Introduction
Hepatocellular carcinoma (HCC) is the third most
common cancer-caused death in the worldwild (1). Despite significant therapeutic
improvement in clinical treatment, long-term therapeutic efficacy
of HCC patients remains poor (2),
mainly because of regional invasion and frequent metastasis.
However, mechanisms underlying HCC metastasis are not clearly
understand, thus it is urgent to identify key biochemical pathways
in metastasis to provide new therapeutic targets for HCC.
E26 transformation-specific (ETS) represent a family
of highly conserved transcription factors that regulate diverse
cellular processes, including proliferation, apoptosis and
migration (3). Previous studies
have proved that aberrant expression of ETS was involved in various
cancers and played critical roles in regulation of tumorigenesis.
Enhanced expression of ETS1 and ETS2 contribute to malignant
progression in breast cancer (4,5). In
addition, ETS1 promotes invasive potential and lymph node
metastasis in human oral squamous cell carcinoma (6).
Friend leukemia virus integration 1 (Fli-1), a newly
identified member of the ETS family, was first isolated in friend
murine leukemia virus (F-MuLV) caused erythroleukemia (7). It emerged as a transcriptional
activator or repressor in embryonic development, hematopoiesis,
aberrant vasculogenesis (8–10).
Emerging evidences have shown that dysregulation of
Fli-1 is a feature of many malignancies, such as breast cancer
(11), melanoma (12), Ewing's sarcoma (13,14),
Nasopharyngeal carcinoma (15). It
modulates different pathological process and functions as an
oncogene or tumor suppressor in different tissue types. However,
studies on elucidating the potential role of Fli-1 in HCC
progression are rare. In this study, we studied the expression
level of Fli-1 in HCC. Furthermore, we examined the effects of
Fli-1 on HCC tumorigenesis and the underlying mechanisms.
Materials and methods
Patients and tissue specimens
A total of 50 matched specimens of cancer-adjacent
tissue and HCC tissue, including 20 females and 30 males (mean age,
58.9±0.14 years) were obtained from patients who underwent surgical
resection at the Guangdong General Hospital (Guangzhou, China).
Each matched cancer-adjacent tissue was obtained at least 10 cm
from the tumor margin. All the samples enrolled in this study were
histologically confirmed by pathologists, and none of the patients
had received chemotherapy or radiotherapy before the surgery.
This study was approved by the Research Ethics
Committee of Guangdong General Hospital and written informed
consent was obtained from each patient.
Cell lines
The human HCC cell line BEL7402, PLC/PRF/5,
SMMC7721, MHCC97H and a normal human liver cell line HL-7702 were
obtained from the Cell Bank of the Chinese Academy of Science
(Shanghai, China) and maintained in DMEM (Gibco, Grand Island, NY,
USA) supplemented with 10% fetal bovine serum (Gibco) in a
humidified chamber with 5% (v/v) CO2 at 37°C.
Construction of plasmid or small
interfering (si)RNA sequences and transfection
The full-length Fli-1 cDNA was amplified and cloned
into the pcDNA3.1 expression vector (Cyagen Biosciences Inc.,
Guangzhou, China). The expression plasmid and control plasmid were
transfected respectively into SMMC7721 and MHCC97H cells with
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the
protocol of the manufacturer.
Primer sequences used for PCR were: Sense,
5′-AAACGAGGGAAATGGGAG-3′ and antisense, 5′-TACCAACGGTGTCAACCTG-3′.
The sequences of siFli-1 were as follows: siFli-1 #1,
5′-AGGAGUGGAUCAAUCAGCCAGUGAG-3′ and siFli-1 #2,
5′-GGGAAAGUUCACUGUUGGCCUAUAA-3′. Negative control (siNC) was:
5′-CAGUACUUUUGUGUAGUACAA-3′. All sequences were synthesized by
Invitrogen. The transfection of siFli-1 and siNC was performed with
RNAiMAX transfection reagent.
RNA extraction and quantitative
PCR
Total RNA extractions from cells and tissue
specimens were performed using the TRIzol reagent (Invitrogen)
according to the manufacturer's instructions. Then the RNA was
reversely transcribed to cDNA with the cDNA synthesis kit (Takara
Bio, Inc., Otsu, Japan) using 1 µg RNA as template. Real-time PCR
primers were synthesized by Invitrogen. GAPDH was used as the
reference for mRNAs. Specific primers used were: Fli-1 forward,
5′-CAGTCGCCTAGCCAACCCTG-3′ and reverse,
5′-GCAATGCCGTGGAAGTCAAAT-3′; GAPDH forward,
5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse,
5′-TGGTGAAGACGCCAGTGGA-3′.
