Introduction
Osteosarcoma (OS), an aggressive bone neoplasm, is
the leading cause of cancer-associated mortality in adolescents and
children (1). OS predominantly
occurs in long bones, especially in the metaphyses, proximal tibia,
proximal humerus and distal femur (2). Currently, treatment modalities for OS
predominantly include chemotherapy, surgery and radiotherapy
(3). Despite the development of
multiple therapeutic strategies, the five-year survival rate and
prognosis of patients with OS remain poor; the majority of patients
eventually die due to local relapse or pulmonary metastases
following surgical resection (4,5). The
five-year survival rate is 60–80% for patients with localized
lesions and 15–30% for those with metastasis (6). The molecular mechanisms underlying
the progression and metastasis of OS remain poorly understood
(7). These mechanisms must be
elucidated to provide novel therapeutic targets or candidates for
treatment of patients with OS.
MicroRNAs (miRNAs) are small, endogenous and
noncoding RNA molecules, of 17–24 nucleotides (8). miRNAs post-transcriptionally modulate
gene expression by base pairing to complementary sites in the
3′-untranslated region (3-UTRs) of their target mRNAs; this
phenomenon leads to mRNA degradation or inhibition of translation
(9). Over 1,000 miRNAs are
predicted to exist in the human genome and may regulate as much as
60% of protein coding genes in humans (10). Increasing evidence has demonstrated
that miRNAs have significant roles in various physiological
processes, including cell proliferation, cycle, differentiation,
apoptosis, angiogenesis, invasion and metastasis (11–14).
Studies have reported that numerous miRNAs are aberrantly expressed
in human cancers and are significantly correlated with
tumorigenesis and tumor development by acting as oncogenes or tumor
suppressors (15–17). Upregulated miRNAs function as
oncogenes by negatively regulating tumor suppressor genes. By
contrast, downregulated miRNAs act as tumor suppressors by blocking
oncogenes in tumor progression (18,19).
Hence, miRNAs are considered putative targets for diagnosis,
prognosis and treatment of patients with tumors.
Researchers have investigated the role of miRNA-625
(miR-625) in several types of human cancer (20–22);
however, miR-625 in OS has not been investigated yet, to the best
of our knowledge. In the present study, the expression levels of
miR-625 in OS tissues and cell lines were determined and the
biological role was examined in OS cells. The molecular mechanisms
underlying the functions of miR-625 in OS cells were also
investigated.
Materials and methods
Tissue samples and cell lines
Between February 2012 and April 2015, 29 paired OS
tissues and associated adjacent non-tumor tissues were obtained
from patients (18 males, 11 females; age range, 19–63 years; mean
age, 34 years) during surgery at the Central Hospital of Enshi
Autonomous Prefecture (Enshi, China). None of these patients had
received chemotherapy or radiotherapy prior to surgery. All tissues
were immediately snap-frozen in liquid nitrogen and then stored at
−80°C. This study was approved by the Ethics Committee of The
Central Hospital of Enshi Autonomous Prefecture. Additionally,
signed informed consent was provided by all patients enrolled in
this study.
OS cell lines (U2OS, Saos-2, and MG-63) and human
normal osteoblasts (hFOB1.19) were purchased from the Institute of
Cell Bank for Biological Sciences (Shanghai, China). Cells were
maintained in Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), 100 U/ml penicillin and 100 U/ml streptomycin
(Gibco; Thermo Fisher Scientific, Inc.), at 37°C in 5%
CO2.
Transfection
Cells were seeded in 6-well plates and cultured to
70% confluence. Cells were transfected with miR-625 mimics,
scramble miRNA negative control (NC; Guangzhou RiboBio Co., Ltd.,
Guangzhou, China), pCDNA3.1-YAP1 or pCDNA3.1 blank vector (Chinese
Academy of Sciences, Shanghai, China) using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), in accordance with
the manufacturer's protocols. The miR-625 mimics sequence was
5′-AGGGGGAAAGUUCUAUAGUCC-3′ and the miR-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. After incubation at 37°C with 5%
CO2 for 8 h, the culture medium was replaced by DMEM
supplemented with 10% FBS. Then, 48 h post transfection, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
performed to determine transfection efficiency. Cell Counting Kit-8
(CCK-8) and Transwell invasion assays were conducted at 24 and 48 h
post transfection. Western blotting analysis was carried out at 72
h following transfection.
RNA extraction and RT-qPCR
Total RNA in tissues and cells was isolated using
TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. The concentration of total RNA was
determined by measuring absorbance at 260 nm using a NanoDrop
spectrophotometer (ND-1000; Thermo Scientific Inc., Wilmington, DE,
USA). For miR-625 detection, RT was performed using TaqMan MicroRNA
Reverse Transcription kit, and qPCR was conducted with a TaqMan
MicroRNA Assay kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) on an ABI7900 Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). U6 was used as an internal control
for miR-625. For the detection of YAP1 and GAPDH, used as the
internal control, cDNA was synthesized using PrimeScript RT reagent
Kit (Takara Biotechnology Co., Ltd., Dalian, China) and qPCR was
performed with SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.,
Dalian, China). The primers were designed as follows: miR-625,
5′-AGGGGGAAAGTTCTATAGTCC-3′ (forward) and 5′-TGGTGTCGTGGAGTCG-3′
(reverse); U6, 5′-CTCGCTTCGGCAGCACA-3′ (forward) and
5′-AACGCTTCACGAATTTGCGT-3′ (reverse); YAP1,
5′-CAACTCCAACCAGCAGCAACA-3′ (forward) and
5′-GCAGCCTCTCCTTCTCCATCTG-3′ (reverse); and GAPDH,
5′-CGGAGTCAACGGATTTGGTCGTAT-3′ (forward) and
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (reverse). Relative expression
levels were calculated using the 2−ΔΔCq method (23).
