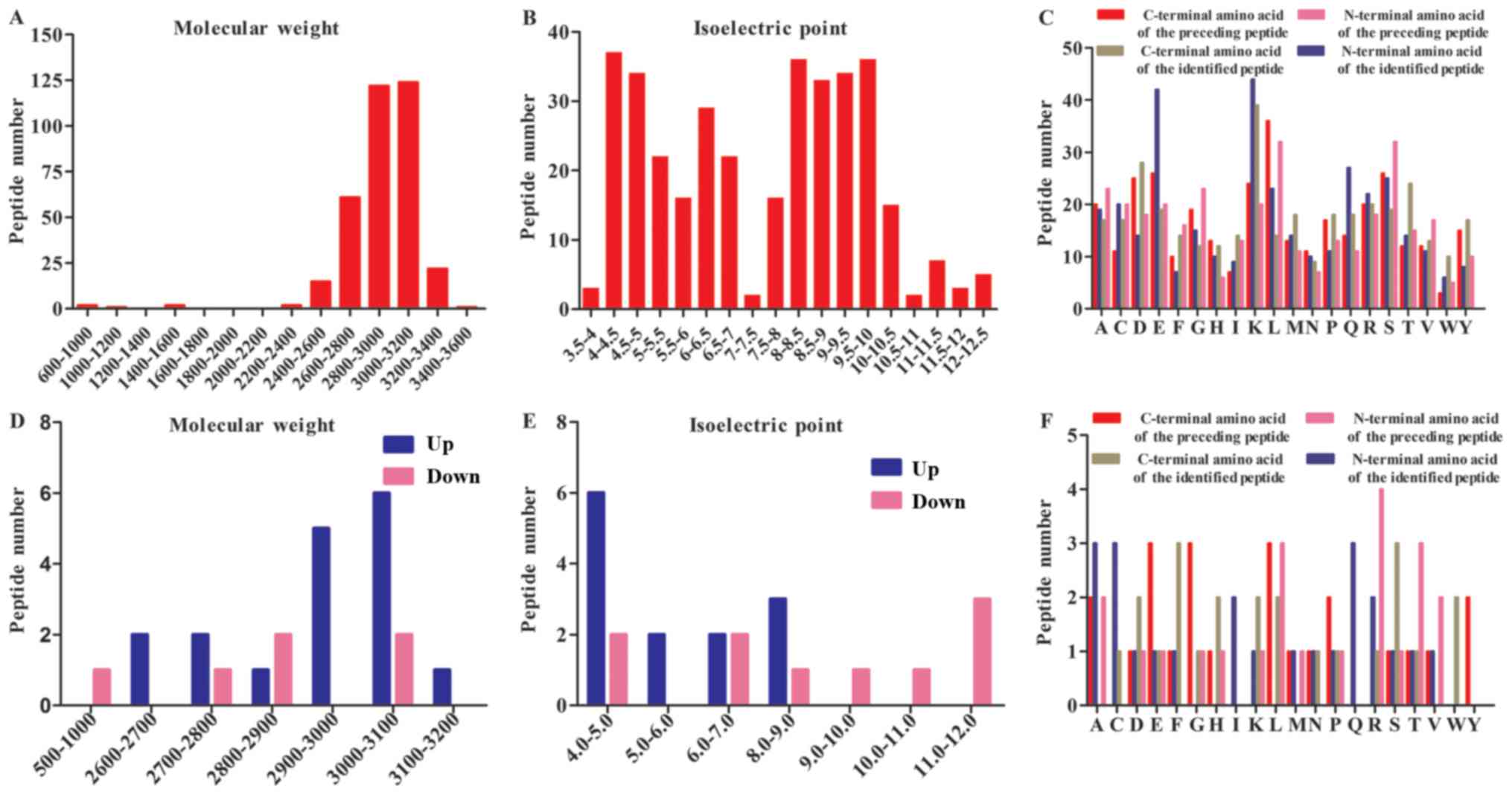
After LC-MS/MS, a total of 352 peptides were
collectively detected in the human amniotic fluid samples (Fig. 1). The peptidome contained peptides
spanning a wide range of molecular weight (Mw) and pI values. In
addition, the analysis revealed that most of the peptides were
between 2800–3200 Da in MW and between 4–6 and 8–10 in pI (Fig. 1A and B). The categories of detected
peptides clustered into two general groups according to the range
of acidic and basic pH. All of the Mw of the peptides analyzed
using LC-MS/MS were under 3500 Da, in agreement with previous
reports.