Colony formation assay
For colony formation assays, 1,000 cells/well were
seeded onto 6-well plates and incubated at 37°C in a humidified
incubator for 2 weeks. When the colonies were visible, fixed with
4% paraformaldehyde and stained with 1% crystal violet. Finally,
the numbers of colonies were calculated.
Migration and invasion assay
Cell migration and invasion assay were performed
with Transwell chambers (Corning Incorporated, Corning, NY, USA).
For invasion assay, the upper chamber was pre-coated with Matrigel.
Cells [5×104 (for migration assay) or 1×105
(for invasion assay)] were resuspended in serum-free DMEM and 200
µl was added to the upper chamber. DMEM (600 µl) supplemented with
10% FBS was added to the lower chamber. After 12 h incubation at
37°C, the membrane was fixed with methanol and stained by crystal
violet, the number of migrating cells was counted under the
microscope.
Wound-healing assay
For wound healing assay, tumor cells were seeded in
12-well plates (12,000 cells per well), scratch wounds were made
using a sterilized 200 µl plastic pipette tip in the middle of the
cell monolayer. The width of wounds was measured under the
microscope and wound-healing percentage was calculated.
Western blotting
Total protein was extracted using the lysis buffer
(Beyotime Institute of Biotechnology, Haimen, China). Next,
proteins were quantified with the Bradford assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA), 20 µg of different proteins
were subjected to 12% sodium dodecylsulfate polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to the polyvinylidene
difluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA).
After blocking in 5% skim milk at room temperature for 2 h, the
membranes were incubated with primary antibodies overnight at 4°C
and followed by matched horseradish peroxidase (HRP)-conjugated
second antibody at room temperature for 1 h. Protein staining was
measured by chemiluminescence (GE Healthcare Life Sciences, Little
Chalfont, UK). The antibodies used were as follows: Rabbit
anti-Fli-1 pAb (11347-1-AP; 1:1,000; ProteinTech Group, Inc.,
Chicago, IL, USA), rabbit anti-matrix metalloproteinase (MMP)2 mAb
(#4022; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA,
USA), GAPDH rabbit mAb (#2118; 1:2,000; Cell Signaling Technology,
Inc.) was used as an internal control protein.
Statistical analysis
Statistically analysis between the two groups was
calculated using Student's t-test and one-way ANOVA followed by
Scheffe test (post hoc) was used for multiple groups. Correlations
between FLI-1 expression level and clinicopathological data were
