Introduction
Macrophages are derived from hematopoietic stem
cells, in particular, from bone marrow myeloid progenitor cells.
Beyond the classical functions of pathogen elimination, tissue
development and wound repair, macrophages are well-recognized key
regulators of both innate and adaptive immunity, as well as
important mediators of systemic metabolism, angiogenesis,
apoptosis, malignancy and reproduction (1–3).
Macrophages display a high degree of plasticity, with the ability
to generate different functional phenotypes (namely M1 and M2) in
response to microenvironmental cues (4,5).
Cytokines and microbial products have been implicated in the
reprogramming of M1 and M2 macrophages: Lipopolysaccharide (LPS)
plus interferon (IFN)-γ induce M1 macrophage activation, while
stimulation of macrophages with interleukin (IL)-4 or IL-13 induces
M2 macrophage activation (6,7). M1
macrophages secrete tumor necrosis factor (TNF)-α, IL12 and IL-23,
as well as large amounts of nitric oxide by expressing inducible
nitric oxide synthase, which are essential for clearing bacterial,
viral and fungal infections and in mediating resistance against
tumors (8). M2 macrophages are
characterized by upregulation of arginase (Arg)1, chitinase 3-like
3 (CHI3L3), resistin-like α (Retnla), mannose receptor C (Mrc)-1
(also known as CD206) and chemokines such as C-C motif chemokine
ligand (CCL)17 and CCL24. They are important in the host response
to parasite infection, tissue remodeling, angiogenesis and tumor
progression (9–12).
Macrophage polarization has been the focus of
previous studies, particularly with regards to transcriptional
regulation. Transcriptional factors, such as nuclear factor-κB, Jun
proto-oncogene AP-1 transcription factor subunit, signal transducer
and activator of transcription (STAT) 1, interferon regulatory
factor (IRF)3, IRF5, IRF8, hypoxia-inducible factor (HIF) 1a,
Kruppel-like factor (KLF) 2 and AKT serine/threonine kinase 1
(AKT1) participate in toll-like receptor (TLR)-induced M1
activation (8,13–17).
In contrast, STAT6, IRF4, HIF2α, peroxisome proliferator-activated
receptor (PPAR)-γ, CCAAT/enhancer-binding protein β, glucocorticoid
receptors, AKT2, and KLF4 are involved in the polarization of
macrophages to the M2 phenotype (8,13–17).
microRNA (miRs), such as miR-27b and miR-155, are involved in M1
polarization, whereas miR-9, miR-21, miR-125b, miR-146a, miR-223,
Let-7i, Let-7c and Let-7e are involved in M2 macrophage
polarization (1,2,6,18).
In addition, enzymes involved in epigenetic regulation, such as
Jumonji domain-containing 3 (JMJD3) and histone deacetylase 3, are
important in M2 macrophage polarization (19–21).
Furthermore, the importance of suppressor of cytokine signaling
(SOCS)2 and SOCS3 proteins in M1 and M2 macrophage polarization has
been recently demonstrated (22).
Microarray and bioinformatics analyses are effective
ways of identifying genes and interactions between genes (23,24).
The present study utilized microarray and bioinformatics approaches
to identify differentially-expressed genes (DEGs) and to analyze
the gene expression features of ex vivo polarized M1 and M2
macrophages. Several molecular markers of each macrophage
polarization phenotype were observed, thereby providing a
theoretical basis for further experimental studies.
Materials and methods
Mice
A total of 20 BALB/c male mice (6–8 weeks old, 25–30
g) were obtained from the Experimental Animal Center of
Qinglongshan (Nanjing, China), and were housed in pathogen-free
mouse colonies with a 12-h light, 12-h dark cycle. Mice received
standard chow diet, with free access to drinking water between 25
and 26°C. Relative humidity was maintained between 60 and 70%, and
padding was changed twice/week. All animal experiments were
performed according to the guidelines for the Care and Use of
Laboratory Animals (Ministry of Health, China, 1998). All
experimental protocols were approved by the Animal Ethics Committee
of Yijishan Hospital (Wuhu, Anhui, China).
Cell culture and stimulation
Bone marrow-derived macrophages (BMDMs) were
isolated from BALB/c mice by flushing the femurs with Dulbecco's
modified Eagle's medium (DMEM; HyClone; GE Healthcare, Chicago, IL,
USA) according to our previous studies (6,25).
Ethical approval was provided by the Animal Ethical Committee of
Yijishan Hospital. Macrophages plated on six-well plates
(1×106 cells/well) were maintained in DMEM supplemented
with 20% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and 20% L929 supernatant at 37°C and 5%
CO2 (26). Following 7
days in culture, the medium was removed, and the cells were
cultured in RPMI-1640 (HyClone; GE Healthcare) supplemented with
10% FBS for an additional 24 h. Macrophages were then stimulated
for 48 h in DMEM/10% FBS containing either 100 ng/ml LPS and 20
ng/ml IFN-γ (for M1 polarization) or 20 ng/ml IL-4 (for M2
polarization), as described previously (6,25).
RNA extraction and purification
BMDMs were collected following 48 h culture with
polarization stimuli, and total RNA was extracted using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. RNA quantity and quality were measured
using a NanoDrop 2000 (Thermo Fisher Scientific, Inc.), and RNA
integrity was assessed using an Agilent Bioanalyzer 2100 (Agilent
Technologies, Inc., Santa Clara, CA, USA) and denaturing agarose
gel electrophoresis. Total RNA was further purified using an RNeasy
Mini kit and RNase-Free DNase set (both from Qiagen GmbH, Hilden,
Germany).
Microarray analysis
Total RNA from each sample was amplified and labeled
by using a Low Input Quick Amp WT Labeling kit (Agilent
Technologies), following the manufacturer's instructions. Labeled
cRNA was purified using an RNeasy Mini kit (Qiagen GmbH). The
concentration and specific activity of the labeled cRNAs (pmol
Cy3/µg cRNA) were measured using a NanoDrop 2000. Each microarray
slide (catalog no. p/n G2534-60011/G2534-60014; Agilent
Technologies Inc.) was hybridized with 1.65 µg Cy3-labeled cRNA
using a gene expression hybridization kit (catalog no. p/n
5188–5242; Agilent Technologies, Inc.) in a hybridization oven
(catalog no. p/n G2545A; Agilent Technologies, Inc.), according to
the manufacturer's protocol. Following 17 h of hybridization, the
slides were washed in staining dishes (Thermo Fisher Scientific,
Inc.) with a gene expression wash buffer kit (catalog no. p/n
5188–5327; Agilent Technologies, Inc.), following the
manufacturer's protocol. Next, the slides were scanned using an
Agilent Microarray Scanner G2565C (Agilent Technologies, Inc.) with
the following settings: Dye channel green, scan resolution 3 µm,
PMT 100% and 20-bit scanning. The Agilent Feature Extraction
software (version 10.7; Agilent Technologies, Inc.) was used to
analyze the acquired array images. Quantile normalization and
subsequent data processing were performed using GeneSpring software
version 11.0 (Agilent Technologies, Inc.). DEGs were identified
through fold change (>2-fold) filtering. Microarray analysis was
performed by Shanghai Biotechnology Corporation (Shanghai, China).