Among the 352 detected peptides, 23 were found to be
differentially expressed (6 were downregulated, and 17 were
upregulated) (P≤0.05) between PE and normal pregnancies. Table II lists these 23 peptides and
their respective precursor proteins. Further analyses revealed that
most of the upregulated peptides were distributed between 2900–3100
Da in Mw and that they were predominantly acidic in pI, whereas the
corresponding distributions of the downregulated peptides were more
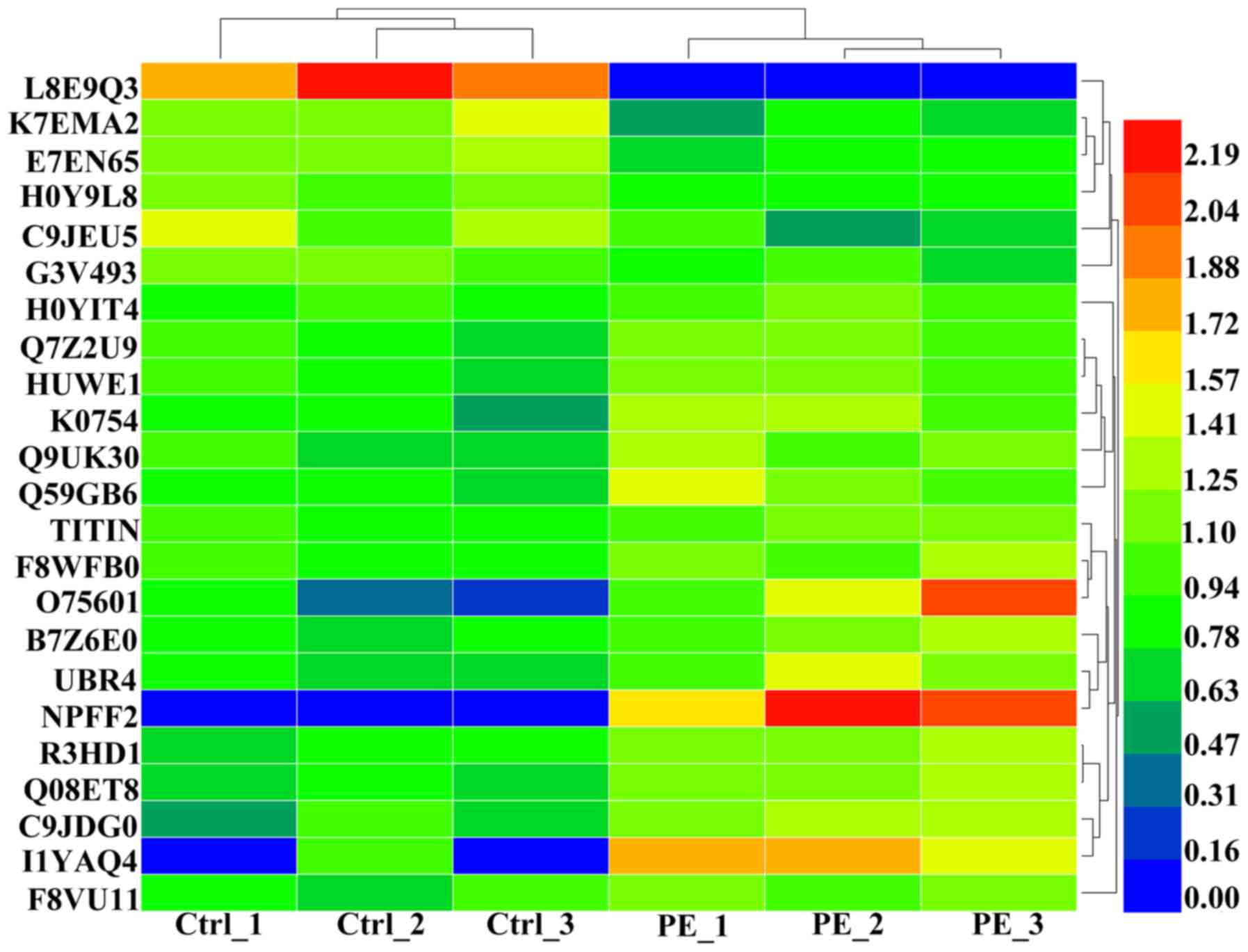
average (Fig. 1D and E). A heat
map of the six individuals is shown to clarify these differences in
expression (Fig. 2).
