Introduction
Gastric cancer is the fifth most frequent malignancy
and fourth third most common cause of cancer-associated mortality
worldwide (1). Gastric cancer
incidence exhibits significant regional differences, particularly
in Asian regions (2). There are
~850,000 newly diagnosed cases of gastric cancer and 650,000
mortalities occur worldwide annually (3). The underlying molecular mechanism of
gastric cancer is complex, and it remains poorly understood despite
extensive clinical and basic research efforts. Several factors,
including oncogene protein expression, Helicobacter pylori
infection, dietary factors, tobacco use, alcohol consumption and
obesity, are involved in gastric cancer occurrence and progression
(4–6). Despite advancements in early
detection, treatment and prevention, advanced-stage gastric cancer
remains incurable, with a remarkably poor 5-year survival rate of
~4–5% (7). Therefore, the
mechanisms underlying gastric cancer carcinogenesis and progression
should be elucidated to identify novel biomarkers for diagnosis and
to develop effective therapeutic strategies.
MicroRNAs (miRNAs) are endogenous, highly conserved,
short (20–25 nucleotides) noncoding RNA molecules (8). These small molecules completely or
partially bind to the 3′-untranslated regions (3′-UTRs) of their
target genes to promote mRNA degradation or inhibit mRNA
translation (9). miRNAs
participate in the modulation of greater than one-third of human
genes (10). Studies have
indicated that miRNAs are dysregulated in various types of human
cancers, such as gastric cancer (11), hepatocellular carcinoma (12), bladder cancer (13), ovarian cancer (14) and breast cancer (15). Abnormally expressed miRNAs have
been implicated in tumour initiation and progression, and may
regulate a variety of important physiological events, including
cell proliferation, cell cycle, apoptosis, angiogenesis, migration
and invasion and metastasis (16–18).
Previous studies have also demonstrated that miRNAs may serve as
oncogenes or tumour suppressors depending on the nature of their
target mRNAs (19–21). Thus, miRNAs may be examined to
identify a novel therapeutic treatment for gastric cancer
patients.
miR-197 is aberrantly expressed in several types of
human cancers, and it serves important roles in tumourigenesis and
tumour development (22–24). However, studies that have
investigated the expression levels and biological roles of miR-197
in gastric cancer are sparse. Therefore, the present study examined
the expression of miR-197 in gastric cancer tissues and cell lines,
and investigated its roles and the underlying mechanisms in the
regulation of aggressive behaviours of gastric cancer cells.
Materials and methods
Human gastric cancer clinical
specimens
The present study was approved by the Ethics
Committee of Renhe Hospital (Shanghai, China), and written informed
consent was obtained from all patients between June 2015 and
December 2016. A total of 45 paired gastric cancer tissues and
adjacent non-tumoural tissues were obtained from patients who
received surgery resection at Department of General Surgery, Renhe
Hospital (Shanghai, China). None of the patients were treated with
chemotherapy or radiotherapy prior to surgery. Tissue samples were
immediately frozen in liquid nitrogen following surgical resection,
and stored at −80°C until use.
Cell lines and culture conditions
Human gastric cancer cell lines (SGC-7901, MGC-803,
AGS and BGC-823), an immortalized gastric epithelial cell line
(GES-1) and the 293T cell line were acquired form the American Type
Culture Collection (Manassas, VA, USA). All cells were grown in
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% foetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 50 U/ml
penicillin and 50 µg/ml streptomycin in a humidified atmosphere
containing 5% CO2 at 37°C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues (1 g) or cells
(1.5×106) using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), following to the manufacturer's protocol.
RNA concentrations were determined using a NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). To detect
miR-197 expression, reverse transcription was conducted using a
TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.), and the TaqMan miRNA assay
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to
examine miR-197 expression, with U6 as an internal control. The
cycling conditions for qPCR were as follows: 50°C for 2 min, 95°C
for 10 min; 40 cycles of denaturation at 95°C for 15 sec and
annealing/extension at 60°C for 60 sec. To quantify MTDH mRNA
expression levels, total RNA was reverse transcribed to cDNA using
a PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.,
Dalian, China). The relative expression levels of MTDH were
determined with SYBR Premix Ex Taq (Takara Biotechnology Co.,
Ltd.), and normalized to GAPDH expression. The amplification was
performed with cycling conditions as follows: 95°C for 5 min,
followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec. The
primers were designed as follows: miR-197,
5′-ATTACTTTGCCCATATTCATTTTGA-3′ (forward) and
5′-ATTCTAGAGGCCGAGGCGGCCGACATGT-3′ (reverse); U6,
5′-CGCTTCGGCAGCACATATAC-3′ (forward), and
5′-TTCACGAATTTGCGTGTCAT-3′ (reverse); MTDH,
5′-AAATAGCCAGCCTATCAAGACTC-3′ (forward) and
5′-TTCAGACTTGGTCTGTGAAGGAG-3′ (reverse); and GAPDH,
5′-GCACCGTCAAGGCTGAGAAC-3′ (forward) and 5′-TGGTGAAGACGCCAGTGGA-3′
(reverse). The relative quantification was calculated using the
2−ΔΔCq method (25).