analyzed using Chi-square test. Data are shown as means ± standard
deviation (SD) from at least three independent experiments,
P<0.05 was considered statistically significant.
Results
Fli-1 was overexpressed in human HCC
tissues and cell lines and was associated with metastasis
Abnormally Fli-1 expression has been reported in
many malignancies, but whether Fli-1 deregulation also exists in
HCC remains unknown. Therefore, quantitative real-time PCR
(qRT-PCR) analysis was first carried out to examine the expression
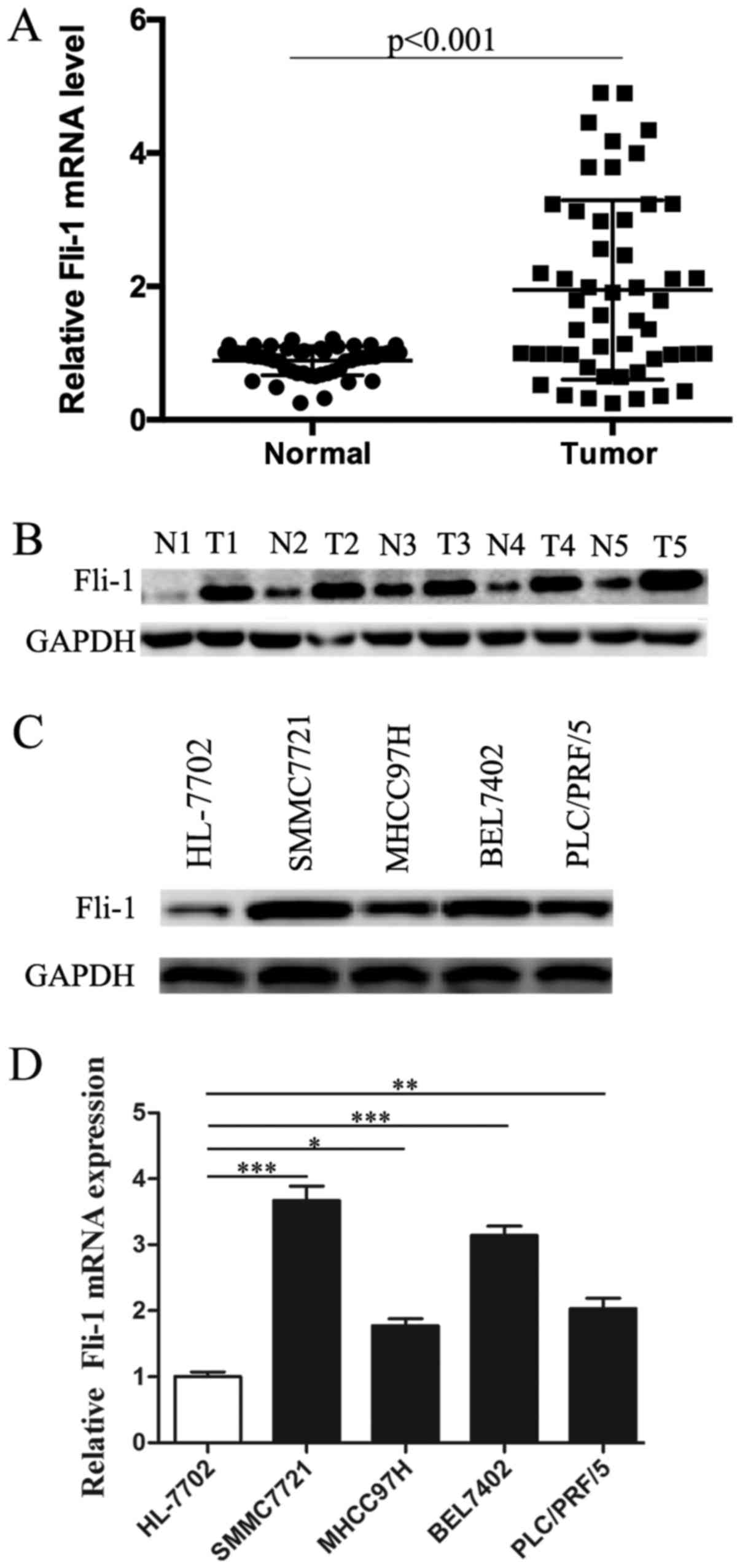
of Fli-1 in 50 matched HCC and cancer-adjacent tissues. As shown in
Fig. 1A, Fli-1 mRNA level was
dramatically elevated in the majority of HCC tissues compared to
cancer-adjacent tissues (P<0.001). We next determined the
expression pattern of Fli-1 in 5 paired HCC samples and adjacent
normal tissues by western blotting (Fig. 1B). Western blot analysis also
showed overexpression of Fli-1 in HCC tissues examined, compared
with matched noncancerous samples. We next examined the correlation
between FLI-1 mRNA expression and clinicopathologic characteristics
in 50 HCC patients, the results were shown in Table I. However, no statistically
significant association were found between FLI-1 expression and
clinicopathologic features except for metastasis.
 | Table I.Correlation between Fli-1 level and
clinicopathologic characteristics. |
Table I.
Correlation between Fli-1 level and
clinicopathologic characteristics.
|
|
| Fli-1 expression
(mean) |
|
|---|
|
|
|
|
|
|---|
| Characteristics | No. of patients
(n=50) | Low (n=12) | High (n=38) | P-value |
|---|
| Sex |
|
|
| 0.362 |
| Male | 40 | 8 | 32 |
|
|
Female | 10 | 4 | 6 |
|
| Age (years) |
|
|
| 0.639 |
| ≤50 | 30 | 6 | 24 |
|
|
>50 | 20 | 6 | 14 |
|
| AFP (ng/ml) |
|
|
| 0.296 |
| ≤400 | 37 | 7 | 30 |
|
|
>400 | 13 | 5 | 8 |
|
| T classification |
|
|
| 0.410 |
| T1-2 | 26 | 5 | 21 |
|
| T3-4 | 24 | 7 | 17 |
|
| TNM |
|
|
| 0.417 |
| I+II | 32 | 6 | 26 |
|
|
III+IV | 18 | 6 | 12 |
|
| Cirrhosis |
|
|
| 0.137 |
| Yes | 39 | 7 | 32 |
|
| No | 11 | 5 | 6 |
|
| Distant
metastasis |
|
|
| 0.028 |
| Yes | 22 | 2 | 20 |
|
| No | 28 | 10 | 18 |
|
In order to further validate our finding, we then
confirmed the expression of Fli-1 in HCC cell lines. Western blot
analysis showed a significant upregulation of Fli-1 in all HCC cell
lines, compared with normal liver cell line (Fig. 1C). Besides, RT-PCR analysis was
performed to detect whether high expression of Fli-1 was also
occurred at transcriptional level. Data in Fig. 1D showed that Fli-1 mRNA expression
also elevated in all HCC cell lines compared with normal liver cell
line.