Array data were deposited at the Gene Expression Omnibus database
of the National Center for Biotechnology Information (accession no.
GSE81922).
Functional enrichment analysis
To further understand the biological relevance and
associated pathways of DEGs, functional enrichment analysis was
performed using the Biological Network Gene Ontology (BiNGO;
v3.0.3) and CluePedia (v1.0.4) web-based tools (27,28).
BiNGO (http://www.psb.ugent.be/cbd/papers/BiNGO) is a tool
that identifies Gene Ontology (GO) terms that are significantly
overrepresented in a set of genes or a subgraph of a biological
network. BiNGO maps the predominant functional themes of the tested
gene set on the GO hierarchy and takes advantage of Cytoscape's
versatile visualization environment to produce an intuitive
molecular interaction network. The CluePediaCytoscape plugin
(v3.0.1; www.ici.upmc.fr/cluepedia) is a search tool for new
markers that are potentially associated to pathways. A pathway-like
visualization can be created using the Cerebral plugin (v2.8.2)
layout (29). The threshold of
hypergeometric distribution of functional annotation was 0.05.
Construction of interaction
networks
Since genes act by interacting with other genes to
accomplish their functions; the interaction networks of the
candidate genes identified were further explored by bioinformatics
analysis. In the present study, 18 macrophage
polarization-associated genes identified by gene expression
profiling (listed in Table I) were
examined for gene interaction networks using the Search for the
Retrieval of Interacting Genes/Proteins (STRING; v9.0) database
(string-db.org) (30). This database provides information
on both experimental and predicted interactions from varied
sources, including computational prediction, literature mining and
knowledge transfer between organisms and information aggregated
from other primary databases. An extended network was constructed
by setting the required confidence score to 0.400.
 | Table I.Differentially-expressed genes in M1
vs. M2 polarized macrophages. |
Table I.
Differentially-expressed genes in M1
vs. M2 polarized macrophages.
| Probe name | Gene symbol | P-value | Fold change | FC (abs) | Regulation |
|---|
| A_51_P257951 | Retnla | 0.0041927 | 0.00014303 | 6991.6038 | Down |
| A_51_P167292 | CHI3L3 | 6.022E-05 | 0.00244865 | 408.38827 | Down |
| A_55_P1988108 | MRC1 | 0.0144366 | 0.01116567 | 89.560221 | Down |
| A_55_P2158741 | NOS2 | 0.0267168 | 80.8592825 | 80.859282 | Up |
| A_66_P116173 | IL23r | 0.00021806 | 60.0522186 | 60.0522186 | Up |
| A_51_P303160 | ARG1 | 0.0001499 | 0.02261723 | 44.214073 | Down |
| A_51_P106799 | PPARG | 0.00702976 | 0.048704658 | 20.531917 | Down |
| A_51_P107362 | SOCS2 | 0.0016812 | 0.048945465 | 20.4309019 | Down |
| A_55_P1992834 | SOCS2 | 0.00505959 | 0.056061637 | 17.8375098 | Down |
| A_51_P322640 | CCL24 | 0.02594911 | 0.067245489 | 14.870886 | Down |
| A_55_P1992838 | SOCS2 | 0.00031572 | 0.072890051 | 13.7192935 | Down |
| A_51_P474459 | SOCS3 | 0.00465443 | 9.357196051 | 9.35719605 | Up |
| A_51_P212782 | IL1b | 0.01326346 | 7.485790577 | 7.48579058 | Up |
| A_55_P1997756 | IL6 | 0.00478943 | 7.184303002 | 7.184303 | Up |
| A_51_P385099 | TNF | 0.0009646 | 6.838318605 | 6.8383186 | Up |