 | Table II.Characteristics of the peptides that
were differentially expressed peptides between PE and normal
pregnancies. |
Table II.
Characteristics of the peptides that
were differentially expressed peptides between PE and normal
pregnancies.
| Sequence | Protein ID | Fragment | Protein names | Cellular
component | Function | Fold change | P-value |
|---|
| Peptides
downregulated in amniotic fluid of pregnancy with PE |
|
|
|
|
|
|
|
| KEDEKAPQLMASY | L8E9Q3 | 61–85 | Alternative
protein | Unknown | Unknown | −∞ |
7.48×10−05 |
| DLYLCQWNIIGW |
|
| CNKSR2 |
|
|
|
|
| AAWGQTVLLLHAL | K7EMA2 | 29–53 | Beclin-1 | Nucleus | Autophagy | 0.5585 |
1.38×10−02 |
| ANKMGLKFQRMD |
|
|
|
|
|
|
|
|
CGIADFLSTYQTKVDKDLQSLEDIL | C9JEU5 | 49–73 | Fibrinogen gamma
chain | Fibrinogen
complex | Platelet
activation; protein polymerization; signal transduction | 0.5711 |
4.19×10−02 |
| CLGKACG | E7EN65 | 704–710 | Elastin | Extracelluar
region | Extracellular | 0.6332 |
4.42×10−03 |
|
IDHYCKHSEEIKDNCRNWKPTCDVF | G3V493 | 33–57 | Protein
SIX6OS1 | unknown | Unknown | 0.7276 |
2.44×10−02 |
|
FMLKPGDLLYFPRGTIHQADTPAGL | H0Y9L8 | 70–94 | Bifunctional
lysine-specific demethylase and histidyl-hydroxylase MINA | Cytoplasm;
nucleus | Unknown | 0.7587 |
8.28×10−03 |
| Peptides
upregulated in amniotic fluid of pregnancy with PE |
|
|
|
|
|
|
|
| VDGKCCKECKCEH
NFYDEYFLWKNK | H0YIT4 | 95–119 | Protein kinase
C-binding protein NELL2 | Unknown | Unknown | 1.1962 |
4.96×10−02 |
| CEKIGKIYEMRMM
MDFNGNNRGYAF | Q7Z2U9 | 76–100 | A1CF protein | Nucleus | Nucleic acid
binding; nucleotide binding | 1.2911 |
4.31×10−02 |
|
EEKAGKESDEKEQEQDKDRELQQAE | F8VU11 | 824–848 | Pre-mRNA-processing
factor 40 homolog B | Unknown | Unknown | 1.2913 |
2.49×10−02 |
|
IKITWFANDREIKESSKHRMSFVES | Q8WZ42 | 5256–5280 | Titin | Cytoplasm;
nucleus | Kinase,
Transferase, Serine/threonine-protein kinase | 1.3004 |
1.12×10−02 |
|
QELEKTLEESKEMDIKRKENKGNDT | Q7Z6Z7 | 1698–1722 | E3
ubiquitin-protein ligase HUWE1 | Cytoplasm;
nucleus | protein
ubiquitination | 1.3191 |
1.19×10−02 |
|
DEAWKSFLENPLTAATKAMMSINGD | F8WFB0 | 30–54 | Grainyhead-like
protein 1 homolog | Unknown | Unknown | 1.3407 |
3.43×10−02 |
| RSRESRLPQSVRHH
LIISHFSSKDF | Q9UK30 | 26–50 | Ras-GRF2 | Unknown | Unknown | 1.3864 |
4.47×10−02 |
|
MDENFISRAFATMGETVMSVKIIRN | B7Z6E0 | 14–38 | cDNA FLJ52201,
mRNA | Nucleus | Nucleic acid
binding; nucleotide binding | 1.4922 |
1.01×10−02 |
|
PSISAAQNALKKQINSVNKFKLRTS | Q15032 | 1060–1084 | R3H
domain-containing protein 1 | Nucleus | Poly(A) RNA
binding | 1.4973 |
2.78×10−03 |
|
