Introduction
Iron is an essential element for almost every
organism, from animals to microorganisms (1). Iron-associated disorders occur in
conditions of iron deficiency or iron overload (2,3).
Hepcidin is a micro-molecule polypeptide that is composed of 25
amino acids (4,5) and is involved in the regulation of
iron metabolism. It is mainly expressed in liver cells, but it is
also expressed in fat cells (6),
pancreatic β cells (7) and bone
marrow cells, including monocytes (8,9),
macrophages (10) and neutrophils.
Hepcidin regulates iron metabolism by binding to ferroportin (FPN),
which is the only route for iron export, leading to its
internalization and degradation (11). As a key element of iron metabolism,
hepcidin is influenced by factors such as iron storage,
inflammation, anoxia and erythropoiesis (12). The most two important signaling
pathways for hepcidin expression are the bone morphogenetic protein
6 (BMP6) (13) and Janus
kinase2/signal transducer and activator of transcription 3
(JAK2/STAT3) signal pathways (14). In the BMP6 signaling pathway, BMP6
binds to its receptor (BMPR) to activate the phosphorylation of
mothers against decapentaplegic homolog (SMAD) 1, 5 and 8, which
subsequently leads to the induction of hepcidin transcription. An
integrated BMP signaling pathway is necessary for maintaining the
expression of hepcidin; mutations or defects in any of the involved
proteins lead to a reduction in hepcidin levels. For example,
deletion of the genes encoding the BMP6 ligand (15), BMPR1α (16), the BMP receptor hemojuvelin
(13) and SMAD 4 (17), or addition of the BMP ligand
inhibitor HJV.Fc (18), BMPR1α-Fc,
and the BMP1Rα-LDN-193189 inhibitor (16) can inhibit the expression of
hepcidin. Furthermore, increase in the levels of interleukin (IL)-6
can also promote hepcidin expression via the JAK2/STAT3 signaling
pathway (19,20). Many studies have demonstrated that
iron metabolism is associated with insulin resistance, diabetes,
and obesity, and that hepcidin serves an important role in these
conditions (21–24). For example, in some types of
obesity (25) or in lean juveniles
(26), abnormality in iron storage
and certain metabolic risk markers are observed. Furthermore, the
hepcidin level in the urine, serum and liver in patients with
dysmetabolic iron overload syndrome was significantly higher than
in patients with hereditary hemochromatosis or insulin resistance
without iron overload (27,28).
It was also observed that hepcidin levels in obese children with
nonalcoholic fatty liver disease was significantly higher than in
obese children without the disease (29). In addition, hepcidin and ferritin
are thought to be associated with the inflammatory status in
obesity and type 2 diabetes (22,23).
One study has reported that iron overload can also induce insulin
resistance in visceral adipose tissue as a result of hepcidin
upregulation (24).
The iron nutritional status is associated with bone
metabolic abnormalities. For example, Weinberg (30,31)
indicated that iron overload was a risk factor for osteoporosis.
Additionally, iron overload was recently reported to be associated
with osteopenia, osteoporosis and osteomalacia (32–35).
In addition, as an important iron regulatory factor, hepcidin is
regarded as a therapeutic target for the treatment of osteoporosis
in menopausal women (36).
However, the molecular mechanism of iron metabolism under
conditions of iron overload in patients with osteoporosis is
unclear, especially regarding the role of hepcidin signaling
pathways.
The insulin receptor substrate (IRS) family of
proteins functions as a substrate for insulin receptors, and they
comprise four members: IRS1, 2, 3 and 4. IRS1 is composed of 1231
amino acids and is mainly distributed in the muscle and liver. It
serves a crucial role in the insulin signaling pathway. In the
present study, a mouse strain carrying a spontaneous mutation that
abolishes IRS-1 activity (IRS−/−) was used. These mice
were lean and the body development in embryonic phase was slow, and
they had a lifespan of <2 weeks on average. These mice carried a
nonsense mutation of the 57th serine (substitution of nucleotide C
with A) of insulin receptor substrate 1 (IRS1). A mutation of the
57th amino acid of this protein led to the disappearance of its
expression. In a previous preliminary study, it was demonstrated
that these IRS−/− mice manifested osteogenesis
imperfecta and adipogenesis imperfecta, and that the expression of
BMPR1α, an important receptor in the hepcidin signaling pathway,
was upregulated (37). Thus, this
mouse model is suitable for investigation of iron metabolism in
insulin metabolic abnormalities, where osteogenesis imperfecta is
manifested as one of the main pathological changes.
Therefore, the present study hypothesized that iron
metabolism is regulated via the insulin signaling pathway, which
also can induce bone metabolism changes. In the present study, the
role of hepcidin in iron metabolism and the signaling pathways
involved in the IRS−/− mouse model were investigated.
Additionally, an attempt was made to provide insight into the
association between iron metabolism and the bone metabolic
abnormalities by inducing an iron overload condition in
osteoblasts, where hepcidin signaling pathway serves an important
role.
Materials and methods
Animal model
The animals used were four 4-week-old wild-type
C57BL/6J female mice (IRS+/+ mice) and four 4-week-old
IRS+/− male mice heterozygous for IRS-1 provided by
Prof. Zhou (Institute of Endocrinology and Metabolism, The Second
Xiang-Ya Hospital of Central South University, Hunan, China). The
present study was approved by the Ethical Committee of the Second
Xiang-Ya Hospital of Central South University. The mice were bred
in a sterile environment at a humidity of 50±10% that was
temperature and light controlled (12-h night/12-h day cycle,
22±1°C), and they were provided adequate food and water. Mice with
different sex and genotypes were bred together due to the research
design. Three weeks after birth, offspring were weaned off the
mothers and their genotype was ascertained. Mice with different
genotypes were used in this study, including wild-type mice
IRS+/+, heterozygote mice IRS+/− and
homozygote mice IRS−/−. Mice were sacrificed 6 months
following breeding, biochemical-associated analyses were then
carried out.
Genotype identification
Tail DNA was extracted via the DNA extraction kit
(Universal Genomic DNA Extraction kit, Takara Bio Inc., Otsu,
Japan), after which polymerase chain reaction (PCR) amplification
and enzyme cleavage were carried out. The enzyme-digested product
was subsequently used for polyacrylamide gel electrophoresis.
Firstly, 10 ml 12% polyacrylamide gel were prepared using 4 ml
polyacrylamide, 3.39 ml double distilled water, 2 ml 5×
tetrabromoethane, 70 µl 10% ammonium persulfate and 3.5 µl
tetramethylethylenediamine, all electrophoresis related reagents
were purchased from Beyotime Institute of Biotechnology (Shanghai,
China). Lastly, the genotype was determined using the gel imaging
analysis system.