CCK-8 assay
Cell proliferation was determined using the CCK-8
assay (Dojindo Molecular Technologies, Inc., Rockville, MD, USA).
Cells were seeded into 96-well plates at a density of
3×103 cells/well, and transfected with miR-625 mimics,
NC, pCDNA3.1-YAP1 or pcDNA3.1. Cells were incubated at 37°C in the
presence of 5% CO2 for 0, 24, 48 or 72 h. Following
incubation, 10 µl CCK-8 solution was added to each well, and the
cells were incubated at 37°C for another 2 h. The absorbance was
read at 450 nm using a microplate reader (Biotek Synergy HT; BioTek
Instruments, Inc., Winooski, VT, USA).
Transwell invasion assay
Cell invasion capacity was assessed using Transwell
chambers (Corning Incorporated, Corning, NY, USA) precoated with
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Transfected
cells suspended in FBS-free culture medium (1×105) were
added into the upper chambers and the lower chambers were filled
with 800 µl DMEM containing 10% FBS. After incubation for 48 h at
37°C in the presence of 5% CO2, the non-invasive cells
on the upper chambers were carefully wiped out by a cotton-tipped
swab. The cells that had invaded to the lower chambers were fixed
with 90% alcohol and stained with 0.5% crystal violet. The number
of cells invading through the membranes were counted in five
randomly selected fields under a light microscope (×200
magnification; Olympus Corporation, Tokyo, Japan).
Bioinformatics analysis and luciferase
reporter assay
To predict the putative targets of miR-625,
bioinformatics analysis was performed using TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/). The
pGL3-YAP1-3′-UTR wild-type (Wt) and pGL3-YAP1-3′-UTR mutant (Mut)
reporter plasmids were synthesized and obtained from Shanghai
GenePharma Co., Ltd. (Shanghai, China). Cells were plated in
24-well plates at a density of 60–70% confluence, and were
transfected with miR-625 mimics or NC, together with
pGL3-YAP1-3′-UTR Wt or pGL3-YAP1-3′-UTR Mut using Lipofectamine
2000. The firefly luciferase activity and Renilla luciferase
activity were detected at 48 h post-transfection using
Dual-Luciferase Reporter Assay System (Promega Corporation,
Madison, WI, USA) in accordance with the manufacturer's guidance.
The firefly luciferase activity was normalized to the
Renilla luciferase activity.
Western blotting
Tissues and cells were solubilized in cold RIPA
Lysis and Extraction Buffer (Beyotime Institute of Biotechnology,
Haimen, China). A BCA Protein Assay kit (Beyotime Institute of
Biotechnology, Haimen, China) was used to determine protein
concentration. Equivalent protein (20 µg) were separated by sodium
dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently
transferred onto polyvinylidene difluoride membranes (EMD
Millipore, Billerica, MA, USA), which were then blocked with
Tris-buffered saline containing 0.05% Tween-20 (TBST; Beyotime
Institute of Biotechnology) containing 5% skimmed milk at room
temperature for 2 h. The membranes were incubated with following
primary antibodies at 4°C overnight: Mouse anti-human monoclonal
Yes associated protein 1 (YAP1; cat. no. ab56701; 1:1,000; Abcam,
Cambridge, UK) and mouse anti-human monoclonal GAPDH (cat. no.
sc-69778; 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA). Following washing three times in TBST, the membranes were
incubated for 2 h at room temperature with goat anti-mouse
horseradish peroxidase-conjugated secondary antibody (cat. no.
sc-2005; 1:3,000; Santa Cruz Biotechnology, Inc.). An ECL kit
(Pierce; Thermo Fisher Scientific, Inc.) was used visualization of
the protein bands. Relative expression of YAP1 protein was analyzed
using Image Pro Plus software (version 6.0; Media Cybernetics,
Inc., Rockville, MD, USA).
Statistical analysis
Data are expressed as the mean ± standard deviation.
Two-tailed Student's t test or one-way analysis of variance was
used to compare difference between groups by using SPSS 19.0
statistical software (IBM Corp., Armonk, NY, USA). The SNK test was
used as a post hoc test following analysis of variance. P<0.05
was considered to indicate a statistically significant
difference.