QGEDKAKLMRAYMQEPLFVEFADCC | Q59GB6 | 93–117 | Thyroid hormone
receptor interactor | Membrane | Receptor | 1.5141 |
2.93×10−02 |
|
ASNPSLQEKKESSSALTESSGHLDH | O94854 | 24–48 | Uncharacterized
protein KIAA0754 | Membrane | Unknown | 1.5746 |
3.18×10−02 |
|
SENELLELFEKMMEDMNLNEDKKAP | C9JDG0 | 118–142 | Deleted | Unknown | Unknown | 1.6752 |
1.22×10−02 |
|
AAQSEGGPKSRDSKRPVATRGAKEH | Q08ET8 | 18–42 | WRAP53 | Unknown | Unknown | 1.7077 |
1.75×10−03 |
|
TESETKAFMAVCIETAKRYNLDDYR | Q5T4S7 | 4302–4326 | E3
ubiquitin-protein ligase UBR4 | Membrane;
cytoplasm; nucleus | Ligase activity;
ubiquitin-protein transferase activity; zinc ion binding | 1.7425 |
1.38×10−02 |
|
NWKEEEKKKYYDAKTEDKVRVMADS | O75601 | 36–60 | Mih1/Tx
protein | Nucleus | Regulation of
apoptotic process | 3.1777 |
3.19×10−02 |
|
QSLWEKACENQRNLNMETARISHWK | I1YAQ4 | 51–75 | LOC340970 | Intracellular | Zinc ion
binding | 4.5037 |
2.83×10−02 |
|
RKKQKIIKMLLIVALLFILSWLPLW | Q9Y5X5 | 369–393 | Neuropeptide FF
receptor 2 | Membrane | G-protein coupled
receptor, receptor, transducer | +∞ |
7.76×10−04 |
Details and cleavage sites of
endogenous peptides
LC-MS/MS identified 352 peptides, and the
bioinformatics analysis showed the specificity of the four cleavage
sites (Fig. 1C). Leucine (L) was
the most common amino acid at the C-terminal cut site of the
preceding peptide, and lysine (K) was most commonly the C-terminal
amino acid of the identified peptide. Glutamic acid (E) was most
commonly observed at the N-terminal of the preceding peptide, and
serine (S) and tryptophan (M) were most commonly observed at the N
terminus of the identified peptides.
The distribution of the cut sites in the
differentially expressed peptides were also variable in the PE
amniotic fluid samples. To further investigate the peptidome in
these samples, we next used bioinformatics analyses to determine
the amino acids at the cleavage sites of both the N- and C-termini
(Fig. 1F). The results of our
investigation of these four cut sites generally reflected the
findings reported in endogenous proteolytic enzymes in human
amniotic fluids. The results revealed that glutamic acid (E),
glycine (G) and leucine (L) were the most common amino acids at the
C-terminal cut site of the preceding peptide, while phenylalanine
(F) and serine (S) were most commonly observed at the C-terminal of
the identified peptide. However, alanine (A), cysteine (C) and
glutamine (G) dominated the cleavage site at the N terminus of the
identified peptide, while arginine (R) was the most common
N-terminal pre-cleavage amino acid.
