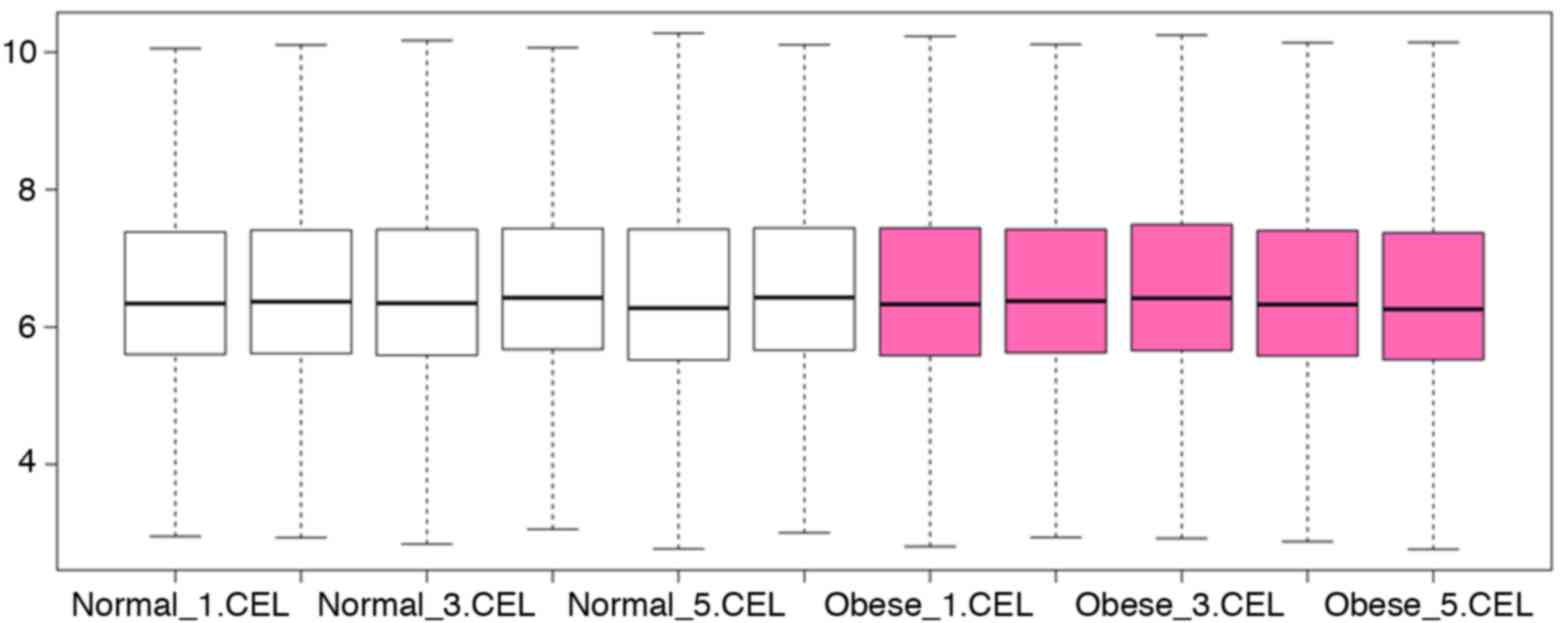
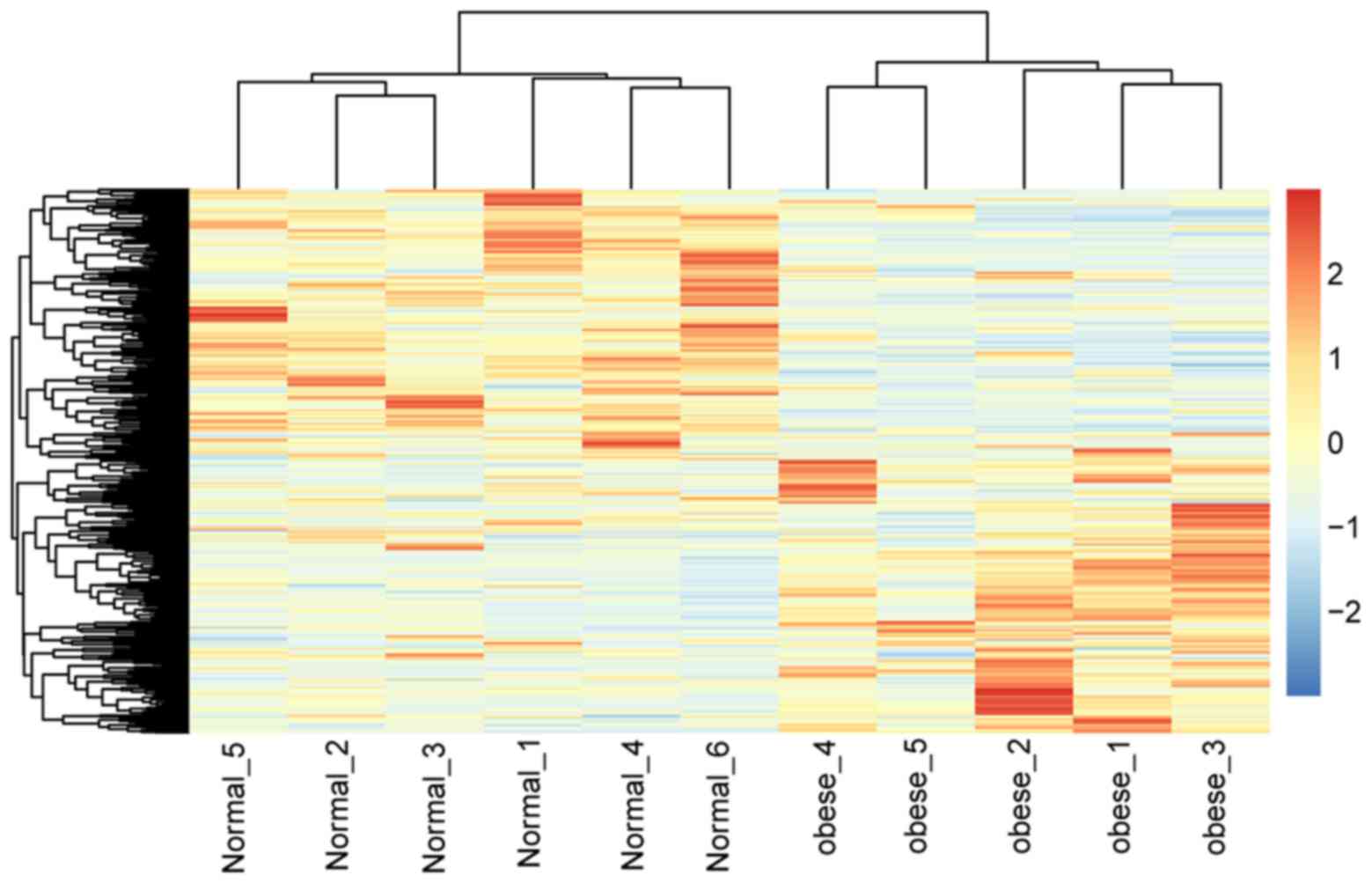
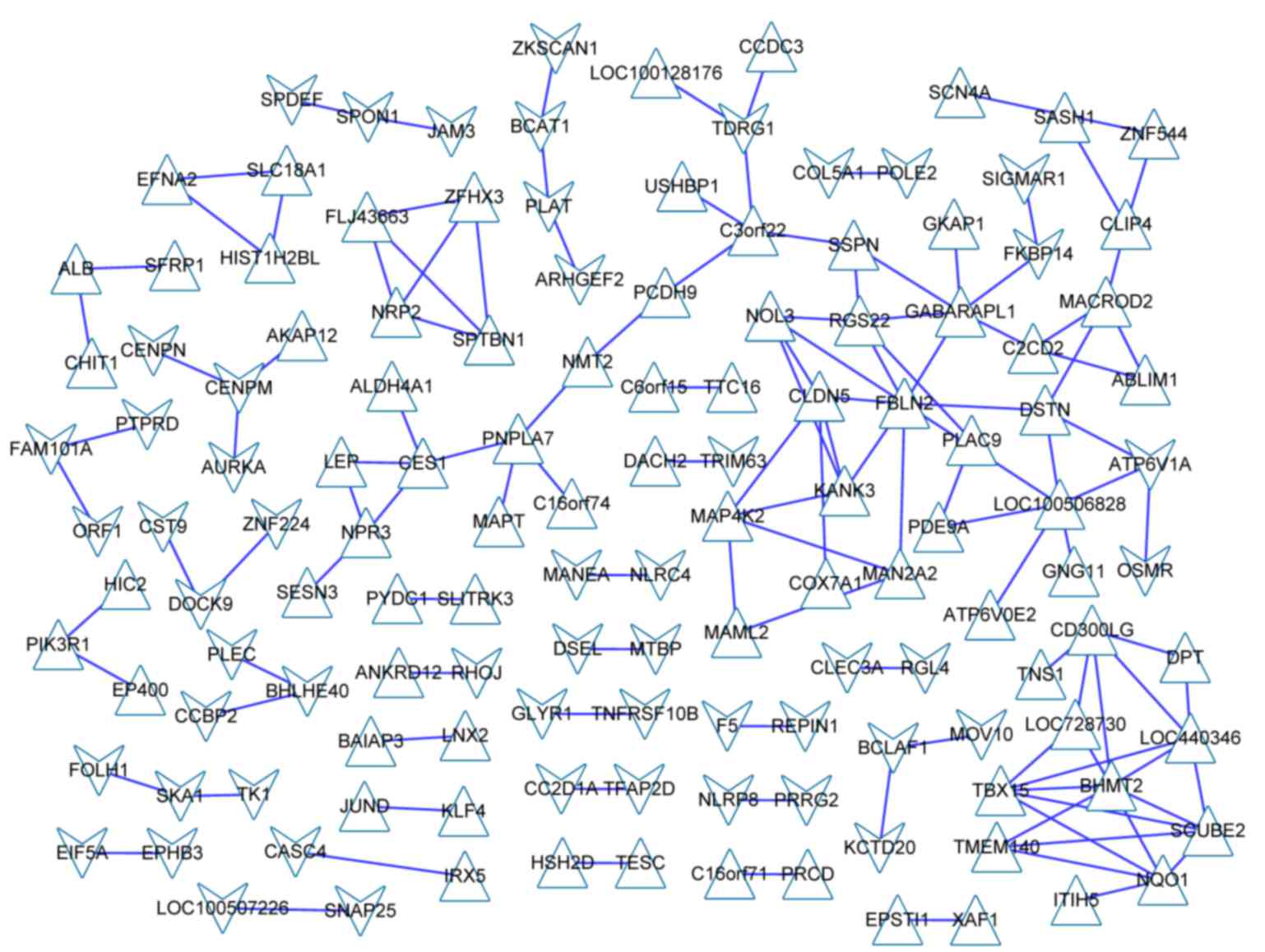