Results
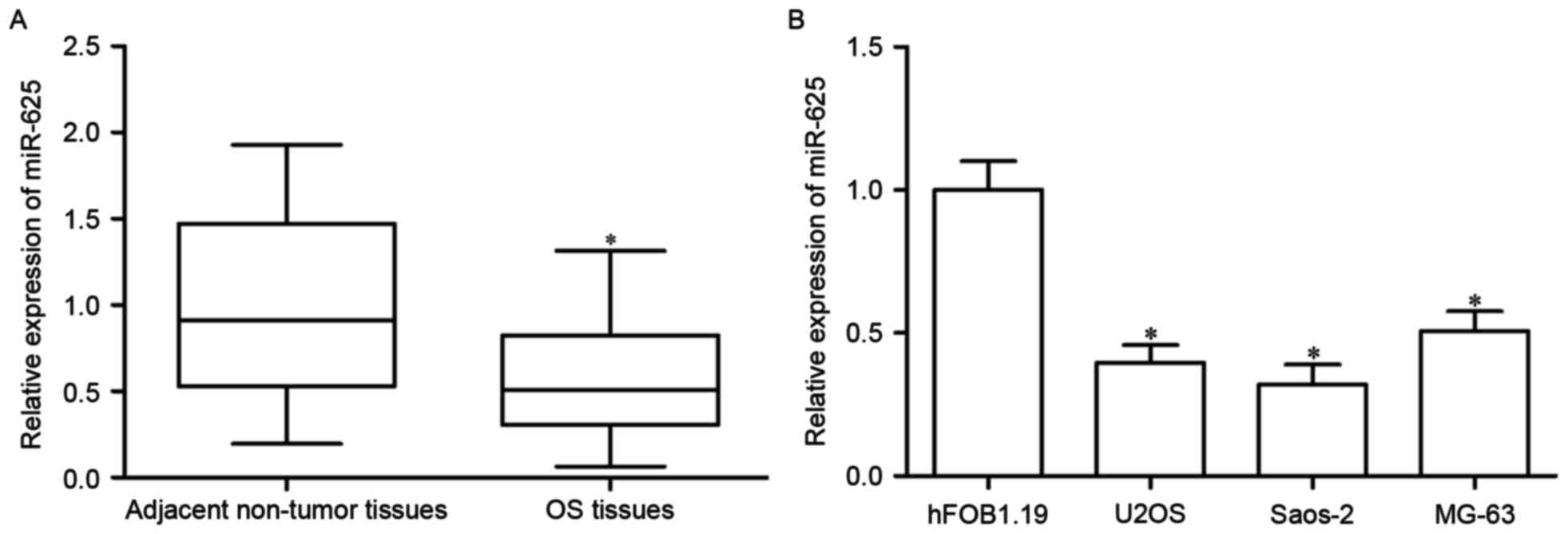
Downregulation of miR-625 in OS
tissues and cell lines
To explore the roles of miR-625 in OS formation and
progression, the expression levels of miR-625 in OS tissues and
associated adjacent non-tumor tissues were determined. The
expression of miR-625 was decreased in OS tissues compared with
those in the adjacent non-tumor tissues (P<0.05; Fig. 1A). As such, the expression levels
of miR-625 in three common OS cell lines (U2OS, Saos-2 and MG-63)
and human normal osteoblasts (hFOB1.19) were then measured. Results
revealed that miR-625 was downregulated in the three OS cell lines
compared with that in hFOB1.19 (P<0.05; Fig. 1B). Thus, miR-625 may have an
important role in OS.
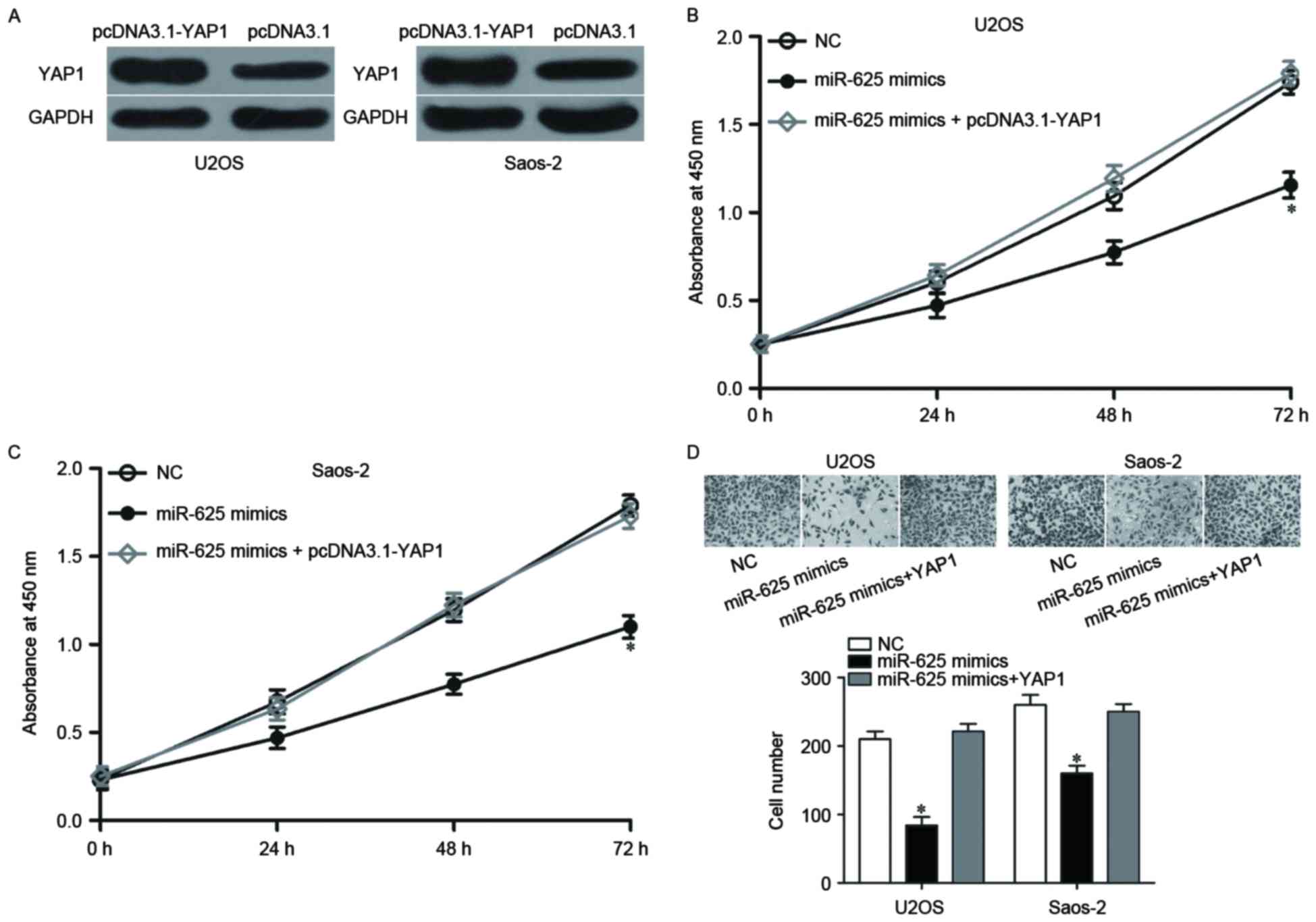
Restoration of miR-625 expression inhibits cell
proliferation and invasion in OS. U2OS and Saos-2 cells expressed
significantly reduced miR-625 levels compared with normal human
osteoblasts; therefore, these cell lines were transfected with
miR-625 mimics to increase the expression of miR-625 (P<0.05;
Fig. 2A and B). The functional
effect of miR-625 on the proliferation of U2OS and Saos-2 cells was
investigated using a CCK-8 assay. As presented in Fig. 2C and D, treatment with miR-625
mimics reduced the proliferation of U2OS and Saos-2 cells compared