| A_51_P473888 | IL6st | 0.003416 | 0.162871741 | 6.13980053 | Down |
| A_55_P2082974 | IRAK2 | 0.02073071 | 2.412076065 | 2.41207607 | Up |
| A_52_P356204 | NOSTRIN | 0.00827602 | 0.419123778 | 2.38593001 | Down |
| A_51_P271503 | IL1r1 | 0.00793288 | 0.450111469 | 2.22167189 | Down |
| A_51_P387608 | HIF1a | 0.01494099 | 2.111818487 | 2.11181849 | Up |
Statistical analysis
The threshold set for significant up- and
downregulated DEGs in microarray data was >2-fold change and
P<0.05. Data were expressed as the mean ± standard error of the
mean. Statistical analysis was performed using a Student's t-test
by using Graphpad Prism v5.0 (GraphPad Software, Inc., La Jolla,
CA, USA) for comparison between two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Overview of DEG profiles in M1 and M2
macrophages
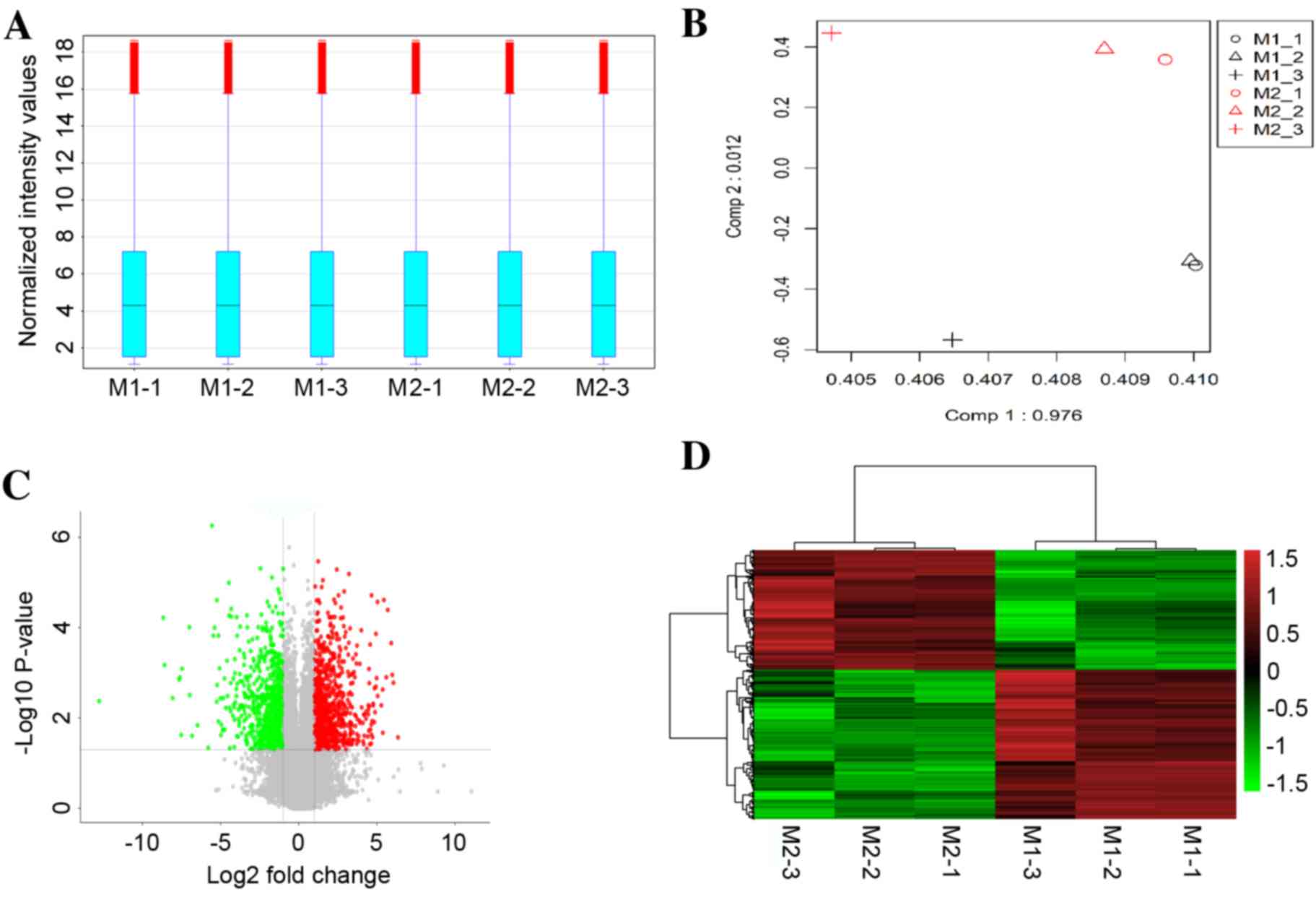
A box-plot was used to visualize the distributions
of the intensities from all samples, and principal component
analysis (PCA) was employed to perform an unsupervised examination
of differences in the signals between M1 macrophages and M2
macrophages. As demonstrated in Fig.
1A, the distribution of the log2-ratio of the microarray
intensity values in the six samples (three repeats for M1 and three
repeats for M2 macrophages) was very similar following quantile
normalization. The M1 macrophage samples were distinctly separated
from the M2 macrophage samples in the PCA plots (Fig. 1B), suggesting a differential gene
expression between M1 and M2 macrophages.
Based on a threshold set at >2-fold change and
P<0.05 for the microarray data, a total of 1,253
differentially-expressed mRNAs were identified in M1 compared with
M2 macrophage samples, of which 696 mRNAs were upregulated and 557
mRNAs were downregulated. A volcano plot illustrated the expression
variance in the number of DEGs at different P-values and fold
changes (Fig. 1C). Independent
hierarchical clustering, visualized by a heat map (Fig. 1D), further confirmed that the
identified DEGs were significantly distinct between the M1 and M2
groups.
GO and pathway analyses of DEGs
To generate insights into the potential biological
functions of DEGs, functional enrichment analysis was performed
using GO and KEGG pathway terms and mapped in functional networks
using the Cytoscape plug-ins, BiNGO and CluePedia. GO identified
three categories: biological process, cellular component, and
molecular function. Through GO analysis, 34 and 40 GO terms were
significantly enriched for up- and downregulated DEGs,
respectively, based on the setting threshold of P<0.05 and false
discovery rate (FDR) <0.05 (Table
II). The main GO categories were: Protein binding, regulation
of biological process, response to stimulus, metabolic process and
cell differentiation (Fig. 2).
Moreover, 15 and four pathways were significantly enriched for up
and downregulated DEGs, respectively, which could be categorized
into 15 and four groups, respectively. The groups were classified
according to their different functions and the function details are
presented in Table III (left
column). Some of the groups shared similar genes. The main pathways
identified by KEGG were the HIF1 signaling pathway, TNF signaling
pathway, innate immune system, apoptosis and cytokine-cytokine
receptor interaction (Fig. 3).