Cell transfection
The miR-197 mimics and corresponding negative
control miRNA (miR-NC) were synthesized and purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). The miR-197 mimic sequence
was 5′-UUCACCACCUUCUCCACCCCAGC-3′ and the miR-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. The MTDH overexpression plasmid
(pcDNA3.1-MTDH) and empty control plasmid (pcDNA3.1) were obtained
from Origene Technologies, Inc. (Rockville, MD, USA). Cells
(7×105 cells/well) were seeded in 6-well plates and
transfected with oligonucleotides (100 pmol) and/or plasmids (2 µg)
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Following a 6 h
incubation at 37°C, the culture medium was replaced with fresh DMEM
containing 10% FBS. Cell Counting Kit-8 assay, Matrigel invasion
assay, RT-qCPR and western blot analysis were performed 24, 48, 48
and 72 h post-transfection, respectively.
Cell Counting kit-8 (CCK-8) assay
Cell proliferation was determined by a CCK-8 assay
(Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according
to the manufacturer's protocol. Briefly, transfected cells were
collected at 24 h post-transfection, and seeded (3×103
cells/well) into 96-well plates. Proliferation was monitored
everyday for 4 days. A total of 10 µl CCK8 solution was added into
each well and cultured for 2 h in a 37°C incubator with 5%
CO2. The absorbance at a wavelength of 450 nm was
measured using a SpectraMAX Plus microplate reader (Molecular
Devices, LLC, Sunnyvale, CA, USA). Each assay was performed in
triplicate and repeated three times.
Matrigel invasion assay
Transwell invasion assays were performed to examine
the cell invasive abilities using Transwell chambers (8 µm; BD
Biosciences, San Jose, CA, USA) coated with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA). Transfected cells were
collected and resuspended in FBS-free DMEM medium. A total of
5×104 transfected cells in FBS-free DMEM medium were
seeded in the top chamber. DMEM supplemented with 10% FBS was added
to the lower chamber and served as a chemoattractant. Following 48
h incubation at 37°C in 5% CO2, the cells remaining on
the upper side were wiped away with cotton swabs and the Transwell
chambers were washed with PBS, fixed in 95% methanol at room
temperature for 15 min, stained with 0.1% crystal violet at room
temperature for 15 min and air-dried. The invasive cells were
imaged and five independent fields were counted with an Olympus
IX53 microscope (Olympus Corporation, Tokyo, Japan). The assay was
repeated at least three times.
Bioinformatics analysis
Bioinformatics analysis was conducted to predict the
candidate genes of miR-197 using TargetScan (http://targetscan.org) and miRanda (http://www.microrna.org/microrna/home.do).
Dual-luciferase reporter assay
MTDH was predicted to be a potential target of
miR-197 by bioinformatics analysis. The luciferase reporter
vectors, pMiR-MTDH-3′UTR wild-type (Wt) and pMiR-MTDH-3′UTR
mutant-type (Mut) were synthesized and confirmed by Shanghai
GenePharma, Co., Ltd. 293T cells were seeded in 24-well plates
until 60–70% confluent. Subsequently, cells were transfected with
either pMiR-MTDH-3′UTR Wt (0.2 µg) or pMiR-MTDH-3′UTR Mut (0.2 µg),
and co-transfected with either miR-197 mimics (50 pmol) or miR-NC
(50 pmol) were transfected into 293T cells using Lipofectamine
2000, following the manufacturer's protocol. Following 48 h
transfection, luciferase activities were detected using the Dual
Luciferase Reporter assay system (Promega Corporation, Madison, WI,
USA). Renilla luciferase activities were used to normalize
firefly luciferase activities. The experiment was repeated three
times, and each was performed in triplicate.
Western blot analysis
Cells (1.5×106) or tissue specimens (1 g)
were lysed in radioimmunoprecipitation lysis buffer (Beyotime
Institute of Biotechnology, Haimen, China) containing protease
inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA).
Protein quantification was performed using a Bicinchoninic Acid
Protein assay (Pierce; Thermo Fisher Scientific, Inc.). Equal
amounts of protein (30 µg) were separated by 10% SDS-PAGE and
electrotransferred to polyvinylidene difluoride membranes (EMD
Millipore, Billerica, MA, USA). Following blocking with 5% non-fat
milk in Tris-buffered saline containing 0.05% Tween-20 (TBST) at
room temperature for 1 h, the membranes were incubated overnight at
4°C with primary antibodies against: MTDH (1:1,000 dilution; cat.
no. sc-517220; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
PTEN (sc-133197; 1:1,000 dilution), p-AKT (sc-271966; 1:1,000
dilution), AKT (sc-81434; 1:1,000 dilution) and GAPDH (sc-32233;
1:1,000 dilution) (all from Santa Cruz Biotechnology, Inc.).
Membranes were washed with TBST and probed with horseradish
peroxidase-conjugated goat-anti mouse secondary antibody (1:5,000
dilution; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) at room
temperature for 2 h. Immunoreactive bands were visualized using an
Enhanced Chemiluminescence Detection kit (Thermo Fisher Scientific,
Inc.). Quantification of band intensity was performed with Quantity
One software (version 4.62; Bio-Rad Laboratories, Inc., Hercules,
CA, USA). GAPDH served as a loading control.