Subcellular localization and
functional clusters of endogenous peptides
To determine whether these peptide precursors were
biased toward specific protein compartments or protein classes, we
combined Pathway and GO analyses to investigate the probable roles
of the endogenous peptidome and the precursor proteins of peptidome
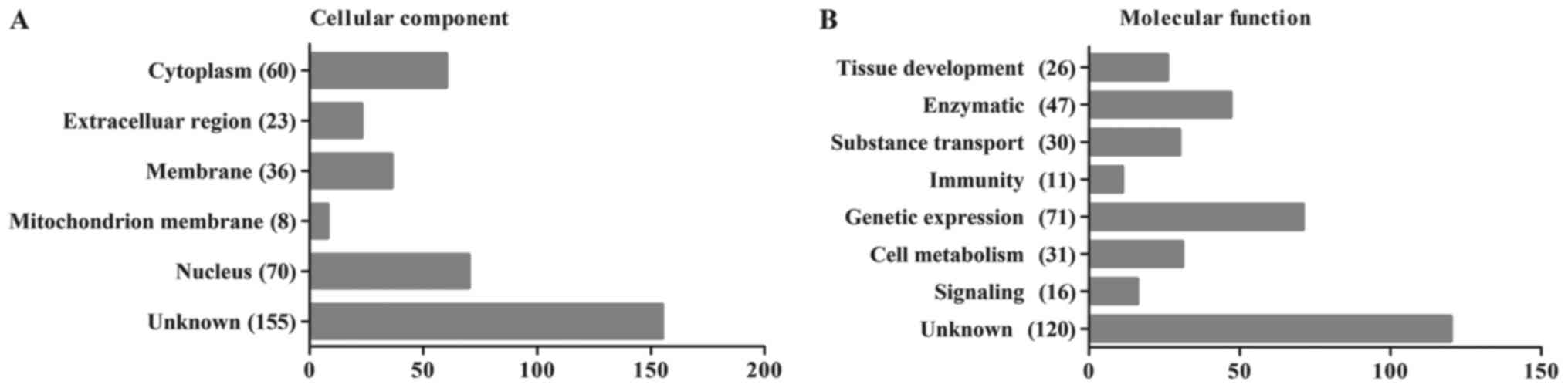
components. The 352 identified peptides were categorized according
to their cellular components and their molecular functions using
the UniProt Database (http://www.uniprot.org/, ©2002–2017, last modified
January 10, 2017). The most likely cellular component and molecular
function categories are shown in Fig.
3A and B, respectively. We found that the majority of the
proteins identified in the present study were cytoplasmic (17%) or
nuclear (20%) proteins and that the most common functions were the
regulation of gene expression (20%) and enzymatic functions (13%).
Because late pregnancy amniotic fluid consists of the metabolites
of a variety of both maternal and neonatal organs, the results of
this analysis also demonstrated the condition of the mothers and
babies.
We also summarized the cellular components and
molecular functions of the peptides found to be differentially
expressed, as shown in Table II.
These peptides were mainly located in the nucleus and have been
shown to play roles in gene expression, consistent with the results
of our analysis of the human amniotic fluid peptidome. The
different categories of the listed protein precursors demonstrated
no existence of the preferential extraction from specific cellular
components or with functions and the successful of the previous
ultrafiltration.
Discussion
PE is a critical complication in pregnancy that has
a reported morbidity rate of 2–8% among all pregnant women
(24). Studies of the pathogenesis
of and pathological changes that occur during PE have attracted the
attention of many researchers. However, the specific factors that
cause PE remain an enigma and the subject of a great deal of debate
(25). It is widely accepted that
early detection and intervention greatly improve perinatal outcomes
in both mothers and infants. Traditional monitoring methods are
performed by detecting blood pressure and urine protein levels,
which exhibit wide fluctuations in response to a variety of
external environmental and many other uncontrollable factors.