with cells transfected with an NC miRNA mimic (P<0.05).
Transwell invasion assay was then performed to evaluate the effects
of miR-625 overexpression on the invasion capacity of U2OS and
Saos-2 cells. The results revealed that miR-625 upregulation
reduced the number of invasive U2OS and Saos-2 cells compared with
that in the NC groups (P<0.05; Fig.
2E). Therefore, miR-625 may suppress OS growth and
metastasis.
miR-625 directly targets YAP1 in
OS
To further explore the molecular mechanism
underlying the tumor-suppressive roles of miR-625 in OS, the
putative target genes of miR-625 were predicted using
bioinformatics analysis. YAP1 was identified as a potential target
of miR-625 (Fig. 3A) and is
involved in the initiation and development of various cancer types
(24). To confirm this hypothesis,
a luciferase reporter assay was performed in U2OS and Saos-2 cells.
The cells were then transfected with miR-625 mimics or NC, along
with luciferase reporter plasmid carrying the wild type or the
mutant 3′-UTR of YAP1. As presented in Fig. 3B and C, miR-625 overexpression
significantly decreased the luciferase activity of the vector
carrying the wild type 3′-UTR of YAP1 (P<0.05); however, the
luciferase activity did not differ between cells co-transfected
with vectors carrying the mutant 3′-UTR of YAP1 and miR-625
mimics.