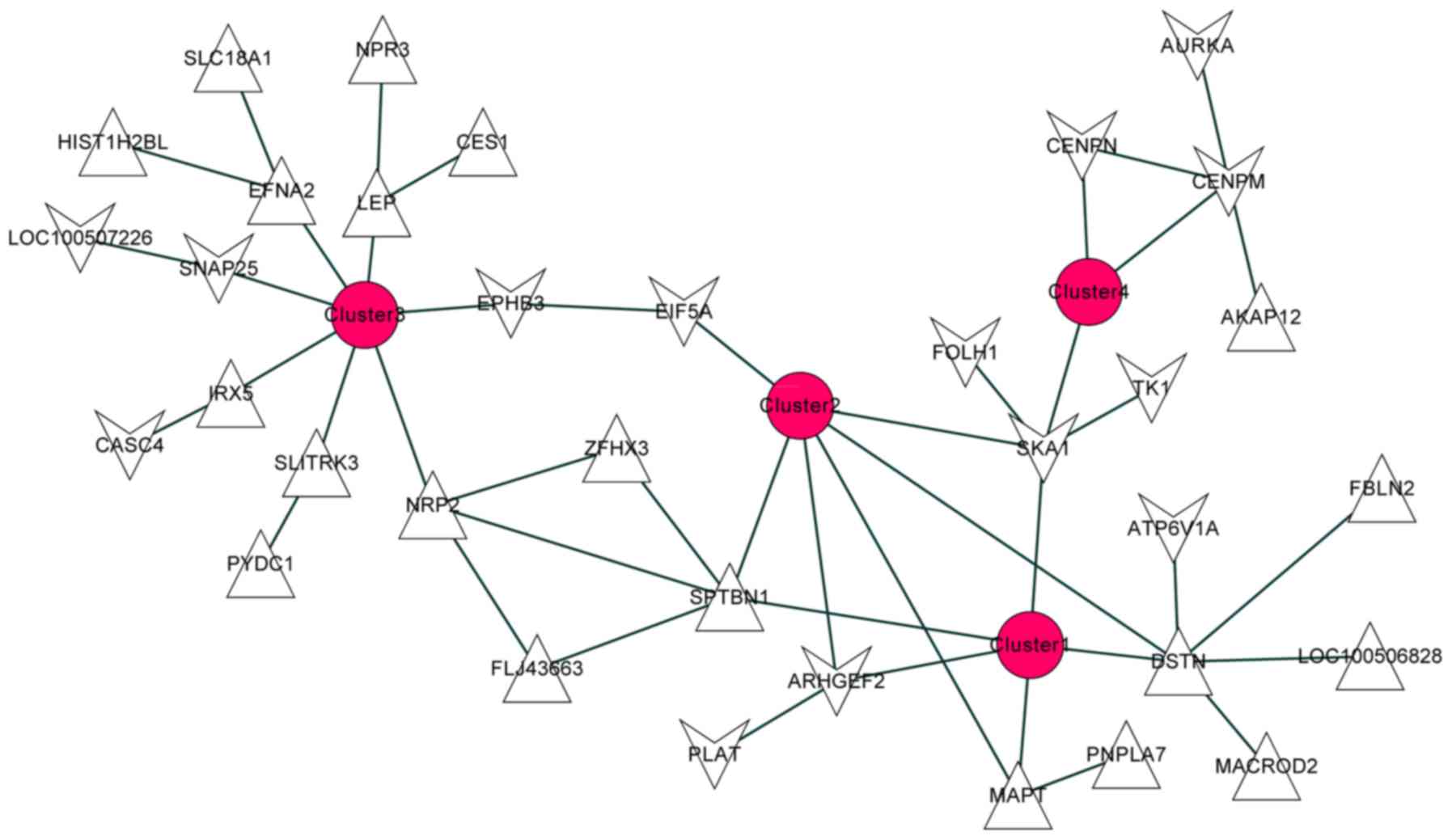
|
1
|
Etelson D, Brand DA, Patrick PA and
Shirali A: Childhood obesity: Do parents recognize this health
risk? Obes Res. 11:1362–1368. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dietz WH: Health consequences of obesity
in youth: Childhood predictors of adult disease. Pediatrics.