Additionally, among the 4 HCC cell lines, Fli-1
expression was much higher in SMMC7721 cells than that in other HCC
cell lines, whereas MHCC97H showed less Fli-1 level. Besides,
SMMC-7721 and MHCC97H cell lines showed high migratory potential.
Thus, SMMC-7721 and MHCC97H cell lines were chosen for subsequent
study.
Silence of Fli-1 suppresses clone
formation of HCC cells
The above data suggest that Fli-1 deregulation may
play a physiological role in HCC tumorigenesis. To ascertain the
link between Fli-1 expression and tumor progression, we reduced
Fli-1 expression in SMMC7721 cells and enhanced Fli-1 expression in
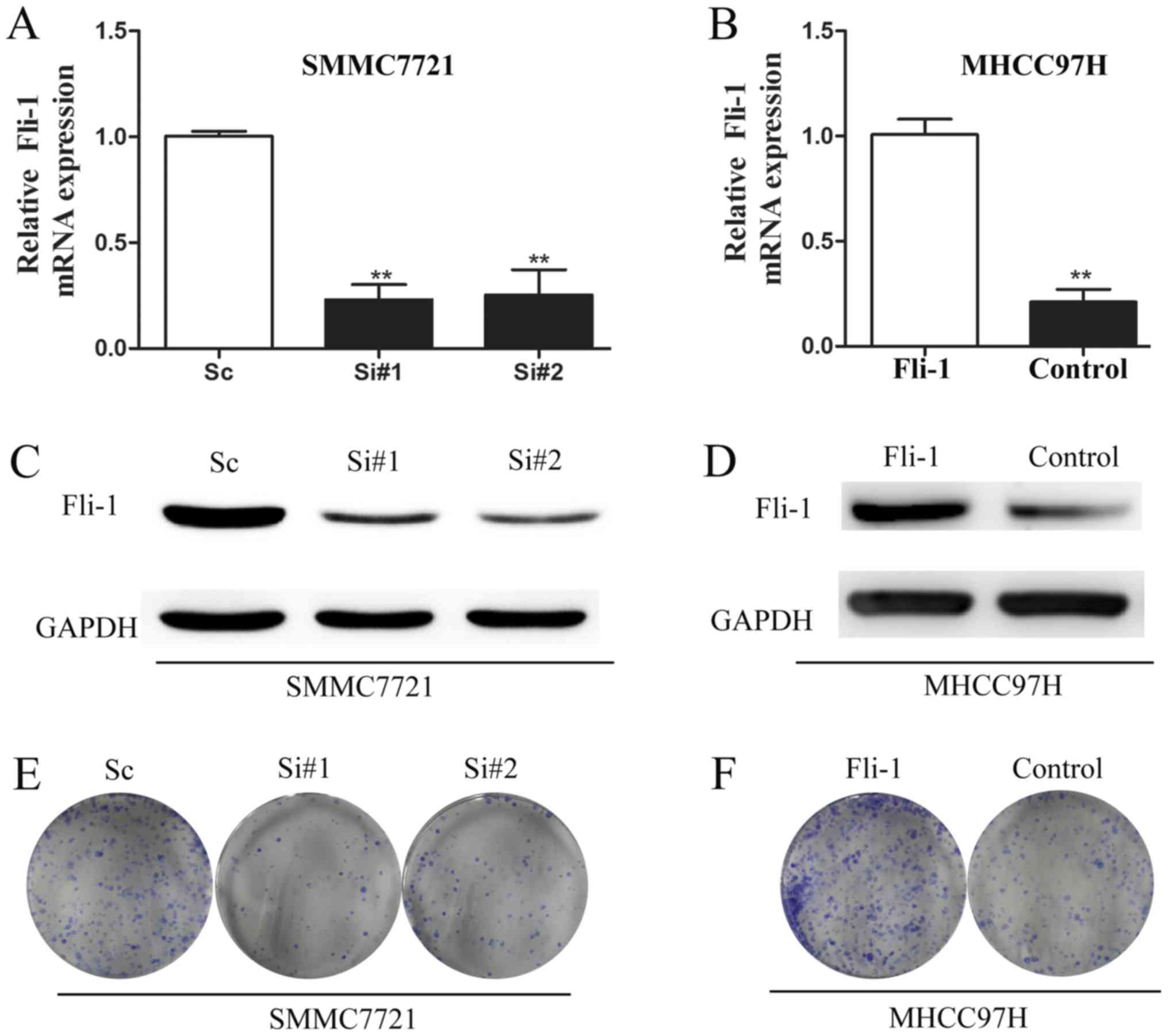
MHCC97H cells. Fig. 2 indicated
each individual siRNA (siRNA #1 or siRNA #2) markedly reduced Fli-1
mRNA expression, compared with siNC (Fig. 2A). Knockdown efficiency was further
verified at the protein level (Fig.