Statistical analysis
Data are expressed as the mean ± standard deviation,
and all statistical analyses were performed with a two-tailed
Student's t test or one-way analysis of variance followed by a
Student-Newman-Keuls test, using SPSS version 16.0 software (SPSS,
Inc., Chicago, IL, USA). Correlation of miR-197 expression with
that of MTDH mRNA expression was conducted with the Spearman's
correlation analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
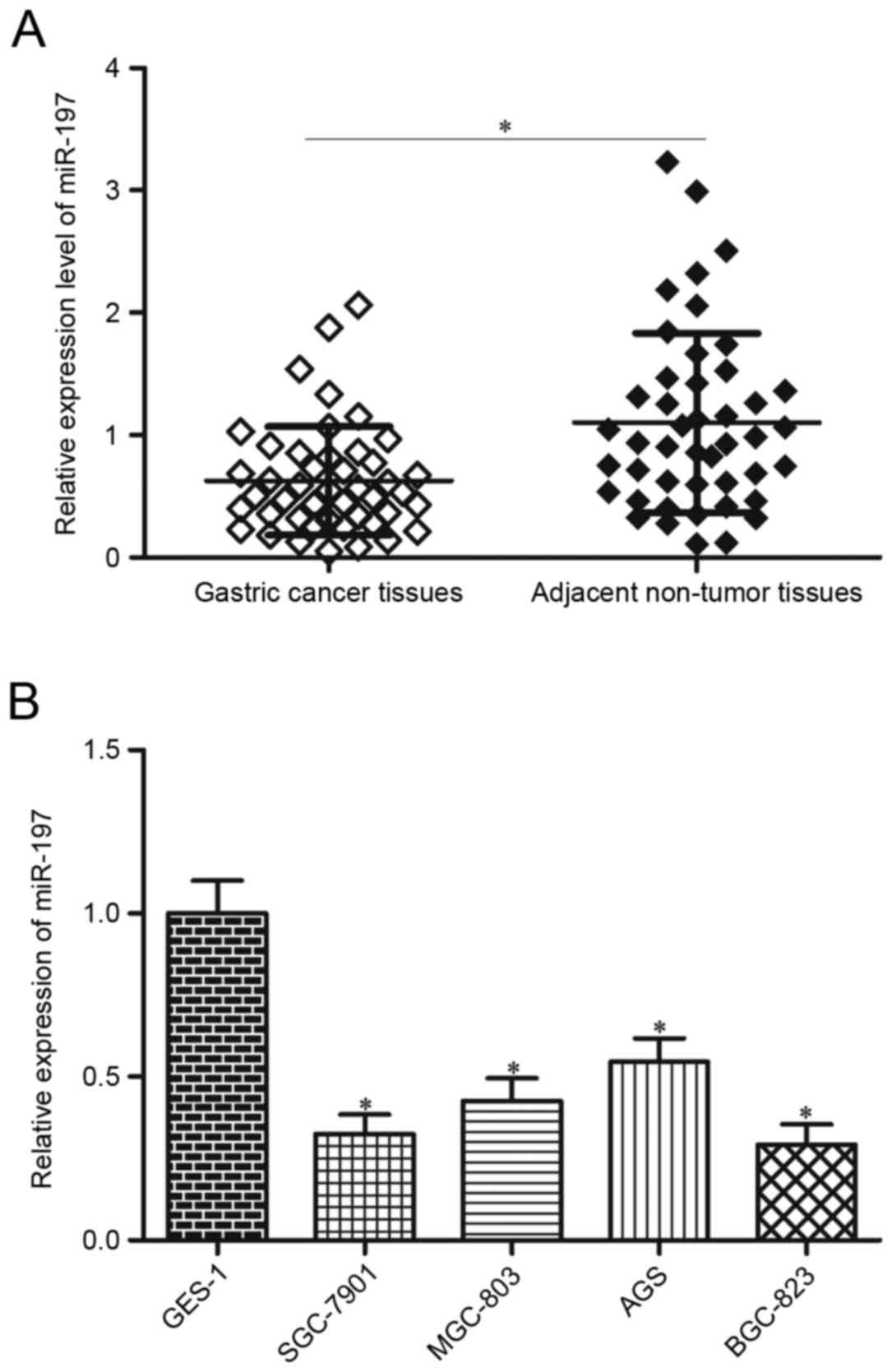
miR-197 is downregulated in gastric
cancer tissues and cell lines
miR-197 expression was measured in 45 pairs of
gastric cancer tissues and adjacent non-tumoural tissues by RT-qPCR
to evaluate the significance of miR-197 in gastric cancer.
Expression of miR-197 was significantly lower in gastric cancer
tissues compared with expression in adjacent non-tumoural tissues
(P<0.05; Fig. 1A). In addition,
associations between miR-197 expression and clinicopathological
factors of patients with gastric cancer were evaluated. Patients
were divided into the following groups according to their median
miR-197 values (Table I):
Low-miR-197 group (n=23) and high-miR-197 group (n=22). Low miR-197
expression was significantly associated with tumour size (P=0.011),
invasive depth (P=0.005), tumour nodes metastasis (TNM) staging
(P=0.011) and lymph node metastasis (P=0.004). However, no
significant association was observed between miR-197 expression and
age (P=0.626), sex (P=0.672) or differentiation (P=0.668).
 | Table I.Association between microRNA-197
expression and clinicopathological factors of patients with gastric
cancer. |
Table I.