Identifying a more reliable and valid method for detecting PE is
therefore urgently needed.
Peptidome analysis is a newly emerging discipline in
proteomics that sometimes outperforms proteomics in its structural
simplicity, operation convenience, study universality and property
stability. In a sense, the peptidome is inherited and developed
from proteomics (26,27). Peptides exist everywhere in the
human body, in both organs and tissues, in both cells and bodily
fluids. Peptide analysis yields a more comprehensive picture of the
nature and molecular functions of proteins by providing information
about the synthesis, modification and degradation of proteins.
Extracting and enriching the peptides in a sample is
one of the key procedures in this type of analysis. Centrifugal
ultrafiltration accompanied with perfect Mw cutoff is a commonly
used method in peptidom analysis (28). We used standard 1000-Da filters in
our study and achieved ideal outcomes that did not include albumin
and Igs, as shown in the MW and PI results (Fig. 1A and B). The products of
ultrafiltration were labeled with isotopomeric dimethyl, allowing
them to be immediately analyzed using LC-MS/MS. A total of 352
peptides were identified in both groups. Of these, 6 were expressed
at significantly lower levels, while 17 were expressed at
significantly higher levels (P≤0.05).
Peptide fragments are created when proteins are
cleaved by proteases. Previous studies have indicated that the
cleavage sites at the N- and C-termini of endogenous peptides are
highly conserved, despite differences in the sample preparation
conditions used to analyze specific serum samples across
individuals. Additionally, the pattern of degradation of proteins
and peptides differs substantially between diseases as a result of
the specificity and activity of specific proteolytic enzymes
(29,14). The cleavage sites in the peptides
that were differentially expressed in our study are summarized in
Fig. 1F. The probability that any
one of the 20 amino acids would be located at the four cutting
sites was dramatically different. Because each protease cuts
proteins according to specific rules, these data reveal that a
distinct set of proteases are active in the amniotic fluid of PE
women. This point deserves further investigation.
Among the precursor proteins identified in our
experiments, some have previously been implicated in the onset of
PE. For example, titin (also known as TTN) has a MW of 4.2 MDa and
is the largest known protein in the human body. Titin functions as
a contractile unit in striated muscle cells, especially in
cardiomyopathies (30–32). Liang et al (33) found that Titin is targeted by
miR-144, which regulates proliferation and invasion in human
chorionic cells via the MAPK and MMPs signaling pathways. Poor
implantation of the placenta and chorionic cells, as well as
insufficient uteroplacental circulation during the early stage of
pregnancy, are key contributors to PE. Additionally, Farina et
al (34) noted that TTN
expression promoted trophoblast invasion by altering the elasticity
of and reconstructing the maternal vascular system and that higher
than normal levels of TTN were observed in PE. These data strongly
support the notion that TTN is a disease-specific biomarker for
PE.
All previous studies support the accuracy and
practicability of the results reported in our study. In 2011, Araki
et al (35) suggested using
BLOTCHIP® analysis as a peptidomic method for biomarker
discovery in PE. However, studies using peptidomic analyses in
disease states have been limited to theoretical studies that lack
large sample populations. Hence, this method should be studies in
more detail in the future.
However, there were certain deficiencies somewhere
in our study. It is necessary to include larger sample sizes to
verify its effectiveness and repeatability and to exploit the
advantages. Another, necessary validation experiment of these
identified peptides would make our discussion more credible.
In conclusion, this study is the first to provide a
peptidomic analysis of the profile of the amniotic fluid in
patients with PE. The data we provide can be used to identify novel
PE biomarkers. We will work hard on picking one significant peptide
for functional verification in future work and exploiting the
advantages associated with using peptides to prevent and more
comprehensively treat diseases.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (nos. 81571444 and 81501341)
and the Nanjing Medical Science and Technology Development
Foundation (no. YKK15163).