2C). Similarly, Fli-1 level was increased in MHCC97H cells
after transfection with pcDNA3.1-Fli-1 plasmid at both mRNA
(Fig. 2B) and protein level
(Fig. 2D).
Since tumor cells growth and clone formation play
crucial roles in tumor metastasis progression (16), clone formation assay was performed
to examine the role of Fli-1 in HCC cell growth. Seven days after
siRNA transfection, we fixed and stained the colonies. As shown in
Fig. 2E, knockdown of Fli-1
sharply suppressed the number and size of colonies, compared with
control. As expected, Fli-1 overexpression in MHCC97H cells led to
a significant enhancement of clone formation (Fig. 2F).
Knockdown of Fli-1 inhibits HCC cell
migration and invasion
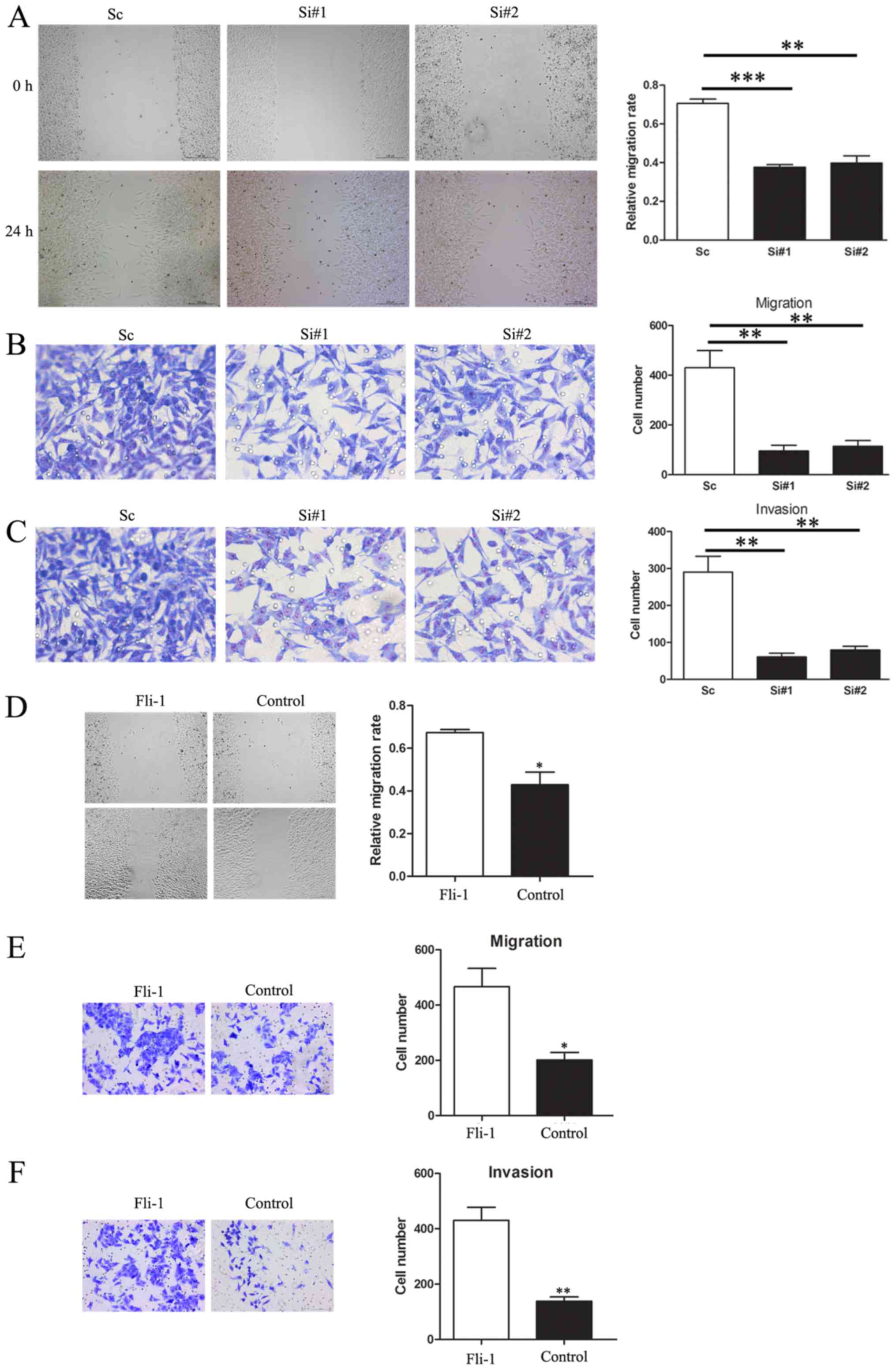
To investigate the role of Fli-1 in tumor cells
migration, wound-healing assay was performed. We observed that
knockdown of Fli-1 expression dramatically inhibited wound closure
in SMMC7721 cells (Fig. 3A).