Association between microRNA-197
expression and clinicopathological factors of patients with gastric
cancer.
| Clinicopathological
factor | n | Low miR-197 | High miR-197 | P-value |
|---|
| Age (years) |
|
|
| 0.626 |
|
<55 | 18 | 10 | 8 |
|
|
≥55 | 27 | 13 | 14 |
|
| Sex |
|
|
| 0.672 |
|
Male | 28 | 15 | 13 |
|
|
Female | 17 | 8 | 9 |
|
| Tumour size
(cm) |
|
|
| 0.011 |
|
<5 | 20 | 6 | 14 |
|
| ≥5 | 25 | 17 | 8 |
|
|
Differentiation |
|
|
| 0.668 |
| Well
and moderate | 19 | 9 | 10 |
|
|
Poor | 26 | 14 | 12 |
|
| Invasive depth |
|
|
| 0.005 |
|
T1+T2 | 21 | 6 | 15 |
|
|
T3+T4 | 24 | 17 | 7 |
|
| TNM staging |
|
|
| 0.011 |
|
I–II | 22 | 7 | 15 |
|
|
III–IV | 23 | 16 | 7 |
|
| Lymph node
metastasis |
|
|
| 0.004 |
| No | 25 | 8 | 17 |
|
|
Yes | 20 | 15 | 5 |
|
miR-197 expression levels were also examined in a
panel of gastric cancer cell lines (SGC-7901, MGC-803, AGS and
BGC-823) and an immortalised normal gastric epithelial cell line
(GES-1). miR-197 expression levels in all of the gastric cancer
cell lines were significantly decreased compared with that of GES-1
(P<0.05; Fig. 1B). These
results suggested that miR-197 may serve important roles in gastric
cancer progression. Notably, SGC-7901 and BGC-823 cells expressed
the lowest levels of miR-197; thus, these two cell lines were
selected as models for subsequent experiments.
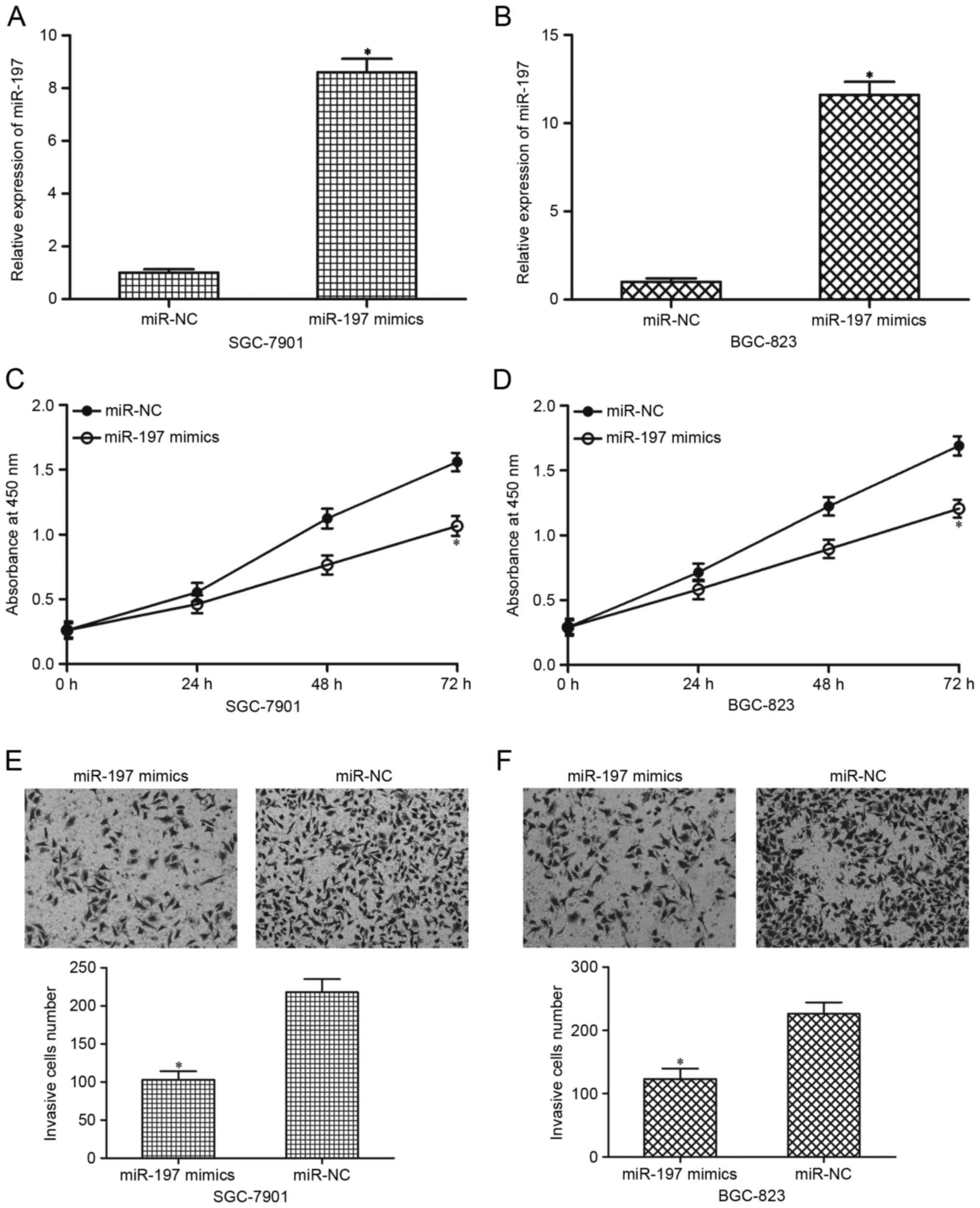
miR-197 overexpression inhibits
proliferation and invasion of gastric cancer cells
miR-197 mimics were transfected into SGC-7901 and
BGC-823 cells to overexpress the total miR-197 levels and to
investigate the biological roles of miR-197 in gastric cancer
progression. RT-qPCR analysis demonstrated that miR-197 was
significantly upregulated in SGC-7901 and BGC-823 cells following
transfection with miR-197 mimics compared with cells transfected
with miR-NC (P<0.05; Fig. 2A and
B, respectively). Subsequently, the effects of miR-197
overexpression on the proliferation and invasion of SGC-7901 and
BGC-823 cells were investigated. CCK-8 assays demonstrated that
miR-197 overexpression suppressed SGC-7901 and BGC-823 cell
proliferation compared with the miR-NC (P<0.05; Fig. 2C and D, respectively). Results from
the Matrigel invasion assays revealed that overexpression of
miR-197 decreased the invasive capacities of the SGC-7901 and
BGC-823 cells compared with the miR-NC (P<0.05; Fig. 2E and F). These results suggested
that miR-197 may act as a tumour suppressor in gastric cancer.