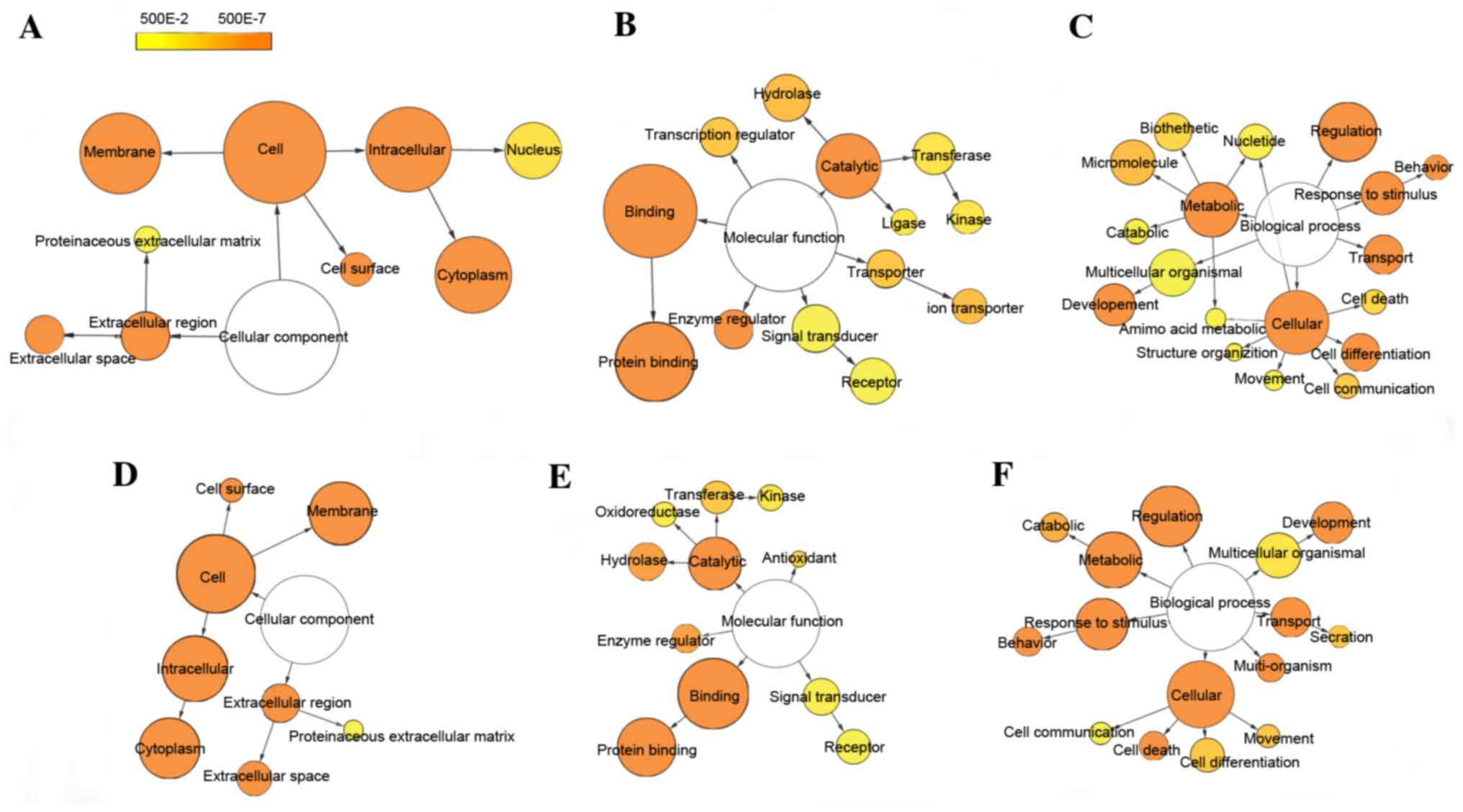 | Figure 2.Differentially-expressed gene GO-term
networks generated using BiNGO. Illustration of downregulated gene
GO enrichment categories (A) CC, (B) MF and (C) BP. Illustration of
upregulated gene GO enrichment categories (D) CC, (E) MF and (F)
BP. Circle size represents GO hierarchy; the larger area of the
circle, the higher hierarchy of the GO-term. Yellow shades
represent enrichment level; the deeper the shade, the more
significant the enrichment level. The threshold of hypergeometric
distribution of the functional annotation was set at P<0.05 and
FDR<0.05. GO, gene ontology; BiNGO, Biological Network Gene
Ontology; FDR, false discovery rate; CC, cellular component; MF,
molecular function; BP, biological process. |
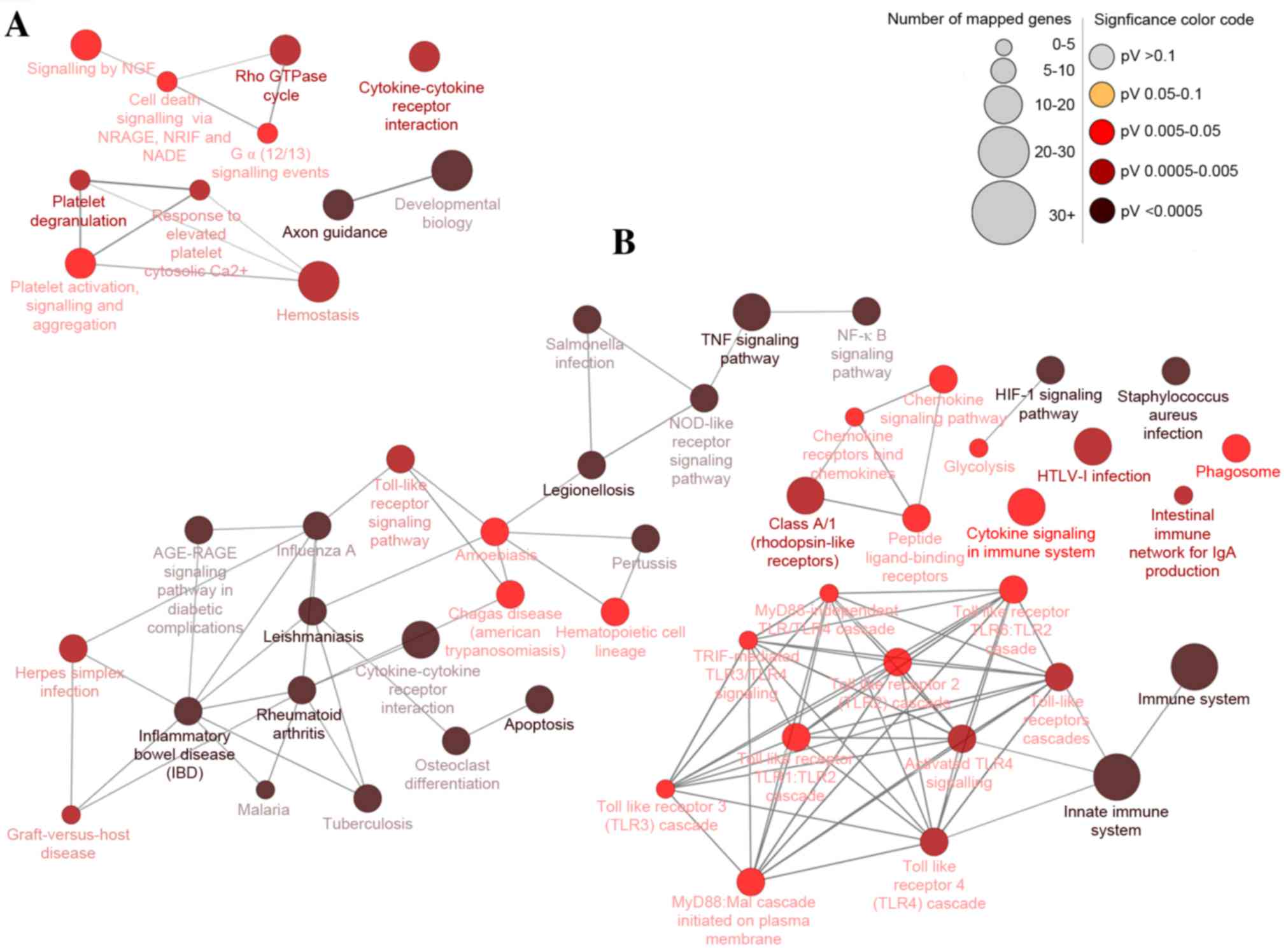 | Figure 3.Differentially-expressed gene pathway
network generated using CluePedia. Interaction pathway networks for
the identified (A) downregulated and (B) upregulated genes. The
size of the circle indicates the number of genes involved in the
pathway, and the color of the circle represents the P-value. The
threshold for the analysis was set at P<0.05 and FDR<0.05.