Moreover, Transwell assay also demonstrated that the migratory
capacity was dramatically decreased in siFli-1-transfected cells
(Fig. 3B).
To further confirm this conclusion, we conducted a
Transwell assay with Matrigel pre-coated to detect the cell
invasion. Compared with the NC group, the number of migrating cells
in Fli-1 knockdown group was sharply decreased (Fig. 3C). In contrast, enhanced Fli-1
expression highly promoted MHCC97H cell migration and invasion
(Fig. 3D-F).
Overall, these results revealed that Fli-1
significantly promoted cell migration and invasion in
vitro.
Decreasing Fli-1 expression alters
MMP2 expression
Given that MMPs were closely associated with tumor
cells metastasis, We next investigated whether Fli-1 mediated
metastasis promoting effect was related to MMPs. MMP2 expression
was examined after transfection with siRNA or pcDNA3.1-Fli-1
plasmid. As shown in Fig. 4A,
downregulating Fli-1 expression was accompanied by decreased MMP2
mRNA level, as compared to the control group. This result was also
reproducible at protein level (Fig.
4B). In the contrast, enhanced MMP2 expression was observed in
Fli-1 overexpressed SMMC7721 cells (Fig. 4C and D). Consistently, Fli-1
overexpression obviously increased MMP2 level in MHCC97H cells
(Fig. 4E).
Discussion
Fli-1 is a newly identified ETS family member and
localized on human chromosome 11q23 (8). It was first discovered in mouse
erythroleukemia, while in human, EWS/Fli-1 fusion oncogene became
characteristic of Ewing sarcoma because of a t(11;22) (q24;q12)
chromosomal translocation (13).
Fli-1 plays vital roles in various biological processes. Aberrant
activation of Fli-1 is associated with inhibition of erythroid
differentiation and apoptosis, and leads to an increase in
proliferation of erythroblasts (8), whereas loss of Fli-1 could impair
vascular homeostasis in endothelial cells (17). Besides, aberrant expression of
Fli-1 has been demonstrated in various malignancies, such as breast
cancer (11), nasopharyngeal
carcinoma (NPC) (15), colon
cancer (18) and osteosarcoma
(19). High expression of Fli-1
contributes to the metastasis and tumorigenesis of cancer (11,15),
while inhibition of Fli-1 leads to growth suppression and apoptosis
of colon tumor cells (18),
indicating that Fli-1 may play a critical role in tumor
progression. However, the physiological function and the underlying
mechanism of Fli-1 in HCC tumorigenesis remain unclear.
In the present study, we demonstrated that Fli-1 is
upregulated both in HCC cancer cell lines and in primary HCC cancer
tissues, suggesting that Fli-1 is involved and may function as an
oncogene in HCC progression.
To further elucidate the role of Fli-1 in HCC
progression, increase and depletion experiments were respectively
conducted in MHCC97H and SMMC7721 cells. Loss of Fli-1 suppresses
the invasion and metastasis of SMMC7721 cells in vitro,
whereas ectopic overexpression of Fli-1 facilitates metastasis,
which is consistent with the effect of Fli-1 in breast cancer and
NPC (11,15) and further support our notion that
Fli-1 is involved in HCC malignant transformation.