Western blot analysis
Mice were sacrificed by cervical dislocation after 6
months of breeding, livers were then extracted for analysis. Liver
tissues were irrigated with sterile PBS to remove residual blood
and cut into small pieces. Liver samples were ground post-fixation
with liquid nitrogen; ultrasonic degradation was then used for
tissue fluid extraction. The protein concentration was determined
using the Bicinchronic Acid protein assay kit (Thermo Fisher
Scientific Inc., Waltham, MA, USA). Proteins (10 µg/well) were
separated by 10% SDS-PAGE and transferred to a nitrocellulose
membrane. After blocking the membrane with 5% bovine serum albumin
in phosphate-buffered saline containing 0.1% Tween 20 at room
temperature for 1 h, the blotted membrane was probed with the
relevant antibodies at 4°C overnight, including IRS-1 (1:1,000;
cat. no. ab40777, Abcam, Cambridge, MA, USA), IL-6 (1:1,000; cat.
no. 12912, Cell Signaling Technologies, Inc., Danvers, MA, USA),
hepcidin-25 (1:100; cat. no. ab30760, Abcam), β-actin (1:1,000;
cat. no. 3700, CST Biological Reagents Co, Ltd, Shanghai, China),
BMPR1α (1:1,000; cat. no. ab38560, Abcam), GAPDH (1:1,000; cat. no.
5174, Cell Signaling Technologies, Inc.) and ferritin (1:2,000;
cat. no. ab75973, Abcam) at specified dilutions. Following this,
membranes were incubated with horseradish peroxidase-conjugated
secondary antibodies at 37°C for 1 h (1:10,000; cat. no.
ab97051/ab6789, Abcam). Blots were detected using enhanced
chemiluminescent western blotting detection reagents (Amersham; GE
Healthcare Life Sciences, Uppsala, Sweden). Membranes were probed
with β-actin and GAPDH (as controls) to ensure equal loading of
proteins.
Immunohistochemistry analysis
After mice sacrifice, liver and jaw bone tissue
samples of the three mouse models were obtained and the jaw bone
tissues were decalcified using 10% EDTA (38). These tissues were deposited in
paraffin-recipient blocks. Subsequently, the paraffin tissue
sections were dewaxed and rehydrated in a descending alcohol series
(100, 95, 90, 80, 70 and 50%). For antigen retrieval, the slides
were heated in 0.01 M sodium citrate buffer at 100°C for 3 min.
After blocking with 5% bovine serum albumin (cat. no. ST023,
Beyotime Institute of Biotechnology) for 1 h at 37°C, the slides
were incubated with anti-hepcidin antibody (1:200; cat. no.
ab30760, Abcam) overnight at 4°C. The slides were subsequently
washed three times with phosphate-buffered saline and incubated
with a horseradish peroxidase-conjugated secondary antibody (1:100;
cat. no. ab97100; Abcam) for 1 h at room temperature. Each slide
was treated with a diaminobenzidine solution (1:25; cat. no. P0203;
Beyotime Institute of Biotechnology) at room temperature for 5 min,
and then prepared for microscope observation.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), and the total RNA concentration
was determined using an ultraviolet spectrophotometer.
Subsequently, the RNA was reverse transcribed to cDNA with an RT
reagent kit (Takara Bio Inc.). RT-qPCR was carried out using SYBR
green (Takara Bio Inc.) using the following primers: GAPDH:
Forward, 5′-TGGCAAAGTGGAGATTGTT-3′ and reverse,
5′-CTTCTGGGTGGCAGTGAT-3′; Hepcidin: Forward,
5′-CACCAACTTCCCCATCTGCATCTT-3′ and reverse,
5′-GAGGGGCTGCAGGGGTGTAGAG-3′; transferrin receptor (TfR)1: Forward,
5′-TGCTATAGGTCCTGAGGGCAT-3′ and reverse,
5′-GGCATACAGCTCAATGGAAGA-3′; TfR2: Forward,
5′-GAGTTGTCCAGGCTCACGTACA-3′ and reverse:
5′-GCTGGGACGGAGGTGACTT-3′; FPN; Forward,
5′-GTCATCCTCTGCGGAATCATCCTGA-3′ and reverse,
5′-GAGACCCATCCATCTCGGAAAGTGC-3′; BMP6: Forward,
5′-AGCACAGAGACTCTGACCTATTTTTG-3′ and reverse,
5′-CCACAGATTGCTAGTTGCTGTGA-3′ and IL-6: Forward,
5′-GTATGAACAACGATGATGCACTTG-3′ and reverse,
5′-ATGGTACTCCAGAAGACCAGAGGA-3′. Thermocycling conditions
constituted 40 cycles, including an initial denaturation at 95°C
for 30 sec, then 95°C for 5 sec, 60°C for 30 sec and 95°C for 5
sec, and was terminated by a final extension at 60°C for 1 min. The
data were analyzed using the 2−∆∆Cq method (39).
Cell culture and treatment
The 3T3-E1 cells were obtained from the Cell Bank of
the Shanghai Infrastructure for Public Research and Development of
the Chinese Academy of Medical Sciences (Shanghai, China). 3T3-E1
cells were incubated in α-minimum essential medium (cat. no.
12571048; Thermo Fisher Scientific, Inc.), containing 10% fetal
bovine serum (cat. no. 10099141; Thermo Fisher Scientific, Inc.),
and the medium was changed every other day. When cells reached
70–80% confluence, they were digested using trypsin-EDTA (cat. no.
25300054; Thermo Fisher Scientific, Inc.). Following this, cells
were harvested and seeded with the same number of cells in each
well of six well plates (100,000 cells/well). Then, complete
culture medium was added per well, and the cells were incubated for
24 h at 37°C. Ferric ammonium citrate (FAC) (F5879, Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) was added at concentrations of 50,
100 and 200 mM to induce iron overload in these cells, and the
plates were incubated for 72 h at 37°C in a 5% CO2
atmosphere. The control group was left untreated.