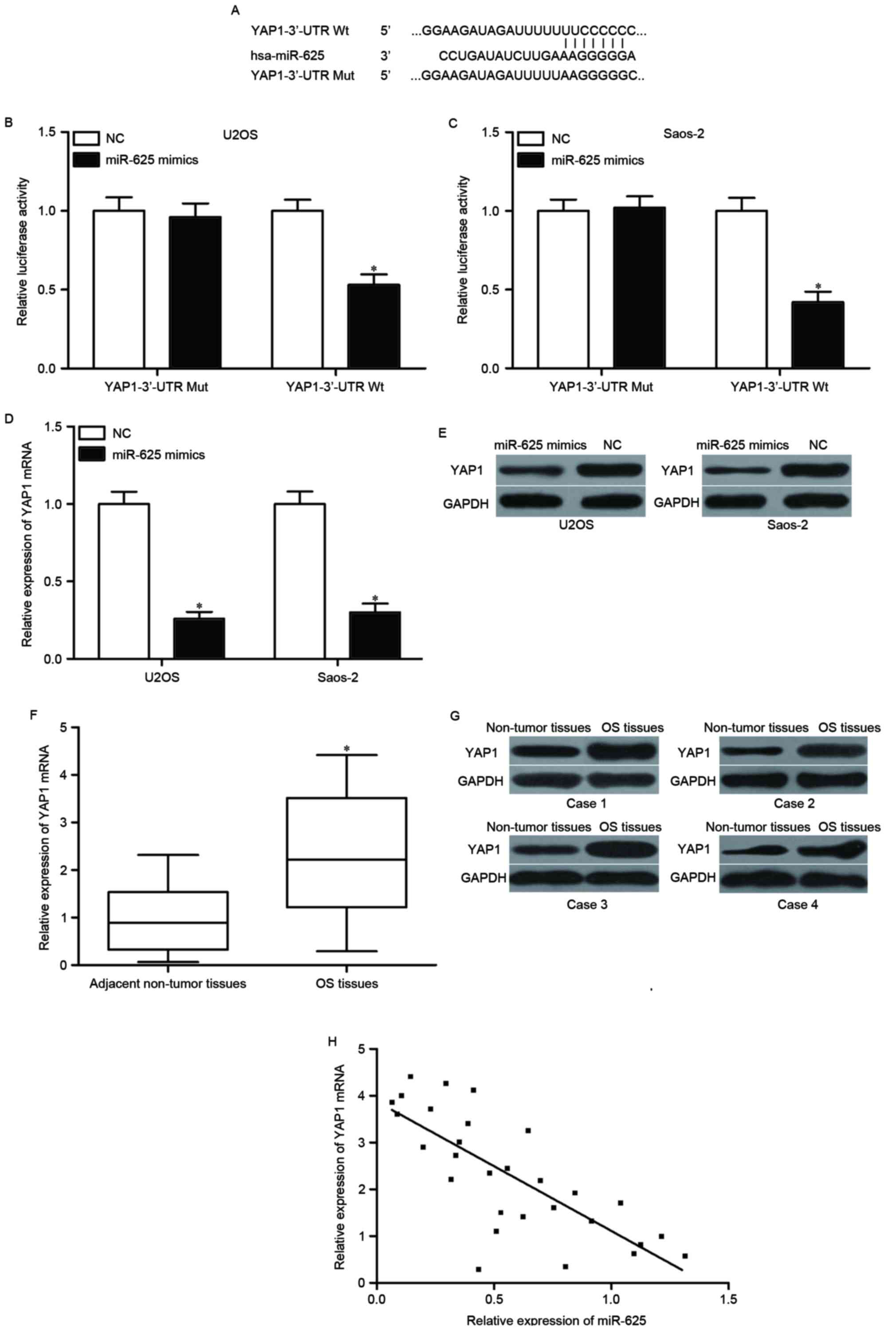 | Figure 3.YAP1 is a direct target of miR-625 in
OS. (A) Schematic representation of miR-625 putative binding sites
in the 3′-UTR of YAP1 mRNA. (B) U2OS and (C) Saos-2 cells were
co-transfected with pGL3-YAP1-3′-UTR Wt or pGL3-YAP1-3′-UTR Mut,
and miR-625 mimics or NC. Luciferase reporter assay was performed
to detect the luciferase activity in each group. Each assay was
repeated three times. (D) RT-qPCR and (E) western blotting were
used to analyze YAP1 mRNA and protein expression in U2OS and Saos-2
cells following transfection with miR-625 mimics or NC. *P<0.05
vs. NC. Each assay was repeated three times (F) YAP1 mRNA level in
29 paired OS tissues and associated adjacent non-tumor tissues was
examined using RT-qPCR. *P<0.05 vs. adjacent non-tumor tissues.
Each assay was repeated three times (G) Western blotting was
performed to measure YAP1 protein expression in OS tissues compared
to adjacent non-tumor tissues. Each assay was repeated three times
(H) A negative correlation between the miR-625 expression and YAP1
mRNA expression was observed in OS tissues, r=−0.7665, P<0.001.
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; YAP1, Yes-associated protein 1; UTR, untranslated region;
Wt, wild-type; miR, microRNA; Mut, mutant; NC, negative control
microRNA; OS, osteosarcoma. |
RT-qPCR and western blot analyses were conducted to
determine the mRNA and protein expression levels of YAP1 in U2OS
and Saos-2 cells transfected with miR-625 mimic or NC. The mRNA and
protein expression levels of YAP1 were suppressed in U2OS and
Saos-2 cells following transfection with the miR-625 mimic
(P<0.05; Fig. 3D and E). The
mRNA and protein expression levels of YAP1 were then measured in OS
tissues and associated adjacent non-tumor tissues. YAP1 was
upregulated at both mRNA and protein level in OS tissues compared
with that in the adjacent non-tumor tissues (P<0.05; Fig. 3F and G). Spearman's correlation
analysis further indicated that YAP1 mRNA expression was inversely
correlated with miR-625 expression in OS tissues (r=−0.7665,
P<0.001; Fig. 3H).
Collectively, these results suggested that YAP1 was directly and
negatively regulated by miR-625 in OS.
Upregulation of YAP1 rescues the
inhibitory effects of miR-625 on cell proliferation and
invasion
To further determine whether YAP1 mediates the
inhibitory effects of miR-625 on OS proliferation and invasion,
rescue experiments were performed. U2OS and Saos-2 cells were
transfected with pcDNA3.1-YAP1 or pcDNA3.1. Following transfection,
YAP1 protein expression was upregulated in
pcDNA3.1-YAP1-transfected U2OS and Saos-2 cells (P<0.05;
Fig. 4A). The rescue experiments
demonstrated that the enforced YAP1 expression rescued the impaired
cell proliferation and invasion induced by miR-625 mimics
(P<0.05; Fig. 4B-D); thus,
YAP1, at least partially, mediated the functional roles of miR-625
in OS.