Metastasis is a heterogeneous process including
cytoskeletal changes, integrin based adhesion, cell-extracellular
matrix (ECM) interactions, localized proteolysis, actin-myosin
contractions, and focal contact disassembly (20), in which active angiogenesis and
disruption of the ECM were the two crucial steps. Aberrant
activation of MMPs have been reported to be involved in the
degradation of ECM and the basal membrane, which assists the
migration and invasion of tumor. MMPs are a family of ECM proteases
(21,22), They can modulate tumor spread and
metastasis by facilitating ECM degradation, remodeling ECM,
maintaining the tumor micro-environment and generating ECM-linked
growth factors. In the MMPs family, MMP2 belongs to a group of
gelatinases. It is a key mediator in ECM degradation and is
involved in angiogenesis and metastasis in various malignancies
(23,24), including HCC (25,26).
This promoted us to investigate the link of MMP2
with Fli-1 in the metastasis of HCC. Here, we found that knockdown
of Fli-1 with siRNAs significantly repressed MMP2 expression, and
inhibited invasion and metastasis of tumor cells in vitro.
Accordingly, enhanced expression of Fli-1 is associated with
increased level of MMP2, accompanied with increased invasion and
migration of tumor cells. It is suggested that Fli-1 exerts its
promoting metastatic effect in HCC through the activation of MMP2
signaling. However, the potential molecular mechanism of Fli-1
exerts its modulation effect on the expression of MMP2 is still not
known, needs to be further explored.
In conclusion, our results demonstrated that high
expression of Fli-1 in HCC was associated with tumor metastasis,
and involved in the HCC tumor progression. These data suggest that
Fli-1 functions as an oncogene in HCC carcinogenesis in
vitro, although its anticancer effect needs to be further
verified in vivo. Our data provide novel insight into the
mechanism of Fli-1/MMP2 pathway in the pathogenesis of HCC.
Furthermore, Fli-1 and relevant downstream signaling may serve as
new therapeutic targets for HCC.
References
|
1
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar
|
|
2
|
Feng M, Gao W, Wang R, Chen W, Man YG,
Figg WD, Wang XW, Dimitrov DS and Ho M: Therapeutically targeting
glypican-3 via a conformation-specific single-domain antibody in
hepatocellular carcinoma. Proc Natl Acad Sci USA. 110:E1083–E1091.
2013. View Article : Google Scholar :
|
|
3
|
Seth A and Watson DK: ETS transcription
factors and their emerging roles in human cancer. Eur J Cancer.
41:2462–2478. 2005. View Article : Google Scholar
|
|
4
|
Buggy Y, Maguire TM, McGreal G, McDermott
E, Hill AD, O'Higgins N and Duffy MJ: Overexpression of the Ets-1
transcription factor in human breast cancer. Br J Cancer.
91:1308–1315. 2004. View Article : Google Scholar :
|
|
5
|
Buggy Y, Maguire TM, McDermott E, Hill AD,
O'Higgins N and Duffy MJ: Ets2 transcription factor in normal and
neoplastic human breast tissue. Eur J Cancer. 42:485–491. 2006.
View Article : Google Scholar
|
|
6
|
Shintani S, Hamakawa H, Nakashiro K,
Shirota T, Hatori M, Tanaka M, Kuroshita Y and Kurokawa Y: Friend
leukaemia insertion (Fli)-1 is a prediction marker candidate for
radiotherapy resistant oral squamous cell carcinoma. Int J Oral
Maxillofac Surg. 39:1115–1119. 2010. View Article : Google Scholar
|
|
7
|
Ben-David Y, Giddens EB, Letwin K and
Bernstein A: Erythroleukemia induction by friend murine leukemia
virus: Insertional activation of a new member of the ets gene
family, Fli-1, closely linked to c-ets-1. Genes Dev. 5:908–918.
1991. View Article : Google Scholar
|
|
8
|
Truong AH and Ben-David Y: The role of
Fli-1 in normal cell function and malignant transformation.
Oncogene. 19:6482–6489. 2000. View Article : Google Scholar
|
|
9
|
Kawada H, Ito T, Pharr PN, Spyropoulos DD,
Watson DK and Ogawa M: Defective megakaryopoiesis and abnormal
erythroid development in Fli-1 gene-targeted mice. Int J Hematol.
73:463–468. 2001. View Article : Google Scholar
|
|
10
|
Spyropoulos DD, Pharr PN, Lavenburg KR,
Jackers P, Papas TS, Ogawa M and Watson DK: Hemorrhage, impaired
hematopoiesis, and lethality in mouse embryos carrying a targeted
disruption of the Fli1 transcription factor. Mol Cell Biol.