Statistical analysis
Data are expressed as the mean ± standard deviation
and were analyzed by one-way analysis of variance followed by the
Bonferroni's post hoc test when equal variance was assumed, or
Dunnett's T3 test when equal variance was not assumed. Statistical
analyses were performed in SPSS version 17.0 (SPSS, Inc., Chicago,
IL, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
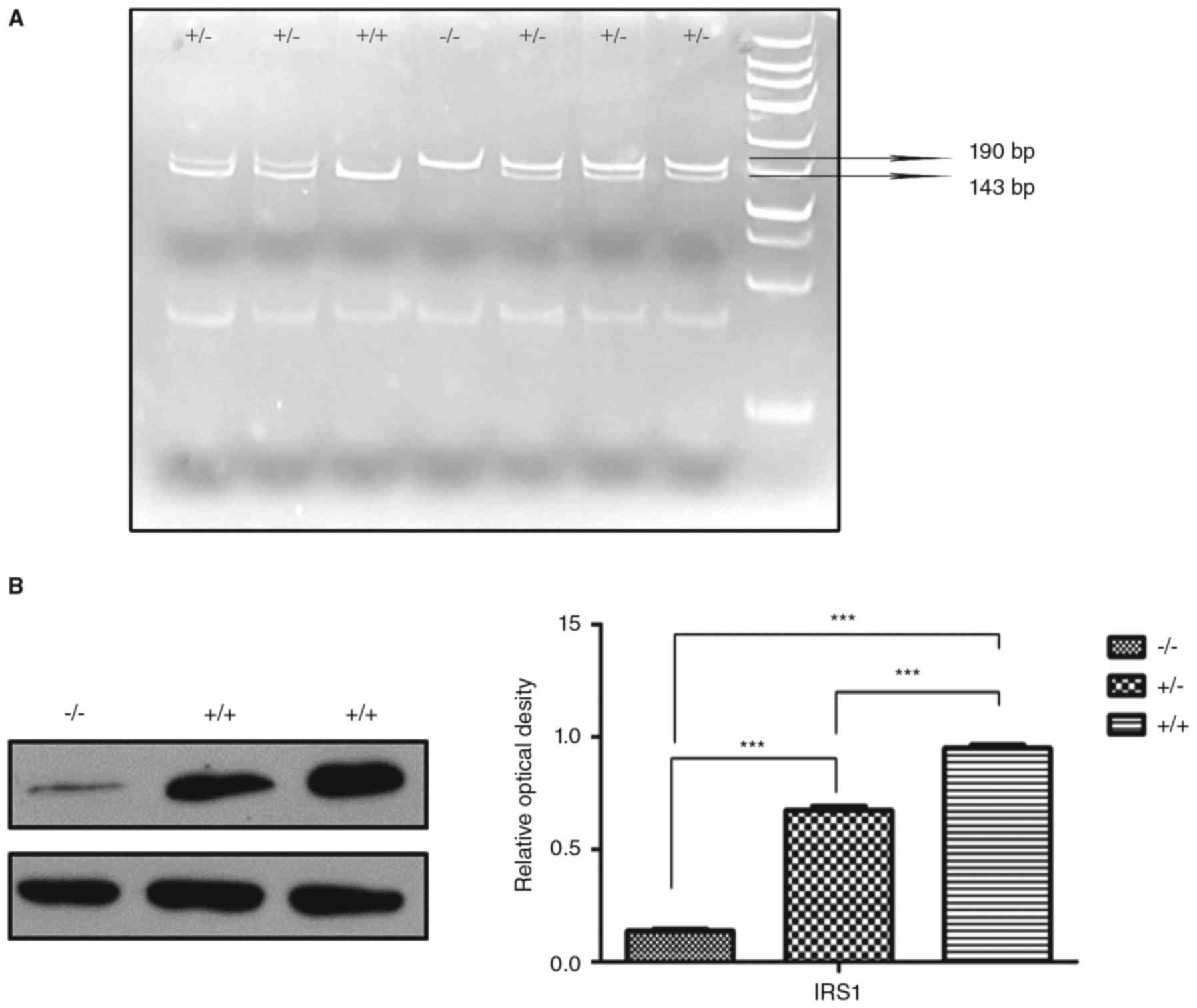
Genotype identification
The IRS−/− mice used in this study were
lean, and their weight was 40–60% of the wild-type mice; this
feature was useful for phenotype identification initially.
Following this, DNA identification was performed using the tail
tissue. The Taq1 enzyme was employed to cut off the DNA at the site
of the C-A nucleotide mutation, and the resulting dissected DNA
fragments were 192 and 143 bp. Gel electrophoresis was employed to
separate the DNA fragments: Wild-type mice only exhibited the
143-bp fragment, the IRS−/− mice only exhibited the
190-bp fragment, and the IRS+/− mice exhibited both the
143 and 190-bp fragments (Fig.
1A). The IRS1 protein levels were detected using western
blotting and were significantly lower in the IRS−/− mice
compared with the IRS+/+ and IRS+/− mice
(Fig. 1B).
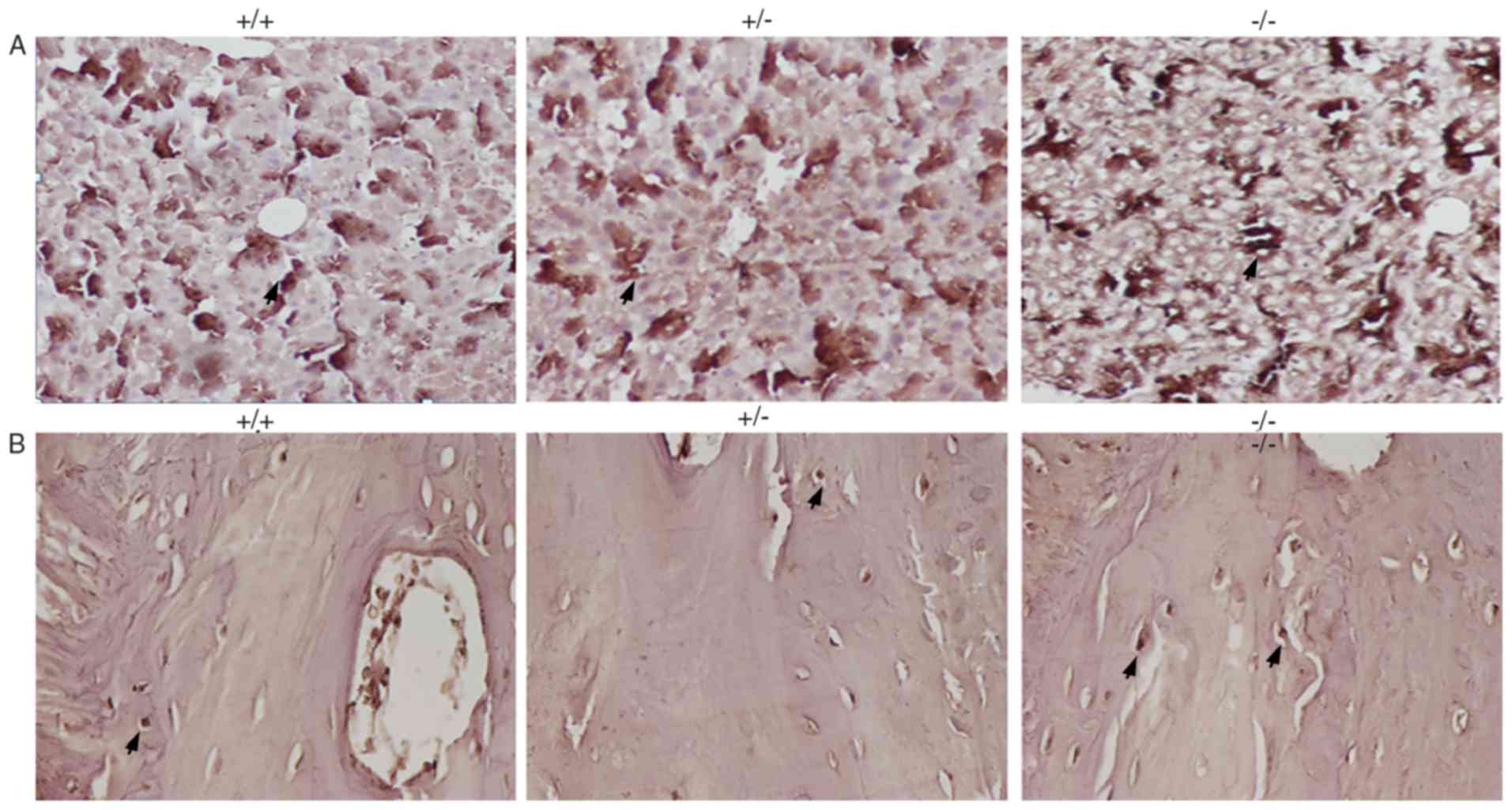
Hepcidin levels in the liver and jaw
bones in the different groups of mice
Hepcidin was widely expressed in the liver (as
indicated by the black arrow in Fig.