Discussion
Accumulated evidence supports the critical roles of
miRNAs in cancer initiation and progression (25–27).
Therefore, OS-associated miRNAs must be identified for use as
therapeutic targets for treatment of OS. In the present study, the
expression and functions of miR-625 in OS were investigated. The
expression of miR-625 was low in OS tissues and cell lines compared
with adjacent normal tissue and a normal osteoblast cell line,
respectively. Functional experiments demonstrated that the ectopic
expression of miR-625 suppressed OS cell proliferation and invasion
in vitro. Mechanistically, YAP1 was identified as the direct
target of miR-625 in OS. These results suggested that miR-625 may
act as a tumor suppressor in the tumorigenesis and development of
OS.
The expression of miR-625 is reduced in several
types of human cancer. For example, miR-625 was reported to be
downregulated in colorectal cancer tissues and cell lines (20–22).
Low expression of miR-625 was previously demonstrated to be
strongly correlated with lymph node metastasis, liver metastasis,
poor overall survival and an unfavorable prognosis for patients
with colorectal cancer (20). In
gastric cancer, miR-625 was expressed at low levels in gastric
cancer and inversely associated with lymph node metastasis
(21). In hepatocellular
carcinoma, miR-625 was reduced in tissues. The low expression level
of miR-625 was closely correlated with lymph node and distant
metastases, the presence of portal venous invasion, TNM staging and
unfavorable overall survival (22). In esophageal squamous cell
carcinoma (ESCC), the expression of miR-625 was decreased in tumor
tissues and was significantly associated with lymph node and
distant metastases, tumor differentiation and TNM staging. In
patients with ESCC, low miR-625 expression levels were associated
with short overall survival. Multivariate Cox regression analysis
indicated miR-625 may be an independent prognostic factor for
overall patient survival in ESCC (28). Thus, miR-625 downregulation may be
a prognostic factor for these cancer types.
Previous studies validated that miR-625 is a tumor
suppressor in numerous human cancers (20–22).
In colorectal cancer, miR-625 re-expression suppressed tumor cell
metastasis in vitro and in vivo (20). Wang et al (21) revealed that miR-625 expression
level restoration inhibited the invasive and metastatic abilities
of gastric cancer cells in vitro and in vivo by
directly targeting integrin linked kinase. Zhou et al
(22) reported that miR-625
upregulation impeded cell motility in hepatocellular carcinoma
in vitro and in vivo through negative regulation of
insulin like growth factor 2 mRNA binding protein 1. Wang et
al (29) demonstrated that the
resumption expression of miR-625 attenuated cell proliferation and
invasion in esophageal cancer by blocking SRY-box 2. Zhou et
al (30) revealed that
enforced expression of miR-625 targeted high mobility group AT-hook
2 to repress the proliferation and invasion of breast cancer cells.
These findings suggest that miR-625 is a potential anticancer
agent.
It is well established that miRNAs exert biological
activities through negative regulation of their target mRNAs
(31). To investigate the
mechanisms by which miR-625 suppressed OS cell proliferation and
invasion, bioinformatics analysis was used to search for potential
targets of miR-625. The analysis indicated that YAP1 mRNA contained
a miR-625 seed match at position 1,896–1,902 of the YAP1 3′-UTR.
Furthermore, a luciferase reporter assay demonstrated that
upregulation of miR-625 decreased the luciferase activity in OS
cells transfected with luciferase reporter plasmid carrying the
wild type 3′-UTR of YAP1, but not affect the luciferase activity of
plasmid carrying the mutant 3′-UTR of YAP1, suggesting that miR-625
directly targeted the 3′-UTR of YAP1. In addition, miR-625
overexpression repressed YAP1 mRNA and protein expression in OS
cells. Furthermore, YAP1 was upregulated in OS tissues and
inversely correlated with miR-625 expression. Additionally,
enforced expression of YAP1 rescued the impaired cell proliferation
and invasion induced by miR-625 in OS, which demonstrated that YAP1
is a direct functional target of miR-625 in OS cells.
YAP1, which is located on chromosome 11q22.1, is
abnormally expressed in various tumor types, including glioma
(32), colorectal cancer (33), gastric cancer (34) and bladder cancer (35). As a member of the Hippo pathway,
YAP1 has important roles in regulating cell proliferation,
invasion, epithelial mesenchymal transition, metastasis,
differentiation and survival (24). In OS, YAP1 was previously reported
to be upregulated in tumor tissues, and significantly correlated
with gender and Enneking staging. Additionally, downregulating YAP1
suppressed OS cell proliferation and invasion in vitro
(36). Yang et al (37) reported that YAP1 knockdown
inhibited cell proliferation, colony formation, cell cycle
progression in vitro and the growth of OS xenograft tumors
in vivo. In the present study, it was demonstrated that
miR-625 targeted YAP1 to inhibit OS cell proliferation and
invasion. Thus, the miR-625/YAP1 axis is a potential target for
inhibiting the rapid growth and metastasis of OS.