FDR, false discovery rate; NGF, nerve growth factor; NRAGE, MAGE
family member D1; NRIF, neurotrophin receptor interacting factor;
NADE, NAD synthetase; TNF, tumor necrosis factor; NFκB, nuclear
factor κB; NOD, atrophin 1; RAGE, receptor for advanced glycation
end products; HIF1, hypoxia-inducible factor 1; HTLV-I, human
T-lymphotropic virus I; MyD88, myeloid differentiation primary
response gene 88; TRIF, toll-like receptor adaptor molecule 2. |
 | Table II.Functional annotation of
differentially-expressed genes via GO enrichment. |
Table II.
Functional annotation of
differentially-expressed genes via GO enrichment.
| GO identifier | Description | Corrected
P-value | Gene count |
|---|
| Upregulated
genes |
|
|
|
|
50896 | Response to
stimulus | 3.55E-35 | 133 |
|
5623 | Cell | 3.29E-29 | 345 |
|
5488 | Binding | 6.81E-27 | 277 |
|
5515 | Protein
binding | 1.23E-24 | 180 |
|
9987 | Cellular
process | 2.79E-22 | 242 |
|
16020 | Membrane | 1.11E-20 | 210 |
|
50789 | Regulation of
biological process | 5.12E-20 | 195 |
|
5615 | Extracellular
space | 9.41E-19 | 46 |
|
5737 | Cytoplasm | 1.07E-17 | 190 |
|
5622 | Intracellular | 6.92E-14 | 233 |
|
3824 | Catalytic
activity | 1.02E-12 | 139 |
|
51704 | Multi-organism
process | 2.10E-12 | 30 |
|
5576 | Extracellular
region | 1.27E-11 | 66 |
|
8219 | Cell death | 1.42E-09 | 31 |
|
8152 | Metabolic
process | 1.69E-09 | 159 |
|
7610 | Behavior | 7.73E-09 | 28 |
|
7275 | Multicellular
organismal development | 8.00E-08 | 79 |
|
6810 | Transport | 1.65E-07 | 71 |
|
9986 | Cell surface | 5.58E-07 | 20 |
|
30234 | Enzyme regulator
activity | 1.98E-06 | 29 |
|
16787 | Hydrolase
activity | 2.57E-06 | 62 |
|
9056 | Catabolic
process | 8.94E-06 | 32 |
|
6928 | Cellular component
movement | 1.30E-04 | 18 |
|
30154 | Cell
differentiation | 1.47E-04 | 48 |
|
46903 | Secretion | 1.49E-04 | 14 |
|
16740 | Transferase
activity | 1.96E-04 | 46 |
|
16209 | Antioxidant
activity | 6.51E-04 | 5 |
|
32501 | Multicellular
organismal process | 2.61E-03 | 96 |
|
16301 | Kinase
activity | 2.78E-03 | 24 |
|
16491 | Oxidoreductase
activity | 4.81E-03 | 21 |
|
4871 | Signal transducer
activity | 8.77E-03 | 61 |
|
5578 | Proteinaceous
extracellular matrix | 2.39E-02 | 10 |
|
4872 | Receptor
activity | 3.77E-02 | 53 |
|
7154 | Cell
communication | 4.28E-02 | 15 |
| Downregulated
genes |
|
|
|
|
5623 | Cell | 3.0026E-32 | 328 |
|
5488 | Binding | 6.1503E-31 | 268 |
|
5515 | Protein
binding | 7.6309E-31 | 182 |
|
50789 | Regulation of
biological process | 9.9221E-20 | 183 |
|
16020 | Membrane | 2.2576E-19 | 194 |
|
9987 | Cellular
process | 3.2737E-19 | 219 |
|
5737 | Cytoplasm | 7.2487E-15 | 171 |
|
50896 | Response to
stimulus | 1.2201E-13 | 87 |
|
5622 | Intracellular | 2.213E-13 | 216 |
|
7275 | Multicellular
organismal development | 2.9385E-11 | 84 |
|
8152 | Metabolic
process | 2.0536E-10 | 152 |
|
30154 | Cell
differentiation | 2.1508E-10 | 61 |
|
5576 | Extracellular
region | 4.247E-09 | 57 |
|
30234 | Enzyme regulator
activity | 1.0124E-08 | 32 |
|
5615 | Extracellular
space | 2.4893E-08 | 29 |
|
3824 | Catalytic
activity | 6.7488E-08 | 115 |
|
6810 | Transport | 1.433E-07 | 67 |
|
9986 | Cell surface | 1.4591E-07 | 20 |
|
32501 | Multicellular
organismal process | 1.8029E-07 | 108 |
|
7610 | Behavior | 4.5276E-06 | 22 |
|
43170 | Macromolecule
metabolic process | 0.00004114 | 96 |
|
15075 | Ion transmembrane
transporter activity | 4.7081E-05 | 24 |
|
16787 | Hydrolase
activity | 4.9943E-05 | 54 |
|
7154 | Cell
communication | 0.00010978 | 21 |
|
30528 | Transcription
regulator activity | 0.00013308 | 33 |
|
5215 | Transporter
activity | 0.00026927 | 30 |
|
8219 | Cell death | 0.00052849 | 19 |
|
9058 | Biosynthetic
process | 0.00075689 | 62 |
|
5634 | Nucleus | 0.0020871 | 83 |
|
16740 | Transferase
activity | 0.0025576 | 39 |
|
6519 | Cellular amino acid
and derivative metabolic process | 0.00291 | 12 |
|
16301 | Kinase
activity | 0.0081283 | 21 |
|
9056 | Catabolic
process | 0.0081283 | 22 |
|
6139 | Nucleobase | 0.010583 | 55 |
|
5578 | Proteinaceous
extracellular matrix | 0.012461 | 10 |
|
43062 | Extracellular
structure organization | 0.013556 | 7 |
|
4871 | Signal transducer
activity | 0.01531 | 55 |
|
6928 | Cellular component
movement | 0.017115 | 12 |
|
4872 | Receptor
activity | 0.034186 | 49 |
|
16874 | Ligase
activity | 0.046244 | 10 |
 | Table III.Functional annotation of
differentially-expressed genes via KEGG Enrichment. |
Table III.