2A). Hepcidin levels were significantly higher in the liver in
the IRS−/− mice compared with the IRS+/− and
IRS+/+ mice. Furthermore, hepcidin levels in the
IRS+/− mice were slightly higher compared with the
IRS+/+ mice (Fig. 2A).
On the other hand, hepcidin levels in the jaw bone were low
compared with in the in mice livers. However, it was observed that
the hepcidin level in the jaw bone of IRS−/− mice was
higher compared with in the other two mouse models (Fig. 2B).
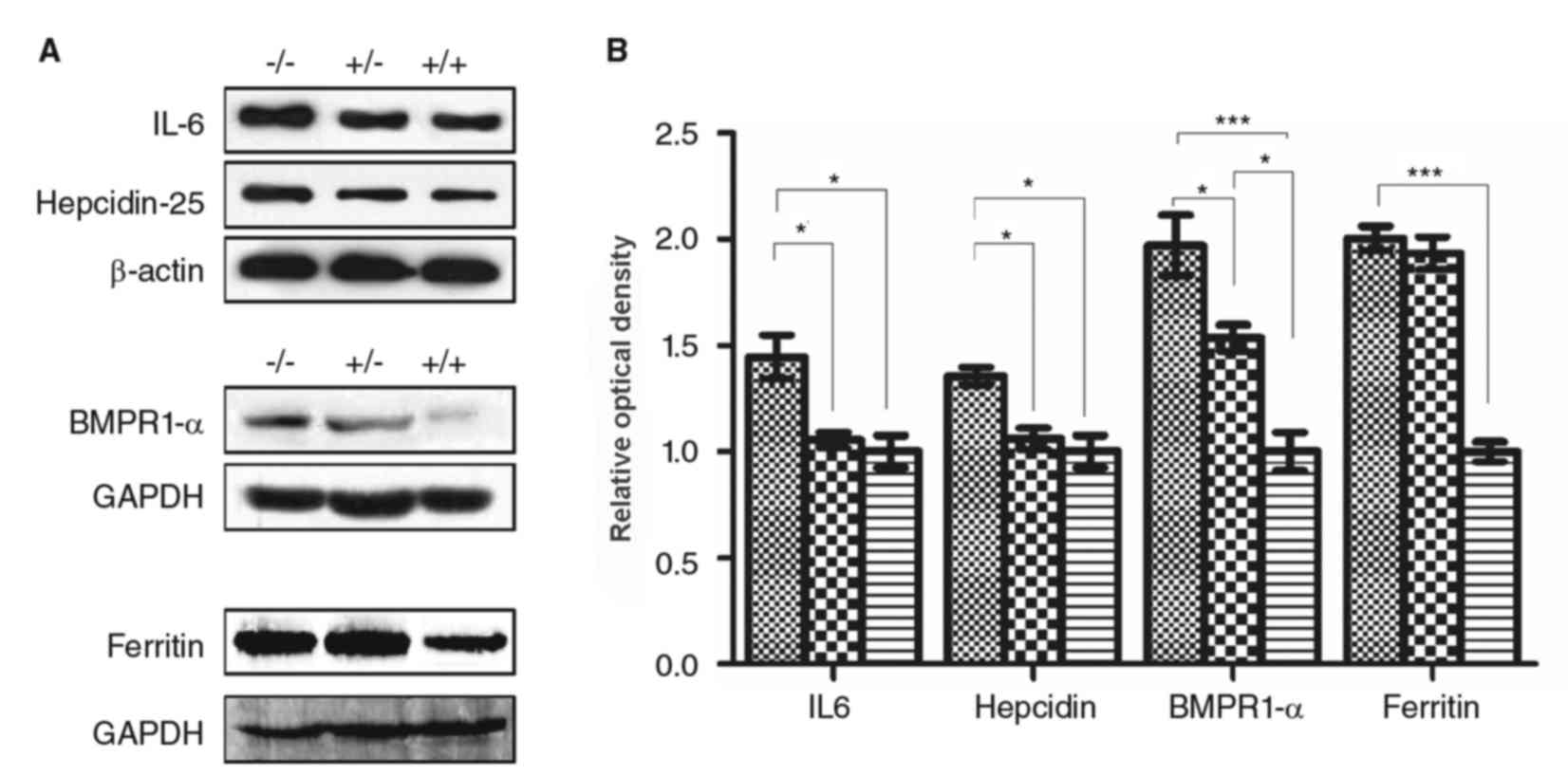
Alterations at the protein and mRNA
levels of hepcidin signaling pathway proteins in the liver of the
different groups of mice
The protein expression levels of hepcidin, IL-6, and
BMPR1 in the liver were higher in the IRS−/− mice
compared with the IRS+/− and IRS+/+ mice
(P<0.05, Fig. 3). Protein
expression levels of BMPR1α and ferritin in IRS−/− mice
were significantly higher compared with the IRS+/+ mice
(P<0.001, Fig. 3).
Additionally, protein levels of BMPR1α in the IRS+/−
mice was significantly higher compared with the IRS+/+
mice (P<0.05, Fig. 3). No
significant differences were observed in the protein expression
levels of hepcidin and IL-6 between IRS+/− and
IRS+/+ groups.