101:518–525. 1998.PubMed/NCBI
|
|
3
|
Dehghan M, Akhtar-Danesh N and Merchant
AT: Childhood obesity, prevalence and prevention. Nutr J. 4:242005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Daniels SR: The consequences of childhood
overweight and obesity. Future Child. 16:47–67. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
French SA, Story M and Perry CL:
Self-esteem and obesity in children and adolescents: A literature
review. Obes Res. 3:479–490. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han JC, Lawlor DA and Kimm SY: Childhood
obesity. Lancet. 375:1737–1748. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Govaerts C, Srinivasan S, Shapiro A, Zhang
S, Picard F, Clement K, Lubrano-Berthelier C and Vaisse C:
Obesity-associated mutations in the melanocortin 4 receptor provide
novel insights into its function. Peptides. 26:1909–1919. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tao YX and Segaloff DL: Functional
characterization of melanocortin-4 receptor mutations associated
with childhood obesity. Endocrinology. 144:4544–4551. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yeo GS, Farooqi IS, Aminian S, Halsall DJ,
Stanhope RG and O'Rahilly S: A frameshift mutation in MC4R
associated with dominantly inherited human obesity. Nat Gene.
20:111–112. 1998. View
Article : Google Scholar
|
|
10
|
Del Giudice Miraglia E, Cirillo G, Nigro
V, Santoro N, D'urso L, Raimondo P, Cozzolino D, Scafato D and
Perrone L: Low frequency of melanocortin-4 receptor (MC4R)
mutations in a Mediterranean population with early-onset obesity.
Int J Obes Relat Metab Disord. 26:647–651. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Challis BG, Pritchard LE, Creemers JW,
Delplanque J, Keogh JM, Luan J, Wareham NJ, Yeo GS, Bhattacharyya
S, Froguel P, et al: A missense mutation disrupting a dibasic
prohormone processing site in pro-opiomelanocortin (POMC) increases
susceptibility to early-onset obesity through a novel molecular
mechanism. Hum Mol Genet. 11:1997–2004. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Santoro N, Perrone L, Cirillo G, Raimondo
P, Amato A, Coppola F, Santarpia M, D'Aniello A and Del Giudice
Miraglia E: Weight loss in obese children carrying the
proopiomelanocortin R236G variant. J Endocrinol Invest. 29:226–230.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hung CN, Poon WT, Lee CY, Law CY and Chan
AY: A case of early-onset obesity, hypocortisolism, and skin
pigmentation problem due to a novel homozygous mutation in the
proopiomelanocortin (POMC) gene in an Indian boy. J Pediatr
Endocrinol Metab. 25:175–179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wardle J, Carnell S, Haworth CM, Farooqi
IS, O'Rahilly S and Plomin R: Obesity associated genetic variation
in FTO is associated with diminished satiety. J Clin Endocrinol
Metab. 93:3640–3643. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gulati P, Cheung MK, Antrobus R, Church
CD, Harding HP, Tung YC, Rimmington D, Ma M, Ron D, Lehner PJ, et
al: Role for the obesity-related FTO gene in the cellular sensing
of amino acids. Proc Natl Acad Sci USA. 110:2557–2562. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu WM, Mei R, Di X, Ryder TB, Hubbell E,
Dee S, Webster TA, Harrington CA, Ho MH, Baid J, et al: Analysis of
high density expression microarrays with signed-rank call
algorithms. Bioinformatics. 18:1593–1599. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smyth GK and Speed T: Normalization of
cDNA microarray data. Methods. 31:265–273. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry
RA and Dudoit S: Springer; New York: pp. 397–420. 2005, View Article : Google Scholar
|
|
19
|
Team RC: R: A Language and Environment for
Statistical ComputingR Foundation for Statistical Computing.
Vienna: 2012
|
|
20
|
Deza MM and Deza E: Encyclopedia of
Distances. 1st edition. Springer-Verlag; Berlin: 2009, View Article : Google Scholar
|
|
21
|
Obayashi T, Kinoshita K, Nakai K, Shibaoka
M, Hayashi S, Saeki M, Shibata D, Saito K and Ohta H: ATTED-II: A
database of co-expressed genes and cis elements for identifying
co-regulated gene groups in Arabidopsis. Nucleic Acids Res.