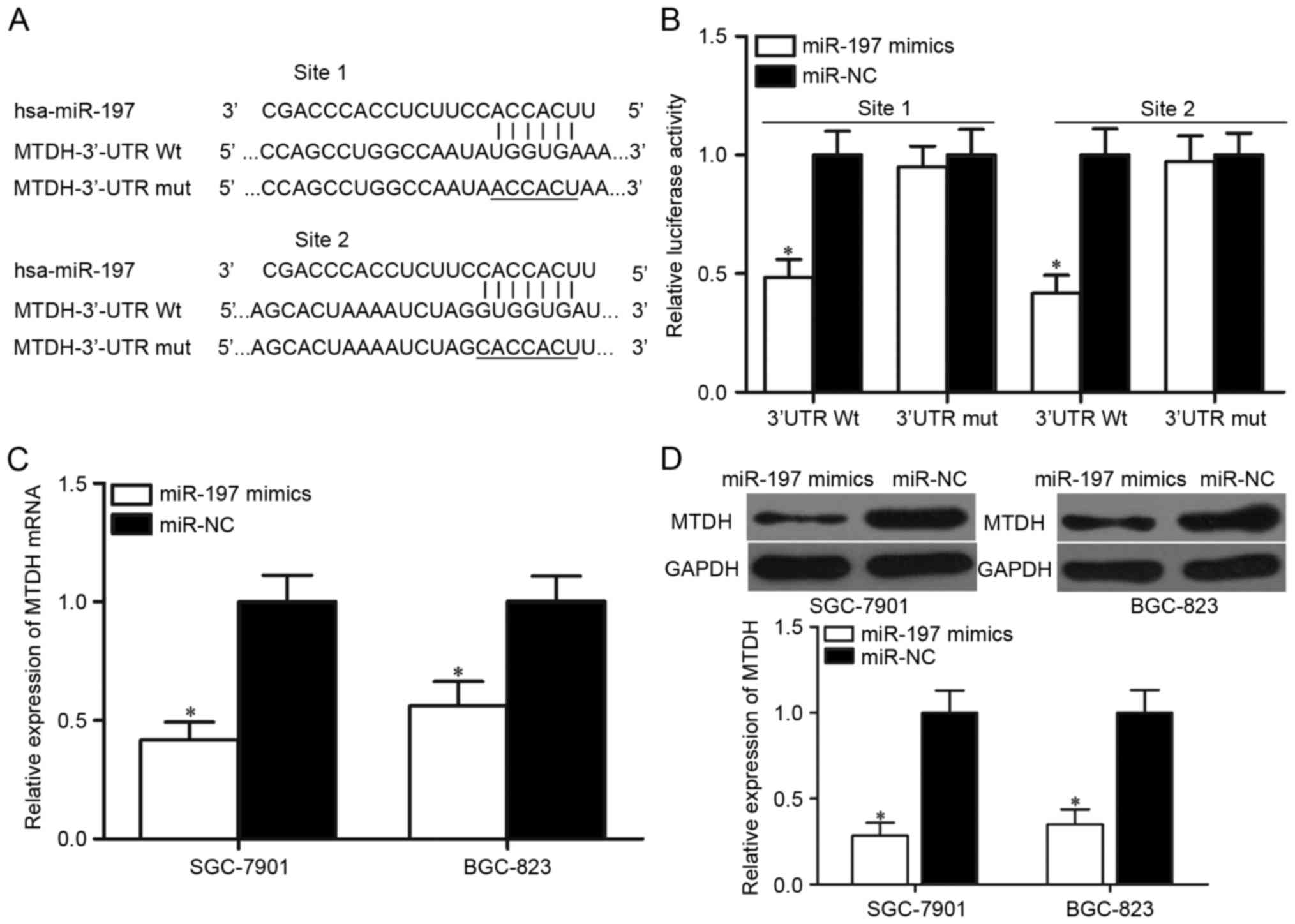
miR-197 directly targets MTDH by
binding to its 3′-UTR to inhibit MTDH expression
Bioinformatics analysis was performed to predict
candidate target genes and to determine the molecular mechanism
underlying miR-197-mediated suppression of gastric cancer cell
proliferation and invasion. Among the predicted targets, MTDH was
selected for further analysis, as it has been reported to be highly
expressed in gastric cancer and may serve oncogenic roles in
gastric cancer initiation and progression (26,27).
Two putative target sites of miR-197 in the 3′-UTR of MTDH were
identified (Fig. 3A).