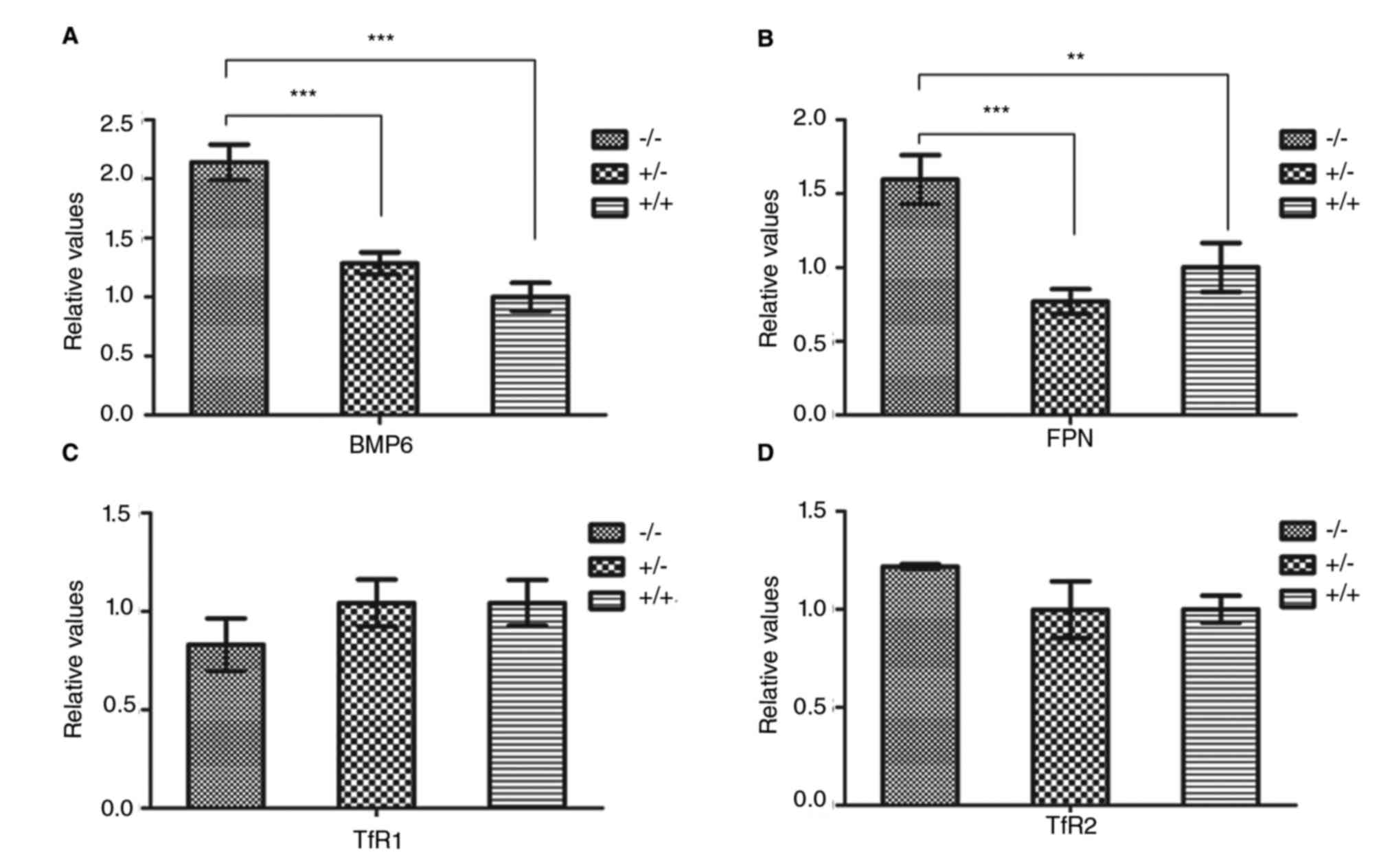
Regarding the mRNA expression levels, the BMP6 mRNA
levels in IRS−/− mice were significantly increased
compared with the other two groups (P<0.001, Fig. 4A). The FPN mRNA levels were
significantly higher in the IRS−/− mice compared with
the IRS+/− (P<0.001, Fig. 4B) and IRS+/+ (P<0.01,
Fig. 4B), while there was no
significant difference between the IRS+/− and
IRS+/+ mice (P<0.05, Fig. 4). There were no significant
differences in the mRNA levels of TfR 1 and 2 among the three mouse
models (P<0.05, Fig. 4C and
D).
Alterations in the mRNA expression
levels of hepcidin signaling pathway proteins in osteoblasts after
ferric ammonium citrate exposure
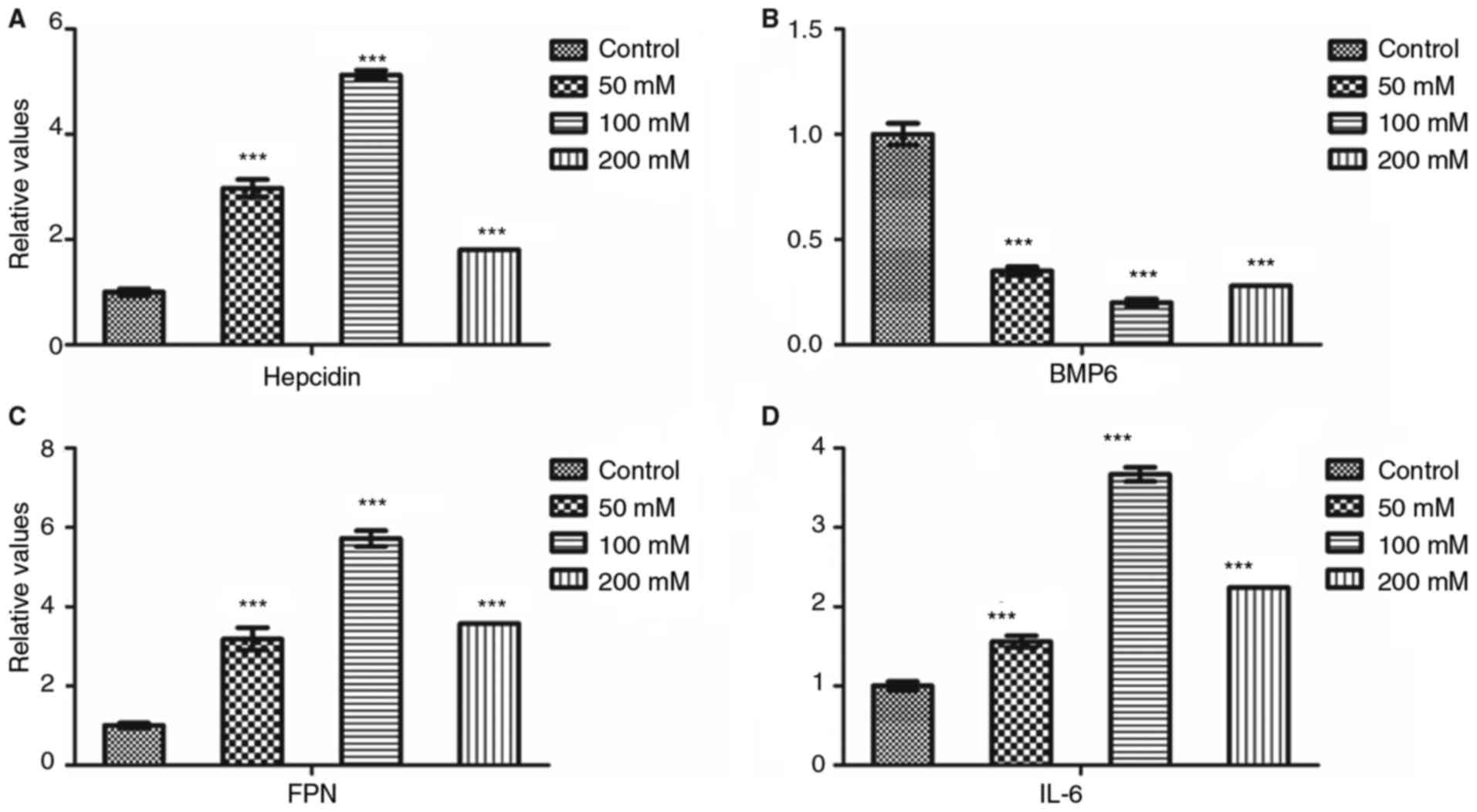
After treatment with FAC at various concentrations,
the mRNA expression levels of hepcidin, FPN and IL-6 significantly
increased compared with the control group (P<0.001, Fig. 5), with the peak mRNA levels
observed at the FAC concentration of 100 mM. However, the BMP6 mRNA
level declined compared with the control group (P<0.001,
Fig. 5), with the minimum level
observed at the concentration of 100 mM.