Functional annotation of
differentially-expressed genes via KEGG Enrichment.
| Function | Groups | Gene count |
|---|
| Upregulated
genes |
|
|
|
Apoptosis | Group 9 | 29 |
| Class
A/1 (Rhodopsin-like receptors) | Group 8 | 30 |
|
Cytokine Signaling in immune
system | None 4 | 21 |
| HIF1
signaling pathway | Group 5 | 17 |
| HTLV-I
infection | None 3 | 22 |
| Immune
system | Group 6 | 62 |
|
Inflammatory bowel disease
(IBD) | Group 4 | 67 |
| Innate
immune system | Group 7 | 36 |
|
Intestinal immune network
for | None 1 | 8 |
| IgA
production |
|
|
|
Legionellosis | Group 3 | 41 |
|
Leishmaniasis | Group 1 | 42 |
|
Phagosome | None 0 | 15 |
|
Rheumatoid arthritis | Group 2 | 32 |
|
Staphylococcus aureus
infection | None 2 | 12 |
| TNF
signaling pathway | Group 0 | 43 |
| Downregulated
genes |
|
|
| Axon
guidance | Group 1 | 24 |
|
Cytokine-cytokine receptor
interaction | None 0 | 18 |
|
Platelet degranulation | Group 0 | 24 |
| Rho
GTPase cycle | Group 2 | 22 |
Interaction network analysis
An interaction network was constructed using STRING
and then visualized using Cytoscape based on the macrophage
polarization-associated genes identified in the present study. The
network comprised 18 genes and 38 interactions (Fig. 4). The main type of gene
associations was co-occurrence. Among these, IL6, TNF, IL1β, nitric
oxide synthase 2 (NOS2) and SOCS3 were the key nodes, displaying
the highest connectivity within the network (Fig. 4).
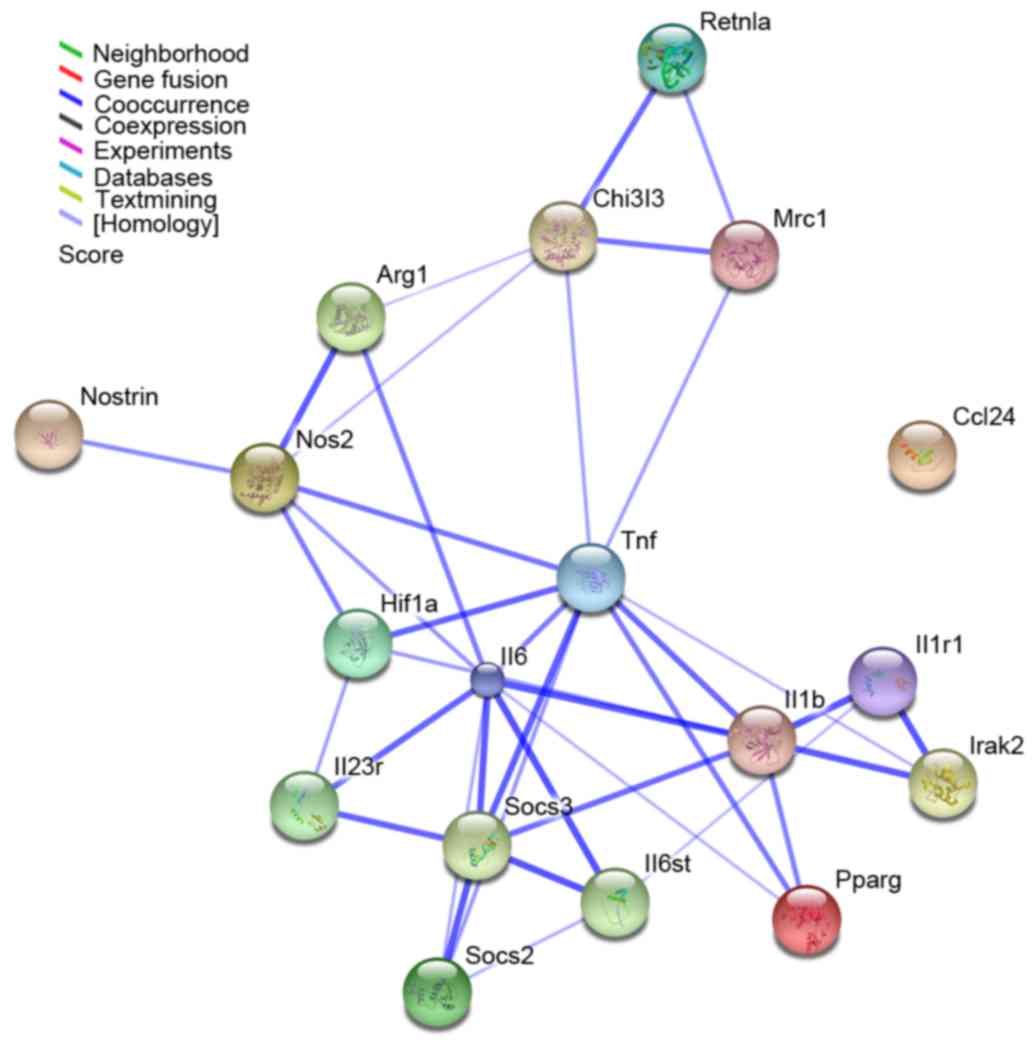 | Figure 4.Interaction network of 18 macrophage
polarization-associated genes as identified by STRING analysis. The
results were expanded to the current network by setting the
required confidence score to 0.400. The nodes represent the genes,
whereas the lines represent interactions between genes. The color
of the line denotes the basis of the predicted interaction
according to the software database. STRING, Search Tool for the
Retrieval of Interacting Genes; Retnla, resistin-like α; Chi3l3,
chitinase 3-like 3; Mrc1, mannose receptor C-type 1; Arg1, arginase
1; Nostrin, nitric oxide synthase trafficker; Nos2, nitric oxide
synthase 2; Hif1, hypoxia-inducible factor 1; Tnf, tumor necrosis
factor; Ccl24, C-C motif chemokine ligand 24; Il, interleukin;
Socs, suppressor of cytokine signaling; Irak2, interleukin 1
receptor associated kinase 2; Pparg, peroxisome
proliferator-activated receptor. |
Discussion
Macrophages, as major innate immune and antigen
presenting cells, are important in infection resistance and
tumorigenesis. Macrophages activated by TLR ligands, such as LPS or
IFN-γ, are called M1 macrophages. In contrast, stimulation of
macrophages with T helper cells type 2 cytokines, such as IL-4 or
IL-13, induces the generation of M2-type macrophages. Treatment of
bone marrow cells with granulocyte-macrophage colony-stimulating
factor (CSF) and macrophage CSF, leads to the generation of M1 and
M2 macrophages, respectively (31). Appropriately activated macrophages
eliminate pathogens and tumors, whereas, activation with
inappropriate stimuli may suppress the immune system, resulting in
tumorigenesis and chronic infections. As the primary cells that
secrete inflammatory cytokines, macrophages (particularly M2-type)
directly mediate the development of inflammatory autoimmune
diseases, tissue damage and inflammatory infiltration in
hypersensitivity reactions (32–35).