Luciferase reporter assays were performed to
determine whether MTDH is a direct target of miR-197. 293T cells
were transfected with MTDH-3′-UTR Wt or MTDH-3′-UTR Mut and
co-transfected with miR-197 mimics or miR-NC. Overexpression of
miR-197 significantly decreased the luciferase activities of
MTDH-3′-UTR Wt site 1 and MTDH-3′-UTR Wt site 2 in 293T cells
compared with cells transfected with the miR-NC (P<0.05;
Fig. 3B). By contrast, no
significant suppression of luciferase activity was observed on the
MTDH-3′-UTR Mut reporter plasmid following miR-197 mimics
transfection. In addition, the influence of miR-197 overexpression
on the levels of MTDH mRNA and protein expression were determined
in SGC-7901 and BGC-823 cells transfected with miR-197 mimics or
miR-NC. RT-qPCR and western blot analysis results indicated that
miR-197 overexpression resulted in decreased MTDH expression in the
SGC-7901 and BGC-823 cells at both the mRNA (P<0.05; Fig. 3C) and the protein (P<0.05;
Fig. 3D) level. These results
suggested that MTDH may be a direct target gene of miR-197 in
gastric cancer.
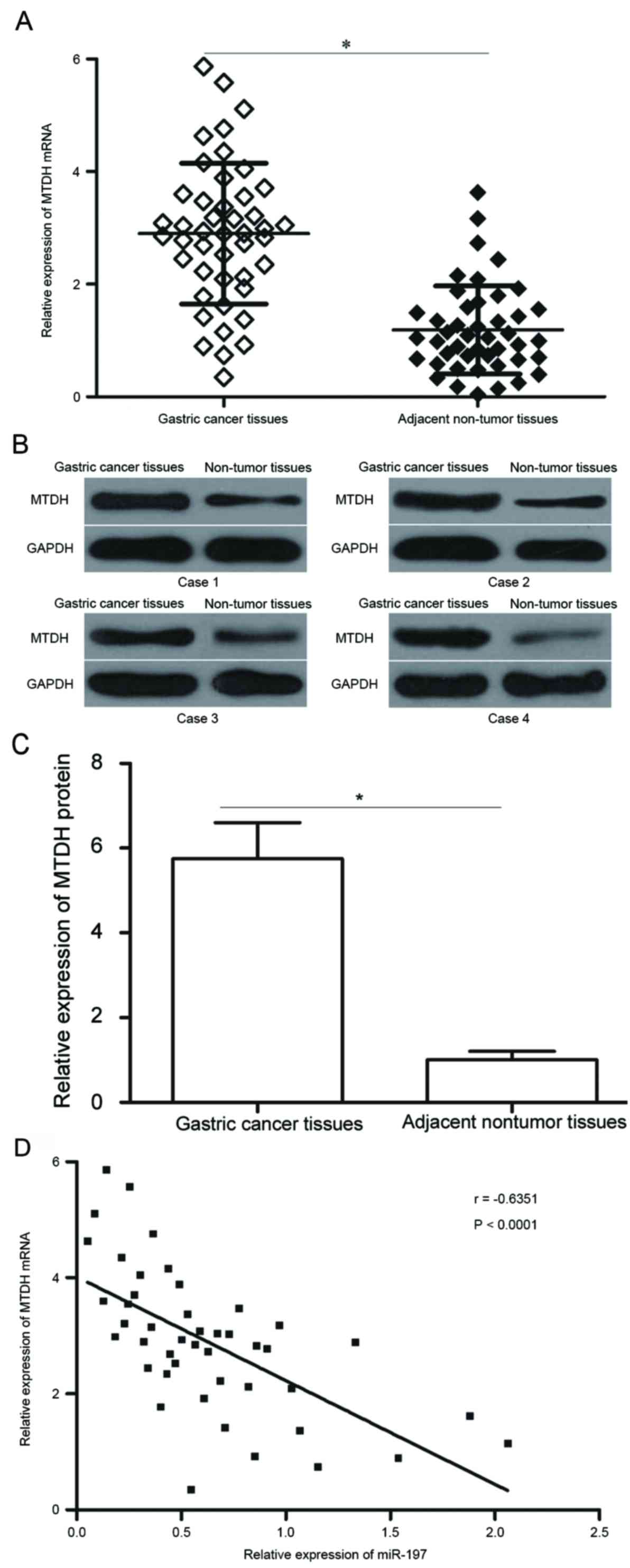
MTDH is upregulated in gastric cancer
tissues and is inversely correlated with miR-197 expression
levels
MTDH mRNA and protein expression levels were
measured in gastric cancer tissues and adjacent non-tumoural
tissues. MTDH mRNA expression levels were significantly higher in
gastric cancer tissues compared with adjacent non-tumoural tissues
(P<0.05; Fig. 4A). Similarly, a
notable increase in the protein expression levels of MTDH were
observed in gastric cancer tissues compared with normal tissue
(P<0.05; Fig. 4B and C). In
addition, an inverse correlation between MTDH mRNA and miR-197
expression levels was observed by Spearman's correlation analysis
in gastric cancer tissues (P<0.0001; r=−0.6351; Fig. 4D).