Discussion
In the present study, the role of hepcidin in iron
metabolism was examined in IRS-deficient mice and osteoblasts with
iron overload induced via exposure to FAC. Furthermore, an attempt
was made to elucidate the signaling pathways involved in the role
of hepcidin in iron metabolism under the aforementioned
conditions.
Hepcidin levels in the liver of IRS−/−
mice was higher compared with in IRS+/− and
IRS+/+ mice. Additionally, the hepatic protein
expression levels of ferritin, which is considered as one of the
main predictors of the iron level, were examined (40,41).
A significant increase in ferritin levels in the IRS−/−
and IRS+/− mice compared with in IRS+/+ mice
was identified. This finding implies that the iron storage levels
in the IRS−/− and IRS+/− mice models are
high, and that iron overload is a possible condition within these
two mouse models. Considering the significantly higher levels of
hepcidin in the IRS−/− mice, hepcidin may be involved
with iron metabolism in this model.
In the present study, the protein expression levels
of BMPR1α were significantly elevated within IRS−/−
mice, in accordance with the data obtained via microRNA and PCR
arrays in a previous study (37).
In order to further understand the role of the BMP signaling
pathway in hepcidin expression within IRS−/− mice, the
mRNA expression level of BMP6 was studied via RT-qPCR analyses. The
BMP6 levels within IRS−/− mice were higher compared with
in the other two genotypes. Thus, upregulation of BMPR1α and BMP6
expression may have induced hepcidin expression in the liver of
this mouse model.
The JAK2/STAT3 signaling pathway is another
important pathway for hepcidin expression, in which IL-6 serves a
key role. A study has reported that the levels of hepcidin and
proinflammatory factors were increased simultaneously in fat tissue
and liver (42). Additionally,
obesity is considered as a moderate inflammatory state
characterized by high levels of hepcidin and TNF-α (28). Furthermore, some studies have
reported that inflammatory factors, including liposaccharide, IL-6,
IL-1-α, and IL-1β can induce hepcidin expression in the liver
(19,43,44).
In the present study, IL-6 expression was significantly increased
in the IRS−/− mice compared with in the other two
models. IL-6 expression may be associated with the inflammatory
status and the presence of adipogenesis imperfecta within
IRS−/− mice. Thus, upregulation of IL-6 expression
probably induced upregulation of hepcidin expression in this
model.
TfR1 and TfR2 levels in the mouse liver were also
examined, in order to understand the iron metabolism in the model
mice. Tf is one of the most important carriers of iron, and many
tissues obtain iron via TfRs. A study has reported that the TfR1
levels in hematochromatosis and non-alcoholic fatty liver disease
(NAFLD) patients with iron overload were lower than those of NAFLD
patients with iron deficiency (28). However, in the present study there
were no significant differences in the TfR levels between the mice
models. Therefore, TfR levels may not be affected within the liver
of IRS−/− mice. Notably, the mRNA expression levels of
FPN, the only iron exportation protein (45), was higher in the IRS−/−
mice than in the IRS+/− and IRS+/+ mice. A
possible reason for this is that increased hepcidin expression in
the IRS−/− mice promoted intracellular iron overload,
leading to higher FPN expression.
The findings of the present study indicated that the
hepcidin and iron levels are increased in the liver of the
IRS−/− mice, and that the BMP6 and IL-6 signaling
pathways are the main factors involved in the alteration of
hepcidin levels. However, alterations in iron status may induce
systemic changes, such as osteoporosis. Previous data have
indicated that iron overload can induce osteoporosis (35,46).
Furthermore, several studies have indicated the direct relationship
between iron overload and osteoporosis, wherein hepcidin serves a
crucial role (47–49). In the present study, osteogenesis
imperfecta was a characteristic of the IRS−/− mice.
Additionally, an increase in the hepcidin levels in the jaw bone of
IRS−/− mice was detected, even though the levels were
low in all animal models. Thus, hepcidin and iron status may be
associated with bone metabolism since abnormal iron metabolism may
induce bone metabolic abnormality, such as osteoporosis.
In order to improve understanding of the association
between hepcidin and bone metabolism the hepcidin signaling
pathways was further investigated in an osteoblast cell line with
iron overload condition. Previous studies have indicated that iron
overload induced biological activity changes to osteoblasts,
including iron metabolism and the osteogenic effect (50–53).
These data indicated that iron may serve an important role in
osteogenesis and that iron deficiency and iron overload may inhibit
osteoblast differentiation and development, respectively.
Furthermore, hepcidin expression is associated with bone
metabolism. Hepcidin promotes osteoblastic differentiation and
mineralization by regulating the expression of alkaline phosphatase
and osteogenic genes (54) or by
increasing intracellular iron levels (48). Hepcidin knockout mice have been
reported to have developed defects in bone microarchitecture and
alterations in bone formation markers (55). In the present study, the mRNA
expression levels of BMP6, an important bone morphogenetic protein
for endochondral ossification, was reduced following FAC treatment
which was not in accordance with the results in the
IRS−/− mice, and this effect was more evident at a
concentration of 100 mM. This may due to the iron overload induced
by FAC, which inhibited the expression of osteogenesis-associated
proteins. Additionally, it has been reported that the expression
level of BMP6 varied across different tissues under iron overload
conditions (56). In the present
study, the IL-6 mRNA expression levels were increased in
osteoblasts after FAC exposure. Excess iron may induce oxidative
stress leading to an inflammatory state, as evidenced by the
increase in the expression of inflammatory factors (57,58).
Thus, IL-6 may be involved in hepcidin induction within osteoblasts
via the JAK2/STAT3 signaling pathway following FAC treatment.
Finally, the increased mRNA expression levels of FPN may due to the
iron overload condition, as FPN promoted iron exportation to
maintain the intracellular iron balance.
In conclusion, the present study indicated that IRS1
deficiency may result in a significant increase in hepcidin
expression and alterations in the expression levels of associated
signaling pathway proteins. The upregulation of hepcidin expression
levels within the liver and jaw bone of IRS efficient mice may due
to the increased expression of BMPR1α and IL-6 via the BMP6 and
JAK2/STAT3 signaling pathways. Therefore, hepcidin may be closely
associated with iron metabolism via the BMP6 and JAK2/STAT3
signaling pathways in the IRS−/− mice model.