References
|
1
|
Phipps E, Prasanna D, Brima W and Jim B:
Preeclampsia: Updates in pathogenesis, definitions and guidelines.
Clin J Am Soc Nephrol. 11:1102–1113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vikse BE, Irgens LM, Leivestad T,
Skjaerven R and Iversen BM: Preeclampsia and the risk of end-stage
renal disease. N Engl J Med. 359:800–809. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Drost JT, Maas AH, van Eyck J and van der
Schouw YT: Preeclampsia as a female-specific risk factor for
chronic hypertension. Maturitas. 67:321–326. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu L, Zhang X, Rong C, Rui C, Ji H, Qian
YJ, Jia R and Sun L: Distinct DNA methylomes of human placentas
between pre-eclampsia and gestational diabetes mellitus. Cell
Physiol Biochem. 34:1877–1889. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ghossein-Doha C, van Neer J, Wissink B,
Breetveld NM, de Windt LJ, van Dijk AP, Van Der Vlugt MJ, Janssen
MC, Heidema WM, Scholten RR and Spaanderman ME: Preeclampsia, an
important female specific risk factor for asymptomatic heart
failure. Ultrasound Obstet. Gynecol. 49:143–149. 2016.
|
|
6
|
Wang A, Rana S and Karumanchi SA:
Preeclampsia: The role of angiogenic factors in its pathogenesis.
Physiology (Bethesda). 24:147–158. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baumwell S and Karumanchi SA:
Pre-eclampsia: Clinical manifestations and molecular mechanisms.
Nephron Clin Pract. 106:C72–C81. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sankaralingam S, Arenas IA, Lalu MM and
Davidge ST: Preeclampsia: Current understanding of the molecular
basis of vascular dysfunction. Expert Rev Mol Med. 8:1–20. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Steegers EA, von Dadelszen P, Duvekot JJ
and Pijnenborg R: Pre-eclampsia. Lancet. 376:631–644. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jia R, Li J, Rui C, Ji H, Ding H, Lu Y, De
W and Sun L: Comparative Proteomic profile of the human umbilical
cord blood exosomes between normal and preeclampsia pregnancies
with high-resolution mass spectrometry. Cell Physiol Biochem.
36:2299–2306. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pillar N, Yoffe L, Hod M and Shomron N:
The possible involvement of microRNAs in preeclampsia and
gestational diabetes mellitus. Best Pract Res Clin Obstet Gynaecol.
29:176–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang YG, Yang HL, Long Y and Li WL:
Circular RNA in blood corpuscles combined with plasma protein
factor for early prediction of pre-eclampsia. BJOG. 123:2113–2118.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qian Y, Lu Y, Rui C, Qian Y, Cai M and Jia
R: Potential Significance of circular RNA in human placental tissue
for patients with preeclampsia. Cell Physiol Biochem. 39:1380–1390.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu L, Ye M and Zou H: Recent advances in
mass spectrometry-based peptidome analysis. Expert Rev Proteomics.
6:433–447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schrader M and Schulz-Knappe P:
Peptidomics technologies for human body fluids. Trends Biotechnol.
19:55–60. 2001. View Article : Google Scholar
|
|
16
|
Xiang Y, Kurokawa MS, Kanke M, Takakuwa Y
and Kato T: Peptidomics: Identification of pathogenic and marker
peptides. Methods Mol Biol. 615:259–271. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Casalegno-Garduño R, Schmitt A and Schmitt
M: Clinical peptide vaccination trials for leukemia patients.
Expert Rev Vaccines. 10:785–799. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tsangaris GT, Karamessinis P, Kolialexi A,
Garbis SD, Antsaklis A, Mavrou A and Fountoulakis M: Proteomic
analysis of amniotic fluid in pregnancies with Down syndrome.