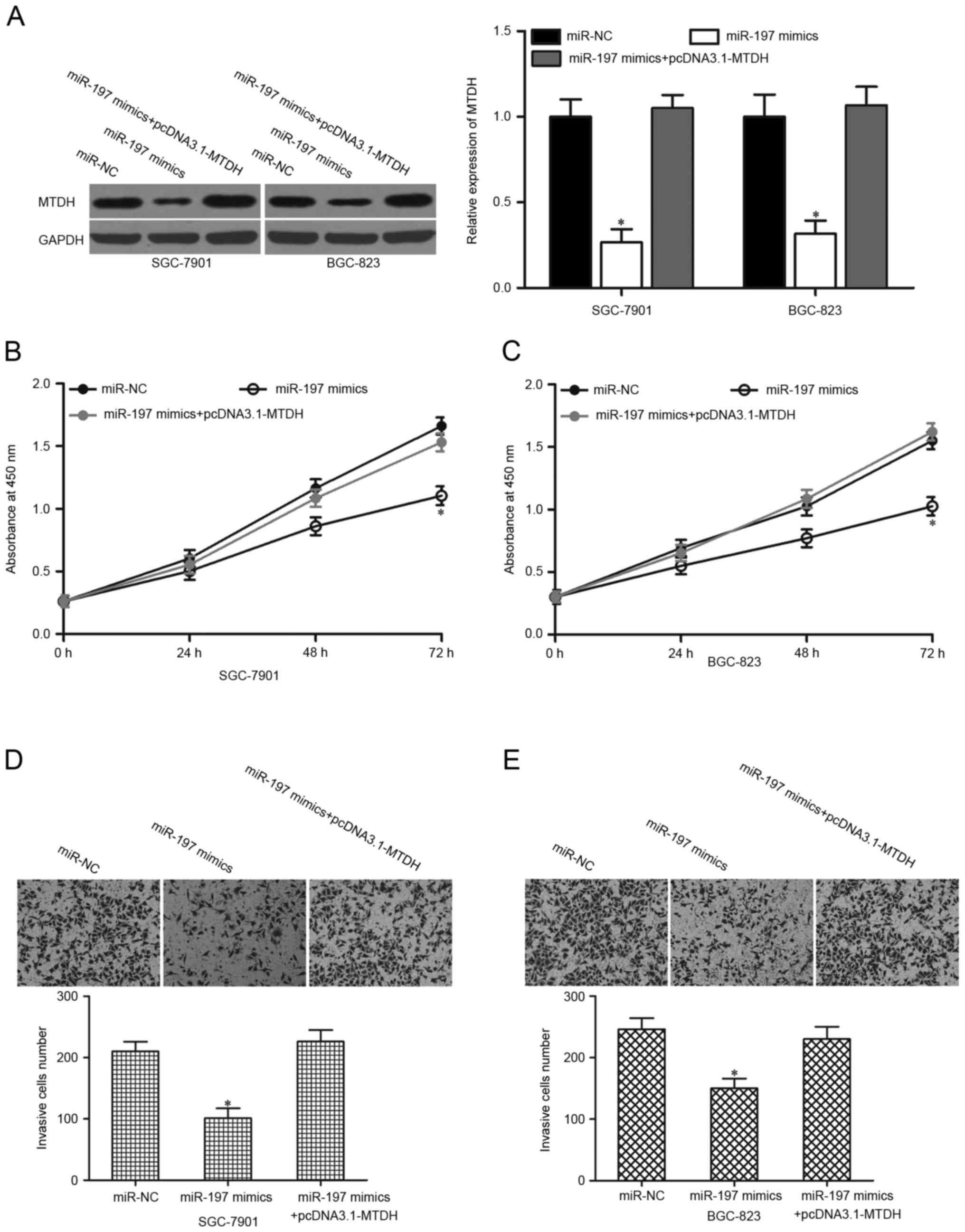
MTDH overexpression reverses the
suppressive roles of miR-197 on the proliferation and invasion of
gastric cancer cells
Following the validation of MTDH as a direct target
of miR-197, the present study examined whether MTDH upregulation
may prevent the suppressive effects of miR-197 on gastric cancer
cell proliferation and invasion. miR-197 mimics were transfected
into SGC-7901 and BGC-823 cells with or without pcDNA3.1-MTDH.
Co-transfection of pCDNA3.1-MTDH and miR-197 mimics enabled the
recovery of MTDH expression (P<0.05; Fig. 5A). Furthermore, MTDH restoration
rescued the proliferation (Fig. 5B and
C; P<0.05) and invasion (Fig.
5D and E; P<0.05) inhibition induced by miR-197
overexpression in SGC-7901 and BGC-823 cells. These results
suggested that MTDH inhibition by miR-197 may partially contribute
to tumour-suppression in human gastric cancer.