Furthermore, IRS1 may serve a role in the hepcidin-associated
signaling pathways; however, further investigation is required.
Finally, iron overload was demonstrated to affect the mRNA
expression levels of the hepcidin-associated genes within
osteoblasts, but this also requires further investigation.
References
|
1
|
Arredondo M and Núñez MT: Iron and copper
metabolism. Mol Aspects Med. 26:313–327. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Doom JR and Georgieff MK: Striking while
the iron is hot: Understanding the biological and
neurodevelopmental effects of iron deficiency to optimize
intervention in early childhood. Curr Pediatr Rep. 2:291–298. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bardou-Jacquet E, de Tayrac M, Mosser J
and Deugnier Y: GNPAT variant associated with severe iron overload
in HFE hemochromatosis. Hepatology. 62:1917–1918. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krause A, Neitz S, Mägert HJ, Schulz A,
Forssmann WG, Schulz-Knappe P and Adermann K: LEAP-1, a novel
highly disulfide-bonded human peptide, exhibits antimicrobial
activity. FEBS Lett. 480:147–150. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park CH, Valore EV, Waring AJ and Ganz T:
Hepcidin, a urinary antimicrobial peptide synthesized in the liver.
J Biol Chem. 276:7806–7810. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bekri S, Gual P, Anty R, Luciani N, Dahman
M, Ramesh B, Iannelli A, Staccini-Myx A, Casanova D, Ben Amor I, et
al: Increased adipose tissue expression of hepcidin in severe
obesity is independent from diabetes and NASH. Gastroenterology.
131:788–796. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aigner E, Felder TK, Oberkofler H, Hahne
P, Auer S, Soyal S, Stadlmayr A, Schwenoha K, Pirich C, Hengster P,
et al: Glucose acts as a regulator of serum iron by increasing
serum hepcidin concentrations. J Nutr Biochem. 24:112–117. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Theurl I, Theurl M, Seifert M, Mair S,
Nairz M, Rumpold H, Zoller H, Bellmann-Weiler R, Niederegger H,
Talasz H and Weiss G: Autocrine formation of hepcidin induces iron
retention in human monocytes. Blood. 111:2392–2399. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang X and Rovin BH: Hepcidin expression
by human monocytes in response to adhesion and pro-inflammatory
cytokines. Biochim Biophys Acta. 1800:1262–1267. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nguyen NB, Callaghan KD, Ghio AJ, Haile DJ
and Yang F: Hepcidin expression and iron transport in alveolar
macrophages. Am J Physiol Lung Cell Mol Physiol. 291:L417–L425.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nemeth E, Tuttle MS, Powelson J, Vaughn
MB, Donovan A, Ward DM, Ganz T and Kaplan J: Hepcidin regulates
cellular iron efflux by binding to ferroportin and inducing its
internalization. Science. 306:2090–2093. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang F, Sun ZZ, Tang YT, Xu C and Jiao
XY: Hepcidin expression and iron parameters change in Type 2
diabetic patients. Diabetes Res Clin Pract. 93:43–48. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Babitt JL, Huang FW, Wrighting DM, Xia Y,
Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, et
al: Bone morphogenetic protein signaling by hemojuvelin regulates
hepcidin expression. Nat Genet. 38:531–539. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wrighting DM and Andrews NC: Interleukin-6
induces hepcidin expression through STAT3. Blood. 108:3204–3209.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meynard D, Kautz L, Darnaud V,
Canonne-Hergaux F, Coppin H and Roth MP: Lack of the bone
morphogenetic protein BMP6 induces massive iron overload. Nat
Genet. 41:478–481. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Steinbicker AU, Bartnikas TB, Lohmeyer LK,
Leyton P, Mayeur C, Kao SM, Pappas AE, Peterson RT, Bloch DB, Yu
PB, et al: Perturbation of hepcidin expression by BMP type I
receptor deletion induces iron overload in mice. Blood.
118:4224–4230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang RH, Li C, Xu X, Zheng Y, Xiao C,
Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L and Deng CX:
A role of SMAD4 in iron metabolism through the positive regulation
of hepcidin expression. Cell Metab. 2:399–409. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Babitt JL, Huang FW, Xia Y, Sidis Y,
Andrews NC and Lin HY: Modulation of bone morphogenetic protein
signaling in vivo regulates systemic iron balance. J Clin Invest.
117:1933–1939. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nemeth E, Rivera S, Gabayan V, Keller C,
Taudorf S, Pedersen BK and Ganz T: IL-6 mediates hypoferremia of
inflammation by inducing the synthesis of the iron regulatory
hormone hepcidin. J Clin Invest. 113:1271–1276. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fleming RE: Iron and inflammation:
Cross-talk between pathways regulating hepcidin. J Mol Med (Berl).
86:491–494. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Al-Hakeim HK, Al-Khakani MM and Al-Kindi
MA: Correlation of hepcidin level with insulin resistance and
endocrine glands function in major thalassemia. Adv Clin Exp Med.
24:69–78. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Andrews M, Soto N and Arredondo-Olguin M:
Association between ferritin and hepcidin levels and inflammatory
status in patients with type 2 diabetes mellitus and obesity.
Nutrition. 31:51–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guzmán Andrews M and Olguín Arredondo M:
Association between ferritin, high sensitivity C-reactive protein
(hsCRP) and relative abundance of Hepcidin mRNA with the risk of
type 2 diabetes in obese subjects. Nutr Hosp. 30:577–584.
2014.PubMed/NCBI
|
|
24
|
Dongiovanni P, Ruscica M, Rametta R,
Recalcati S, Steffani L, Gatti S, Girelli D, Cairo G, Magni P,
Fargion S and Valenti L: Dietary iron overload induces visceral
adipose tissue insulin resistance. Am J Pathol. 182:2254–2263.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dubern B, Girardet JP and Tounian P:
Insulin resistance and ferritin as major determinants of abnormal
serum aminotransferase in severely obese children. Int J Pediatr
Obes. 1:77–82. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aigner E, Hinz C, Steiner K, Rossmann B,
Pfleger J, Hohla F, Steger B, Stadlmayr A, Patsch W and Datz C:
Iron stores, liver transaminase levels and metabolic risk in
healthy teenagers. Eur J Clin Invest. 40:155–163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barisani D, Pelucchi S, Mariani R,
Galimberti S, Trombini P, Fumagalli D, Meneveri R, Nemeth E, Ganz T
and Piperno A: Hepcidin and iron-related gene expression in
subjects with dysmetabolic hepatic iron overload. J Hepatol.
49:123–133. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aigner E, Theurl I, Theurl M, Lederer D,
Haufe H, Dietze O, Strasser M, Datz C and Weiss G: Pathways
underlying iron accumulation in human nonalcoholic fatty liver
disease. Am J Clin Nutr. 87:1374–1383. 2008.PubMed/NCBI
|
|
29
|
Demircioğlu F, Görünmez G, Dağıstan E,
Göksügür SB, Bekdaş M, Tosun M, Kızıldağ B and Kısmet E: Serum
hepcidin levels and iron metabolism in obese children with and
without fatty liver: Case-control study. Eur J Pediatr.