In conclusion, the results of the current study
demonstrate that miR-625 was downregulated in OS tissues and cell
lines. Ectopic expression of miR-625 attenuated the proliferation
and invasion of OS cells by directly targeting YAP1. The abnormal
miR-625 expression may potentially be used as a therapeutic target
for treatment of patients with OS.
References
|
1
|
Guillon MA, Mary PM, Brugière L,
Marec-Bérard P, Pacquement HD, Schmitt C, Guinebretière JM and
Tabone MD: Clinical characteristics and prognosis of osteosarcoma
in young children: A retrospective series of 15 cases. BMC Cancer.
11:4072011. View Article : Google Scholar :
|
|
2
|
Ritter J and Bielack SS: Osteosarcoma. Ann
Oncol. 21 Suppl 7:vii320–vii325. 2010. View Article : Google Scholar
|
|
3
|
Williams SA, Maecker HL, French DM, Liu J,
Gregg A, Silverstein LB, Cao TC, Carano RA and Dixit VM: USP1
deubiquitinates ID proteins to preserve a mesenchymal stem cell
program in osteosarcoma. Cell. 146:918–930. 2011. View Article : Google Scholar
|
|
4
|
Bacci G, Briccoli A, Rocca M, Ferrari S,
Donati D, Longhi A, Bertoni F, Bacchini P, Giacomini S, Forni C, et
al: Neoadjuvant chemotherapy for osteosarcoma of the extremities
with metastases at presentation: Recent experience at the Rizzoli
Institute in 57 patients treated with cisplatin, doxorubicin, and a
high dose of methotrexate and ifosfamide. Ann Oncol. 14:1126–1134.
2003. View Article : Google Scholar
|
|
5
|
Marina N, Gebhardt M, Teot L and Gorlick
R: Biology and therapeutic advances for pediatric osteosarcoma.
Oncologist. 9:422–441. 2004. View Article : Google Scholar
|
|
6
|
Benjamin RS: Osteosarcoma: Better
treatment through better trial design. Lancet Oncol. 16:12–13.
2015. View Article : Google Scholar
|
|
7
|
Letson GD and Muro-Cacho CA: Genetic and
molecular abnormalities in tumors of the bone and soft tissues.
Cancer Control. 8:239–251. 2001. View Article : Google Scholar
|
|
8
|
Haghikia A, Hoch M, Stapel B and
Hilfiker-Kleiner D: STAT3 regulation of and by microRNAs in
development and disease. JAKSTAT. 1:143–150. 2012.
|
|
9
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:(Database Issue).
D140–D144. 2006. View Article : Google Scholar
|
|
10
|
Namløs HM, Meza-Zepeda LA, Barøy T,
Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H,
Cleton-Jansen AM and Myklebost O: Modulation of the osteosarcoma
expression phenotype by microRNAs. PLoS One. 7:e480862012.
View Article : Google Scholar :
|
|
11
|
Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang
Y, Zou C, Zhang X, Liu S, Wang X, et al: Analysis of MiR-195 and
MiR-497 expression, regulation and role in breast cancer. Clin
Cancer Res. 17:1722–1730. 2011. View Article : Google Scholar
|
|
12
|
Li Z, Yu X, Shen J and Jiang Y: MicroRNA
dysregulation in uveal melanoma: A new player enters the game.
Oncotarget. 6:4562–4568. 2015. View Article : Google Scholar :
|
|
13
|
Li Z, Yu X, Wang Y, Shen J, Wu WK, Liang J
and Feng F: By downregulating TIAM1 expression, microRNA-329
suppresses gastric cancer invasion and growth. Oncotarget.
6:17559–17569. 2015. View Article : Google Scholar :
|
|
14
|
Li Z, Yu X, Shen J, Law PT, Chan MT and Wu
WK: MicroRNA expression and its implications for diagnosis and
therapy of gallbladder cancer. Oncotarget. 6:13914–13921. 2015.
View Article : Google Scholar :
|
|
15
|
Iorio MV and Croce CM: Causes and
consequences of microRNA dysregulation. Cancer J. 18:215–222. 2012.
View Article : Google Scholar :
|
|
16
|
Garzon R, Liu S, Fabbri M, Liu Z, Heaphy
CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, et al:
MicroRNA-29b induces global DNA hypomethylation and tumor
suppressor gene reexpression in acute myeloid leukemia by targeting
directly DNMT3A and 3B and indirectly DNMT1. Blood. 113:6411–6418.
2009. View Article : Google Scholar :
|
|
17
|
Fenger JM, Roberts RD, Iwenofu OH, Bear
MD, Zhang X, Couto JI, Modiano JF, Kisseberth WC and London CA:
MiR-9 is overexpressed in spontaneous canine osteosarcoma and
promotes a metastatic phenotype including invasion and migration in
osteoblasts and osteosarcoma cell lines. BMC Cancer. 16:7842016.