35:(Database Issue). D863–D869. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fröhlich H, Speer N, Poustka A and
Beissbarth T: GOSim-an R-package for computation of information
theoretic GO similarities between terms and gene products. BMC
Bioinformatics. 8:1662007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Q, Dou S, Ji Z and Xue Q: Synonymous
codon usage and gene function are strongly related in Oryza sativa.
Bio Systems. 80:123–131. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
McInerney JO: GCUA: General codon usage
analysis. Bioinformatics. 14:372–373. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rice P, Longden I and Bleasby A: EMBOSS:
The European molecular biology open software suite. Trends Genet.
16:276–277. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ebbeling CB, Pawlak DB and Ludwig DS:
Childhood obesity: Public-health crisis, common sense cure. Lancet.
360:473–482. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Welburn JP, Grishchuk EL, Backer CB,
Wilson-Kubalek EM, Yates JR III and Cheeseman IM: The human
kinetochore Ska1 complex facilitates microtubule
depolymerization-coupled motility. Dev Cell. 16:374–385. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Welburn JP, Grishchuk EL, Backer CB,
Wilson-Kubalek EM, Yates JR III and Cheeseman IM: The human
kinetochore Ska1 complex facilitates microtubule
depolymerization-coupled motility. Dev Cell. 16:374–385. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schmidt JC, Arthanari H, Boeszoermenyi A,
Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M,
Milligan RA, Bathe M, et al: The kinetochore-bound Ska1 complex
tracks depolymerizing microtubules and binds to curved
protofilaments. Dev Cell. 23:968–980. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bochukova EG, Huang N, Keogh J, Henning E,
Purmann C, Blaszczyk K, Saeed S, Hamilton-Shield J, Clayton-Smith
J, O'Rahilly S, et al: Large, rare chromosomal deletions associated
with severe early-onset obesity. Nature. 463:666–670. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Farooqi IS: Genetic and hereditary aspects
of childhood obesity. Best Pract Res Clin Endocrinol Metab.
19:359–374. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Walters RG, Jacquemont S, Valsesia A, de
Smith AJ, Martinet D, Andersson J, Falchi M, Chen F, Andrieux J,
Lobbens S, et al: A new highly penetrant form of obesity due to
deletions on chromosome 16p11.2. Nature. 463:671–675. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bachmann-Gagescu R, Mefford HC, Cowan C,
Glew GM, Hing AV, Wallace S, Bader PI, Hamati A, Reitnauer PJ,
Smith R, et al: Recurrent 200-kb deletions of 16p11. 2 that include
the SH2B1 gene are associated with developmental delay and obesity.
Genet Med. 12:641–647. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Siesser PF, Motolese M, Walker MP,
Goldfarb D, Gewain K, Yan F, Kulikauskas RM, Chien AJ, Wordeman L
and Major MB: FAM123A binds to microtubules and inhibits the
guanine nucleotide exchange factor ARHGEF2 to decrease actomyosin
contractility. Sci Signal. 5:ra642012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brajenovic M, Joberty G, Küster B,
Bouwmeester T and Drewes G: Comprehensive proteomic analysis of
human Par protein complexes reveals an interconnected protein
network. J Biol Chem. 279:12804–12811. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Poroyko V and Birukova A: ARHGEF2 (rho/rac
guanine nucleotide exchange factor (GEF) 2). Atlas Genet Cytogenet
Oncol Haematol. 4:582007.