173:947–951. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Weinberg ED: Iron loading: A risk factor
for osteoporosis. Biometals. 19:633–635. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Weinberg ED: Role of iron in osteoporosis.
Pediatr Endocrinol Rev. 6 Suppl 1:S81–S85. 2008.
|
|
32
|
Matsushima S, Hoshimoto M, Torii M, Ozaki
K and Narama I: Iron lactate-induced osteopenia in male
Sprague-Dawley rats. Toxicol Pathol. 29:623–629. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guggenbuhl P, Deugnier Y, Boisdet JF,
Rolland Y, Perdriger A, Pawlotsky Y and Chalès G: Bone mineral
density in men with genetic hemochromatosis and HFE gene mutation.
Osteoporos Int. 16:1809–1814. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mahachoklertwattana P, Sirikulchayanonta
V, Chuansumrit A, Karnsombat P, Choubtum L, Sriphrapradang A,
Domrongkitchaiporn S, Sirisriro R and Rajatanavin R: Bone
histomorphometry in children and adolescents with beta-thalassemia
disease: Iron-associated focal osteomalacia. J Clin Endocrinol
Metab. 88:3966–3972. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Isomura H, Fujie K, Shibata K, Inoue N,
Iizuka T, Takebe G, Takahashi K, Nishihira J, Izumi H and Sakamoto
W: Bone metabolism and oxidative stress in postmenopausal rats with
iron overload. Toxicology. 197:93–100. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang XI: Treatment of osteoporosis in
peri- and post-menopausal women with hepcidin. US Patent
20100204122 A1. Filed Feb 11, 2010; issued April 7, 2015.
|
|
37
|
Guo Y, Tang CY, Man XF, Tang HN, Tang J,
Wang F, Zhou CL, Tan SW, Feng YZ and Zhou HD: Insulin receptor
substrate-1 time-dependently regulates bone formation by
controlling collagen Iα2 expression via miR-342. FASEB J.
30:4214–4226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Park S, Kanayama K, Kaur K, Tseng HC,
Banankhah S, Quje DT, Sayre JW, Jewett A and Nishimura I:
Osteonecrosis of the jaw developed in mice: Disease variants
regulated by γδ T cells in oral mucosal barrier immunity. J Biol
Chem. 290:17349–17366. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ramanathan G, Olynyk JK and Ferrari P:
Diagnosing and preventing iron overload. Hemodial Int. 21 Suppl
1:S58–S67. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ratcliffe LE, Thomas W, Glen J, Padhi S,
Pordes BA, Wonderling D, Connell R, Stephens S, Mikhail AI, Fogarty
DG, et al: Diagnosis and management of iron deficiency in CKD: A
summary of the NICE guideline recommendations and their rationale.
Am J Kidney Dis. 67:548–558. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dallalio G, Law E and Means RT Jr:
Hepcidin inhibits in vitro erythroid colony formation at reduced
erythropoietin concentrations. Blood. 107:2702–2704. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nemeth E, Valore EV, Territo M, Schiller
G, Lichtenstein A and Ganz T: Hepcidin, a putative mediator of
anemia of inflammation, is a type II acute-phase protein. Blood.
101:2461–2463. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lee P, Peng H, Gelbart T, Wang L and
Beutler E: Regulation of hepcidin transcription by interleukin-1
and interleukin-6. Proc Natl Acad Sci USA. 102:1906–1910. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bogdan AR, Miyazawa M, Hashimoto K and
Tsuji Y: Regulators of iron homeostasis: New players in metabolism,
cell death, and disease. Trends Biochem Sci. 41:274–286. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tsay J, Yang Z, Ross FP,
Cunningham-Rundles S, Lin H, Coleman R, Mayer-Kuckuk P, Doty SB,
Grady RW, Giardina PJ, et al: Bone loss caused by iron overload in
a murine model: Importance of oxidative stress. Blood.
116:2582–2589. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen L, Zhu Z, Peng X, Wang Y, Wang Y,
Chen M, Wang Q and Jin J: Hepatic magnetic resonance imaging with
T2* mapping of ovariectomized rats: Correlation between iron
overload and postmenopausal osteoporosis. Eur Radiol. 24:1715–1724.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhao GY, Di DH, Wang B, Huang X and Xu YJ:
Effects of mouse hepcidin 1 treatment on osteoclast differentiation
and intracellular iron concentration. Inflammation. 38:718–727.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rossi F, Perrotta S, Bellini G, Luongo L,
Tortora C, Siniscalco D, Francese M, Torella M, Nobili B, Di Marzo
V and Maione S: Iron overload causes osteoporosis in thalassemia
major patients through interaction with transient receptor
potential vanilloid type 1 (TRPV1) channels. Haematologica.
99:1876–1884. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yamasaki K and Hagiwara H: Excess iron
inhibits osteoblast metabolism. Toxicol Lett. 191:211–215. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Messer JG, Kilbarger AK, Erikson KM and
Kipp DE: Iron overload alters iron-regulatory genes and proteins,
down-regulates osteoblastic phenotype, and is associated with
apoptosis in fetal rat calvaria cultures. Bone. 45:972–979. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Messer JG, Cooney PT and Kipp DE: Iron
chelator deferoxamine alters iron-regulatory genes and proteins and
suppresses osteoblast phenotype in fetal rat calvaria cells. Bone.
46:1408–1415. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
He YF, Ma Y, Gao C, Zhao GY, Zhang LL, Li
GF, Pan YZ, Li K and Xu YJ: Iron overload inhibits osteoblast
biological activity through oxidative stress. Biol Trace Elem Res.
152:292–296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li GF, Xu YJ, He YF, Du BC, Zhang P, Zhao
DY, Yu C, Qin CH and Li K: Effect of hepcidin on intracellular
calcium in human osteoblasts. Mol Cell Biochem. 366:169–174. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Shen GS, Yang Q, Jian JL, Zhao GY, Liu LL,
Wang X, Zhang W, Huang X and Xu YJ: Hepcidin1 knockout mice display
defects in bone microarchitecture and changes of bone formation
markers. Calcif Tissue Int. 94:632–639. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kautz L, Besson-Fournier C, Meynard D,
Latour C, Roth MP and Coppin H: Iron overload induces BMP6
expression in the liver but not in the duodenum. Haematologica.
96:199–203. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chang JS, Li YL, Lu CH, Owaga E, Chen WY
and Chiou HY: Interleukin-10 as a potential regulator of hepcidin
homeostasis in overweight and obese children: A cross-sectional
study in Taiwan. Nutrition. 30:1165–1170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Barbosa MC, Dos Santos TE, de Souza GF, de
Assis LC, Freitas MV and Gonçalves RP: Impact of iron overload on
interleukin-10 levels, biochemical parameters and oxidative stress
in patients with sickle cell anemia. Rev Bras Hematol Hemoter.
35:29–34. 2013. View Article : Google Scholar : PubMed/NCBI
|