Macrophage polarization has been a topic of intense
interest in macrophage research. Early studies identified a number
of genes involved in macrophage polarization. For example, previous
studies have demonstrated that the JMJD3-interferon regulatory
factor (Irf) 4 axis regulates M2 macrophage polarization and host
responses against helminth infections (21). SOCS2 and SOCS3 diametrically
control macrophage polarization (22). Formyl peptide receptor (FPR) 2
promotes antitumor host defense by limiting M2 polarization of
macrophages (36). IRF5 and IRF8
promote M1 macrophage polarization (14,15),
while KLF4 is involved in M2 macrophage polarization (16). Akt1 and Akt2 protein kinases
differentially contribute to macrophage polarization (17). However, although several genes
associated with macrophage polarization have been identified, the
interaction among genes and the mechanism of this constellation of
genes in the response of macrophages to polarizing conditions
remain elusive.
The accessibility of microarray data and gene
profiling has facilitated a better understanding of the underlying
mechanisms of complex biological processes and responses. In the
present study, mRNA-based microarray methods were employed to
analyze RNA samples from ex vivo programmed M1 and M2
macrophages isolated from BALB/c mice. Bioinformatics analysis
identified a total of 1,253 DEGs in M1 macrophages, including 696
upregulated genes and 557 downregulated genes relative to M2
macrophages. Previous studies have examined the gene expression
profiles of M1 and M2 macrophages derived from C57BL/6J mice and
from human blood samples (37,38).
In the present microarray study, all 8 genes corresponding to
canonical M1 markers (NOS2, IL23 receptor, SOCS3, IL-1β, IL-6, TNF,
interleukin 1 receptor associated kinase 2 and HIF1a) and the M1
markers CD38, G-protein coupled receptor (Gpr)18 and Fpr2,
identified in C57BL/6 murine macrophages (37), were demonstrated to be upregulated
in M1 compared with M2 macrophages (Table I). In addition, 10 genes
corresponding to canonical M2 markers (including Retnla, Chi313,
MRC1, ARG1 and PPARG), and the M2 markers early growth response 2
and c-myc identified in C57BL/6 murine macrophages (37), were demonstrated to be up-regulated
in M2 compared with M1 macrophages in the present study (Table I). These data validate the
robustness of the microarray results presented in the current
study.
A better understanding of the gene functions and
molecular pathways associated with different macrophage subtypes is
necessary for further progress in the macrophage field. In the
present study, a gene expression analysis of M1 and M2 macrophages
derived from BALB/c mice was performed. The bioinformatics analysis
demonstrated that, for the upregulated genes, GO functional
analysis identified 34 enriched terms, including eight cellular
components, 11 molecular functions and 15 biological process terms.
Biological process terms comprised of response to stimulus, cell
differentiation and regulation of biological process. KEGG
functional analysis identified 15 enriched terms, which included
apoptosis, cytokine signaling in immune system, HIF1 signaling
pathway, innate immune system, and TNF signaling pathway. For the
downregulated genes, GO functional analysis identified 40 enriched
terms, which consisted of nine cellular components, 13 molecular
functions and 18 biological process terms. KEGG functional analysis
identified four enriched terms, namely, axon guidance,
cytokine-cytokine receptor interaction, platelet degranulation and
Rho GTPase cycle. Interaction network analysis of the screened
DEGs, generated by STRING, indicated that genes including TNF,
IL-6, IL-1β, SOCS3, NOS2 and HIF1a may serve key roles in
macrophage polarization.
In summary, the current study identified 1,253 DEGs
and analyzed their functions through GO and KEGG pathway enrichment
analyses. Subsequently, an interaction network was constructed to
analyze the overlapping DEGs with known genes associated with
macrophage polarization. The present study may thus provide novel
insights into the role of genes in macrophage differentiation and
polarization. Further experimental studies will be needed in the
future in order to confirm these findings and further explore the
molecular mechanisms of macrophage polarization.
Acknowledgements
The National Natural Science Foundation of China
(grant nos. 81300172, 81301497 and 81472017), Natural Science
Foundation of Anhui Province (grant no. 1408085QH148), Key projects
of Natural Science Research of universities in Anhui Province
(grant no. KJ2016A721) and Program for Excellent Young Talents in
College and University of Anhui Province supported the present
study.
References
|
1
|
Liu G and Abraham E: MicroRNAs in immune
response and macrophage polarization. Arterioscler Thromb Vasc
Biol. 33:170–177. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tugal D, Liao X and Jain MK: Transcription
control of macrophage polarization. Arterioscler Thromb Vasc Biol.
33:1135–1144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stefater JA III, Ren S, Lang RA and
Duffield JS: Metchnikoff's policemen: Macrophages in development,
homeostasis and regeneration. Trends Mol Med. 17:743–752. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lawrence T and Natoli G: Transcriptional
regulation of macrophage polarization: Enabling diversity with
identity. Nat Rev Immunol. 11:750–761. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Murray PJ and Wynn TA: Obstacles and
opportunities for understanding macrophage polarization. J Leukoc
Biol. 89:557–563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Zhang M, Zhong M, Suo Q and Lv K:
Expression profiles of miRNAs in polarized macrophages. Int J Mol
Med. 31:797–802. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mosser DM: The many faces of macrophage
activation. J Leukoc Biol. 73:209–212. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Murray PJ, Allen JE, Biswas SK, Fisher EA,
Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence
T, et al: Macrophage activation and polarization: Nomenclature and
experimental guidelines. Immunity. 41:14–20. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bronte V and Zanovello P: Regulation of
immune responses by L-arginine metabolism. Nat Rev Immunol.