20:5643–5652. 2000. View Article : Google Scholar :
|
|
11
|
Song W, Li W, Li L, Zhang S, Yan X, Wen X,
Zhang X, Tian H, Li A, Hu JF and Cui J: Friend leukemia virus
integration 1 activates the Rho GTPase pathway and is associated
with metastasis in breast cancer. Oncotarget. 6:23764–23775. 2015.
View Article : Google Scholar :
|
|
12
|
Torlakovic EE, Slipicevic A, Flørenes VA,
Chibbar R, DeCoteau JF and Bilalovic N: Fli-1 expression in
malignant melanoma. Histol Histopathol. 23:1309–1314. 2008.
|
|
13
|
Ladanyi M: EWS-FLI1 and Ewing's sarcoma:
Recent molecular data and new insights. Cancer Biol Ther.
1:330–336. 2002. View Article : Google Scholar
|
|
14
|
Riggi N, Suvà ML, Suvà D, Cironi L,
Provero P, Tercier S, Joseph JM, Stehle JC, Baumer K, Kindler V and
Stamenkovic I: EWS-FLI-1 expression triggers a Ewing's sarcoma
initiation program in primary human mesenchymal stem cells. Cancer
Res. 68:2176–2185. 2008. View Article : Google Scholar
|
|
15
|
Liang X, Shi D, Yun J, Mao Y, Ouyang P, Su
Z, Fu J, Hou J, Deng W and Xie F: Friend leukemia virus integration
1 expression has prognostic significance in nasopharyngeal
carcinoma. Transl Oncol. 7:493–502. 2014. View Article : Google Scholar :
|
|
16
|
Jia D, Yan M, Wang X, Hao X, Liang L, Liu
L, Kong H, He X, Li J and Yao M: Development of a highly metastatic
model that reveals a crucial role of fibronectin in lung cancer
cell migration and invasion. BMC Cancer. 10:3642010. View Article : Google Scholar :
|
|
17
|
Asano Y, Stawski L, Hant F, Highland K,
Silver R, Szalai G, Watson DK and Trojanowska M: Endothelial Fli1
deficiency impairs vascular homeostasis: A role in scleroderma
vasculopathy. Am J Pathol. 176:1983–1998. 2010. View Article : Google Scholar :
|
|
18
|
Zhang J, Guo H, Zhang H, Wang H, Qian G,
Fan X, Hoffman AR, Hu JF and Ge S: Putative tumor suppressor
miR-145 inhibits colon cancer cell growth by targeting oncogene
Friend leukemia virus integration 1 gene. Cancer. 117:86–95. 2011.
View Article : Google Scholar
|
|
19
|
Wu P, Liang J, Yu F, Zhou Z, Tang J and Li
K: MiR-145 promotes osteosarcoma growth by reducing expression of
the transcription factor friend leukemia virus integration 1.
Oncotarget. 7:42241–42251. 2016. View Article : Google Scholar :
|
|
20
|
Friedl P and Wolf K: Tumour-cell invasion
and migration: Diversity and escape mechanisms. Nat Rev Cancer.
3:362–374. 2003. View
Article : Google Scholar
|
|
21
|
Kleiner DE and Stetler-Stevenson WG:
Matrix metalloproteinases and metastasis. Cancer Chemother
Pharmacol. 43 Suppl:S42–S51. 1999. View Article : Google Scholar
|
|
22
|
Curran S and Murray GI: Matrix
metalloproteinases in tumour invasion and metastasis. J Pathol.
189:300–308. 1999. View Article : Google Scholar
|
|
23
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: Regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar :
|
|
24
|
Itoh T, Tanioka M, Yoshida H, Yoshioka T,
Nishimoto H and Itohara S: Reduced angiogenesis and tumor
progression in gelatinase A-deficient mice. Cancer Res.
58:1048–1051. 1998.
|
|
25
|
Ogasawara S, Yano H, Momosaki S, Nishida
N, Takemoto Y, Kojiro S and Kojiro M: Expression of matrix
metalloproteinases (MMPs) in cultured hepatocellular carcinoma
(HCC) cells and surgically resected HCC tissues. Oncol Rep.
13:1043–1048. 2005.
|
|
26
|
Giannelli G, Bergamini C, Marinosci F,
Fransvea E, Quaranta M, Lupo L, Schiraldi O and Antonaci S:
Clinical role of MMP-2/TIMP-2 imbalance in hepatocellular
carcinoma. Int J Cancer. 97:425–431. 2002. View Article : Google Scholar
|