View Article : Google Scholar :
|
|
18
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar :
|
|
19
|
Yang T, Thakur A, Chen T, Yang L, Lei G,
Liang Y, Zhang S, Ren H and Chen M: MicroRNA-15a induces cell
apoptosis and inhibits metastasis by targeting BCL2L2 in non-small
cell lung cancer. Tumour Biol. 36:4357–4365. 2015. View Article : Google Scholar
|
|
20
|
Lou X, Qi X, Zhang Y, Long H and Yang J:
Decreased expression of microRNA-625 is associated with tumor
metastasis and poor prognosis in patients with colorectal cancer. J
Surg Oncol. 108:230–235. 2013. View Article : Google Scholar
|
|
21
|
Wang M, Li C, Nie H, Lv X, Qu Y, Yu B, Su
L, Li J, Chen X, Ju J, et al: Down-regulated miR-625 suppresses
invasion and metastasis of gastric cancer by targeting ILK. FEBS
Lett. 586:2382–2388. 2012. View Article : Google Scholar
|
|
22
|
Zhou X, Zhang CZ, Lu SX, Chen GG, Li LZ,
Liu LL, Yi C, Fu J, Hu W, Wen JM and Yun JP: miR-625 suppresses
tumour migration and invasion by targeting IGF2BP1 in
hepatocellular carcinoma. Oncogene. 34:965–977. 2015. View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Zeng Q and Hong W: The emerging role of
the hippo pathway in cell contact inhibition, organ size control,
and cancer development in mammals. Cancer Cell. 13:188–192. 2008.
View Article : Google Scholar
|
|
25
|
Liu H, Li W, Chen C, Pei Y and Long X:
MiR-335 acts as a potential tumor suppressor miRNA via
downregulating ROCK1 expression in hepatocellular carcinoma. Tumour
Biol. 36:6313–6319. 2015. View Article : Google Scholar
|
|
26
|
Tu Y, Liu L, Zhao D, Liu Y, Ma X, Fan Y,
Wan L, Huang T, Cheng Z and Shen B: Overexpression of miRNA-497
inhibits tumor angiogenesis by targeting VEGFR2. Sci Rep.
5:138272015. View Article : Google Scholar :
|
|
27
|
Shin JY, Kim YI, Cho SJ, Lee MK, Kook MC,
Lee JH, Lee SS, Ashktorab H, Smoot DT, Ryu KW, et al: MicroRNA 135a
suppresses lymph node metastasis through down-regulation of ROCK1
in early gastric cancer. PLoS One. 9:e852052014. View Article : Google Scholar :
|
|
28
|
Li C, Li DC, Che SS, Ma K, Wang YJ, Xia
LH, Dai XM, Zhang GT, Shen Y, Jiao WJ and Tian KH: The decreased
expression of miR-625 predicts poor prognosis of esophageal
squamous cell carcinoma. Int J Clin Exp Med. 8:9560–9564. 2015.
|
|
29
|
Wang Z, Qiao Q, Chen M, Li X, Wang Z, Liu
C and Xie Z: miR-625 down-regulation promotes proliferation and
invasion in esophageal cancer by targeting Sox2. FEBS Lett.
588:915–921. 2014. View Article : Google Scholar
|
|
30
|
Zhou WB, Zhong CN, Luo XP, Zhang YY, Zhang
GY, Zhou DX and Liu LP: miR-625 suppresses cell proliferation and
migration by targeting HMGA1 in breast cancer. Biochem Biophys Res
Commun. 470:838–844. 2016. View Article : Google Scholar
|
|
31
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer. 96
Suppl:R40–R44. 2007.
|
|
32
|
Orr BA, Bai H, Odia Y, Jain D, Anders RA
and Eberhart CG: Yes-associated protein 1 is widely expressed in
human brain tumors and promotes glioblastoma growth. J Neuropathol
Exp Neurol. 70:568–577. 2011. View Article : Google Scholar :
|
|
33
|
Lee KW, Lee SS, Kim SB, Sohn BH, Lee HS,
Jang HJ, Park YY, Kopetz S, Kim SS, Oh SC and Lee JS: Significant
association of oncogene YAP1 with poor prognosis and cetuximab
resistance in colorectal cancer patients. Clin Cancer Res.
21:357–364. 2015. View Article : Google Scholar
|
|
34
|
Kang W, Tong JH, Chan AW, Lee TL, Lung RW,
Leung PP, So KK, Wu K, Fan D, Yu J, et al: Yes-associated protein 1
exhibits oncogenic property in gastric cancer and its nuclear
accumulation associates with poor prognosis. Clin Cancer Res.
17:2130–2139. 2011. View Article : Google Scholar
|
|
35
|
Li S, Yu Z, Chen SS, Li F, Lei CY, Chen
XX, Bao JM, Luo Y, Lin GZ, Pang SY and Tan WL: The YAP1 oncogene
contributes to bladder cancer cell proliferation and migration by
regulating the H19 long noncoding RNA. Urol Oncol. 33(427): e1–10.
2015.
|
|
36
|
Zhang YH, Li B, Shen L, Shen Y and Chen
XD: The role and clinical significance of YES-associated protein 1
in human osteosarcoma. Int J Immunopathol Pharmacol. 26:157–167.
2013. View Article : Google Scholar
|
|
37
|
Yang Z, Zhang M, Xu K, Liu L, Hou WK, Cai
YZ, Xu P and Yao JF: Knockdown of YAP1 inhibits the proliferation
of osteosarcoma cells in vitro and in vivo. Oncol Rep.
32:1265–1272. 2014. View Article : Google Scholar
|