Proteomics. 6:4410–4419. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Wu LJ, Gu M, Chen YM, Zhang QJ, Li
H, Cheng ZJ, Hu P, Han SP, Yu ZB and Qian LM: Peptidomic analysis
of amniotic fluid for identification of putative bioactive peptides
in ventricular septal defect. Cell Physiol Biochem. 38:1999–2014.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hsu JL, Huang SY, Chow NH and Chen SH:
Stable-isotope dimethyl labeling for quantitative proteomics. Anal
Chem. 75:6843–6852. 2004. View Article : Google Scholar
|
|
21
|
Liu F, Zhao C, Liu L, Ding H, Huo R and
Shi Z: Peptidome profiling of umbilical cord plasma associated with
gestational diabetes-induced fetal macrosomia. J Proteomics.
139:38–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wan J, Cui XW, Zhang J, Fu ZY, Guo XR, Sun
LZ and Ji CB: Peptidome analysis of human skim milk in term and
preterm milk. Biochem Biophys Res Commun. 438:236–241. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cui X, Li Y, Yang L, You L, Wang X, Shi C,
Ji C and Guo X: Peptidome analysis of human milk from women
delivering macrosomic fetuses reveals multiple means of protection
for infants. Oncotarget. 7:63514–63525. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ghulmiyyah L and Sibai B: Maternal
mortality from preeclampsia/eclampsia. Semin Perinatol. 36:56–59.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shah DM: Preeclampsia: New insights. Curr
Opin Nephrol Hypertens. 16:213–220. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ivanov VT and Yatskin ON: Peptidomics: A
logical sequel to proteomics. Expert Rev Proteomics. 2:463–473.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dallas DC, Guerrero A, Parker EA, Robinson
RC, Gan J, German JB, Barile D and Lebrilla CB: Current
peptidomics: Applications, purification, identification,
quantification, and functional analysis. Proteomics. 15:1026–1038.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu RN, Fan L, Rieser MJ and El-Shourbagy
TA: Recent advances in high-throughput quantitative bioanalysis by
LC-MS/MS. J Pharm Biomed Anal. 44:342–355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang F, Zhu J, Hu L, Qin H, Ye M and Zou
H: Comprehensive analysis of the N and C terminus of endogenous
serum peptides reveals a highly conserved cleavage site pattern
derived from proteolytic enzymes. Protein Cell. 3:669–674. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gigli M, Begay RL, Morea G, Graw SL,
Sinagra G, Taylor MRG, Granzier H and Mestroni L: A review of the
giant protein titin in clinical molecular diagnostics of
cardiomyopathies. Front Cardiovasc Med. 3:212016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lindstedt S and Nishikawa K: Huxleys'
missing filament: Form and function of titin in vertebrate striated
muscle. Annu Rev Physiol. 79:145–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Walker JS and de Tombe PP: Titin and the
developing heart. Circ Res. 94:860–862. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liang Y, Lin Q, Luo F, Wu W, Yang T and
Wan S: Requirement of miR-144 in CsA induced proliferation and
invasion of human trophoblast cells by targeting titin. J Cell
Biochem. 115:690–696. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Farina A, Morano D, Arcelli D, De Sanctis
P, Sekizawa A, Purwosunu Y, Zucchini C, Simonazzi G, Okai T and
Rizzo N: Gene expression in chorionic villous samples at 11 weeks
of gestation in women who develop preeclampsia later in pregnancy:
Implications for screening. Prenat Diagn. 29:1038–1044. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Araki Y, Nonaka D, Tajima A, Maruyama M,
Nitto T, Ishikawa H, Yoshitake H, Yoshida E, Kuronaka N, Asada K,
et al: Quantitative peptidomic analysis by a newly developed
one-step direct transfer technology without depletion of major
blood proteins: Its potential utility for monitoring of
pathophysiological status in pregnancy-induced hypertension.
Proteomics. 11:2727–2737. 2011. View Article : Google Scholar : PubMed/NCBI
|