5:641–654. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nair MG, Gallagher IJ, Taylor MD, Loke P,
Coulson PS, Wilson RA, Maizels RM and Allen JE: Chitinase and Fizz
family members are a generalized feature of nematode infection with
selective upregulation of Ym1 and Fizz1 by antigen-presenting
cells. Infect Immun. 73:385–394. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stein M, Keshav S, Harris N and Gordon S:
Interleukin 4 potently enhances murine macrophage mannose receptor
activity: A marker of alternative immunologic macrophage
activation. J Exp Med. 176:287–292. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mantovani A, Sica A, Sozzani S, Allavena
P, Vecchi A and Locati M: The chemokine system in diverse forms of
macrophage activation and polarization. Trends Immunol. 25:677–686.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang N, Liang H and Zen K: Molecular
mechanisms that influence the macrophage m1-m2 polarization
balance. Front Immunol. 5:6142014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Krausgruber T, Blazek K, Smallie T,
Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M and Udalova
IA: IRF5 promotes inflammatory macrophage polarization and TH1-TH17
responses. Nat Immunol. 12:231–238. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu H, Zhu J, Smith S, Foldi J, Zhao B,
Chung AY, Outtz H, Kitajewski J, Shi C, Weber S, et al: Notch-RBP-J
signaling regulates the transcription factor IRF8 to promote
inflammatory macrophage polarization. Nat Immunol. 13:642–650.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liao X, Sharma N, Kapadia F, Zhou G, Lu Y,
Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, et
al: Krüppel-like factor 4 regulates macrophage polarization. J Clin
Invest. 121:2736–2749. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arranz A, Doxaki C, Vergadi E, de la Torre
Martinez Y, Vaporidi K, Lagoudaki ED, Ieronymaki E, Androulidaki A,
Venihaki M, Margioris AN, et al: Akt1 and Akt2 protein kinases
differentially contribute to macrophage polarization. Proc Natl
Acad Sci USA. 109:9517–9522. 2012; View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Zhang M, Li X, Tang Z, Wang X,
Zhong M, Suo Q, Zhang Y and Lv K: Silencing microRNA-155 attenuates
cardiac injury and dysfunction in viral myocarditis via promotion
of M2 phenotype polarization of macrophages. Sci Rep. 6:226132016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takeuch O and Akira S: Epigenetic control
of macrophage polarization. Eur J Immunol. 41:2490–2493. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mullican SE, Gaddis CA, Alenghat T, Nair
MG, Giacomin PR, Everett LJ, Feng D, Steger DJ, Schug J, Artis D
and Lazar MA: Histone deacetylase 3 is an epigenomic brake in
macrophage alternative activation. Genes Dev. 25:2480–2488. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Satoh T, Takeuchi O, Vandenbon A, Yasuda
K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh
T, et al: The Jmjd3-Irf4 axis regulates M2 macrophage polarization
and host responses against helminth infection. Nat Immunol.
11:936–944. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Spence S, Fitzsimons A, Boyd CR, Kessler
J, Fitzgerald D, Elliott J, Gabhann JN, Smith S, Sica A, Hams E, et
al: Suppressors of cytokine signaling 2 and 3 diametrically control
macrophage polarization. Immunity. 38:66–78. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu C, Fei HD, Sun ZY and Tian JW:
Bioinformatic analysis of the microarray gene expression profile in
degenerative intervertebral disc cells exposed to TNF-α. Eur Rev
Med Pharmacol Sci. 19:3332–3339. 2015.PubMed/NCBI
|
|
24
|
Zhao L, Zhang J, Tan H, Wang W, Liu Y,
Song R and Wang L: Gene function analysis in osteosarcoma based on
microarray gene expression profiling. Int J Clin Exp Med.
8:10401–10410. 2015.PubMed/NCBI
|
|
25
|
Zhang Y, Zhang Y, Li X, Zhang M and Lv K:
Microarray analysis of circular RNA expression patterns in
polarized macrophages. Int J Mol Med. 39:373–379. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Boltz-Nitulescu G, Wiltschke C, Holzinger
C, Fellinger A, Scheiner O, Gessl A and Förster O: Differentiation
of rat bone marrow cells into macrophages under the influence of
mouse L929 cell supernatant. J Leukoc Biol. 41:83–91.
1987.PubMed/NCBI
|
|
27
|
Maere S, Heymans K and Kuiper M: BiNGO: A
Cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bindea G, Galon J and Mlecnik B: CluePedia
Cytoscape plugin: Pathway insights using integrated experimental
and in silico data. Bioinformatics. 29:661–663. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barsky A, Gardy JL, Hancock RE and Munzner
T: Cerebral: A Cytoscape plugin for layout of and interaction with
biological networks using subcellular localization annotation.
Bioinformatics. 23:1040–1042. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39(Database Issue): D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Banerjee S, Cui H, Xie N, Tan Z, Yang S,
Icyuz M, Thannickal VJ, Abraham E and Liu G: miR-125a-5p regulates
differential activation of macrophages and inflammation. J Biol
Chem. 288:35428–35436. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li K, Xu W, Guo Q, Jiang Z, Wang P, Yue Y
and Xiong S: Differential macrophage polarization in male and
female BALB/c mice infected with coxsackievirus B3 defines
susceptibility to viral myocarditis. Circ Res. 105:353–364. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chacón-Salinas R, Serafín-López J,
Ramos-Payán R, Méndez-Aragón P, Hernández-Pando R, Van-Soolingen D,
Flores-Romo L, Estrada-Parra S and Estrada-García I: Differential
pattern of cytokine expression by macrophages infected in vitro
with different Mycobacterium tuberculosis genotypes. Clin Exp
Immunol. 140:443–449. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Verreck FA, de Boer T, Langenberg DM, van
der Zanden L and Ottenhoff TH: Phenotypic and functional profiling
of human proinflammatory type-1 and anti-inflammatory type-2
macrophages in response to microbial antigens and IFN-gamma and
CD40L-mediated costimulation. J Leukoc Biol. 79:285–293. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang W, Xu W and Xiong S: Blockade of
Notch1 signaling alleviates murine lupus via blunting macrophage
activation and M2b polarization. J Immunol. 184:6465–6478. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Y, Chen K, Wang C, Gong W, Yoshimura
T, Liu M and Wang JM: Cell surface receptor FPR2 promotes antitumor
host defense by limiting M2 polarization of macrophages. Cancer
Res. 73:550–560. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jablonski KA, Amici SA, Webb LM,
Ruiz-Rosado Jde D, Popovich PG, Partida-Sanchez S and
Guerau-de-Arellano M: Novel markers to delineate murine M1 and M2
macrophages. PLoS One. 10:e01453422015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Martinez FO, Gordon S, Locati M and
Mantovani A: Transcriptional profiling of the human monocyte-to
macrophage differentiation and polarization: New molecules and
patterns of gene expression. J Immunol. 177:7303–7311. 2006.
View Article : Google Scholar : PubMed/NCBI
|