|
|
38
|
Conde C and Cáceres A: Microtubule
assembly, organization and dynamics in axons and dendrites. Nat Rev
Neurosci. 10:319–332. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kuznetsov SA, Rodionov VI, Gelfand VI and
Rosenblat VA: Microtubule-associated protein MAP1 promotes
microtubule assembly in vitro. FEBS Lett. 135:241–244. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Emoto M, Langille SE and Czech MP: A role
for kinesin in insulin-stimulated GLUT4 glucose transporter
translocation in 3T3-L1 adipocytes. J Biol Chem. 276:10677–10682.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Patki V, Buxton J, Chawla A, Lifshitz L,
Fogarty K, Carrington W, Tuft R and Corvera S: Insulin action on
GLUT4 traffic visualized in single 3T3-l1 adipocytes by using
ultra-fast microscopy. Mol Biol Cell. 12:129–141. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kanzaki M and Pessin JE:
Insulin-stimulated GLUT4 translocation in adipocytes is dependent
upon cortical actin remodeling. J Biol Chem. 276:42436–42444. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hillen N, Mester G, Lemmel C, Weinzierl
AO, Müller M, Wernet D, Hennenlotter J, Stenzl A, Rammensee HG and
Stevanović S: Essential differences in ligand presentation and T
cell epitope recognition among HLA molecules of the HLA-B44
supertype. Eur J Immunol. 38:2993–3003. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guilherme R, Guimiot F, Tabet AC,
Khung-Savatovsky S, Gauthier E, Nouchy M, Benzacken B, Verloes A,
Oury JF, Delezoide AL and Aboura A: Abnormal muscle development of
the diaphragm in a fetus with 2p14-p16 duplication. Am J Med Genet
A. 149A:2892–2897. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Aksu S, Koczan D, Renne U, Thiesen HJ and
Brockmann GA: Differentially expressed genes in adipose tissues of
high body weight-selected (obese) and unselected (lean) mouse
lines. J Appl Genet. 48:133–143. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Moriyama K, Yonezawa N, Sakai H, Yahara I
and Nishida E: Mutational analysis of an actin-binding site of
cofilin and characterization of chimeric proteins between cofilin
and destrin. J Biol Chem. 267:7240–7244. 1992.PubMed/NCBI
|
|
47
|
Yahara I, Aizawa H, Moriyama K, Iida K,
Yonezawa N, Nishida E, Hatanaka H and Inagaki F: A role of
cofilin/destrin in reorganization of actin cytoskeleton in response
to stresses and cell stimuli. Cell Struct Funct. 21:421–424. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Agrawal PB, Greenleaf RS, Tomczak KK,
Lehtokari VL, Wallgren-Pettersson C, Wallefeld W, Laing NG, Darras
BT, Maciver SK, Dormitzer PR and Beggs AH: Nemaline Myopathy with
minicores caused by mutation of the CFL2 gene encoding the
skeletal muscle actin-binding protein, cofilin-2. Am J Hum Genet.
80:162–167. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
49
|
Verdoni AM, Aoyama N, Ikeda A and Ikeda S:
Effect of destrin mutations on the gene expression profile in vivo.
Physiol Genomics. 34:9–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Maciver SK and Hussey PJ: The ADF/cofilin
family: Actin-remodeling proteins. Genome Biol. 3:30072002.
View Article : Google Scholar
|
|
51
|
Hawkins M, Pope B, Maciver SK and Weeds
AG: Human actin depolymerizing factor mediates a pH-sensitive
destruction of actin filaments. Biochemistry. 32:9985–9993. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wiche G: Role of plectin in cytoskeleton
organization and dynamics. J Cell Sci. 111:2477–2486.
1998.PubMed/NCBI
|
|
53
|
Kawaguchi N, Sundberg C, Kveiborg M,
Moghadaszadeh B, Asmar M, Dietrich N, Thodeti CK, Nielsen FC,
Möller P, Mercurio AM, et al: ADAM12 induces actin cytoskeleton and
extracellular matrix reorganization during early adipocyte
differentiation by regulating beta1 integrin function. J Cell Sci.
116:3893–3904. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bost F, Aouadi M, Caron L and Binétruy B:
The role of MAPKs in adipocyte differentiation and obesity.
Biochimie. 87:51–56. 2005. View Article : Google Scholar : PubMed/NCBI
|