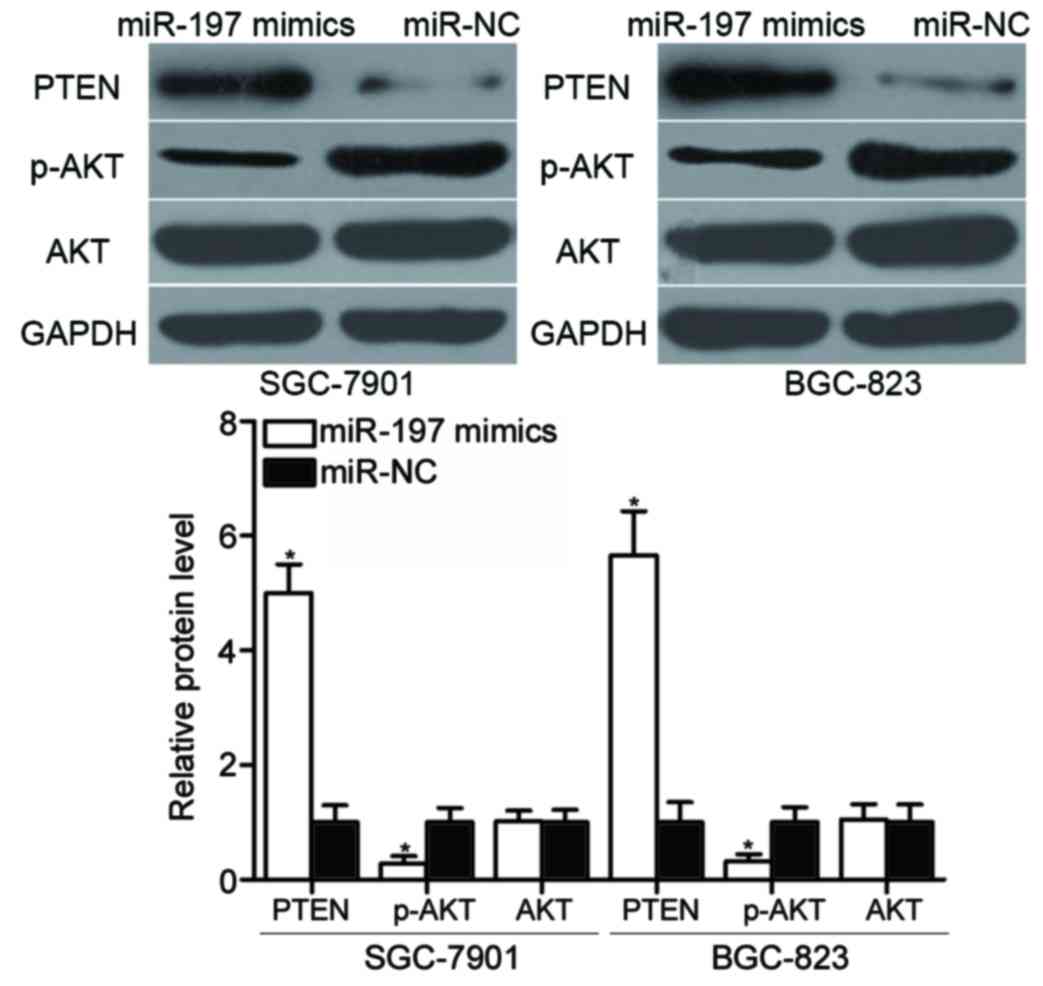
miR-197 regulates the phosphatase and
tensin homolog (PTEN)/AKT pathway in gastric cancer
MTDH negatively regulates PTEN expression by
blocking PTEN transcription (28,29).
Western blot analysis was conducted to detect PTEN, AKT and
phosphorylated (p)-AKT protein expression levels in SGC-7901 and
BGC-823 cells following transfection with miR-197 mimics or miR-NC.
The results revealed that PTEN protein levels were notably
increased in cells transfected with miR-197 mimics, compared with
miR-NC-transfected cells (Fig. 6).
p-AKT expression was reduced by overexpression of miR-197, and AKT
expression levels were unaltered by miR-197 overexpression in
SGC-7901 and BGC-823 cells (Fig.
6; P<0.05). These results suggested that miR-197 may
indirectly regulate the PTEN/AKT signalling pathway in gastric
cancer by regulating MTDH expression.
Discussion
miRNAs are a group of small RNAs that regulate
several cellular processes (30,31).
A large number of miRNAs are involved in gastric cancer formation
and progression and thus may be used as effective diagnostic
biomarkers for gastric cancer therapy (32). In the present study, miR-197
expression was demonstrated to be downregulated in the gastric
cancer tissues and cell lines. Low miR-197 expression was
significantly associated with tumour size, invasive depth, TNM
staging and lymph node metastasis in gastric cancer. The results
also demonstrated that miR-197 overexpression significantly
inhibited gastric cancer cell proliferation and invasion in
vitro. Notably, MTDH was confirmed as a direct target of
miR-197. These findings may contribute to the establishment of
effective therapeutic targets for gastric cancer treatment.
MiR-197 is downregulated in multiple malignant
tumours. For example, expression levels of miR-197 is decreased in
patients with oesophageal cancer with poor prognosis, and
Kaplan-Meier analysis indicated that miR-197 expression is
associated with survival rate of oesophageal cancer patients
(33). Furthermore, patients with
oesophageal cancer with low miR-197 levels commonly had short
survival outcomes (33). Low
miR-197 expression was also reported in colorectal cancer (22), glioblastoma (23) and multiple myeloma (24). However, in hepatocellular
carcinoma, miR-197 expression levels are increased in tumour
tissues and cell lines (34). In
lung cancer, miR-197 is upregulated and significantly associated
with large tumours and squamous cell carcinoma histotype (35). In addition, high expression of
miR-197 is a poor prognosis marker for patients with non-small cell
lung cancer (35). Notably,
miR-197 is highly expressed in breast cancer (36), Wilms tumour (37) and pancreatic cancer (38). These conflicting results suggested
that miR-197 expression is varied and tissue-dependent in human
cancers.
miR-197 serves as a tumour suppressor in
tumourigenicity and tumour progression. It has been reported that
miR-197 inhibits cell growth and metastasis of glioblastoma
(23,39). A previous study revealed that
miR-197 upregulation increased the sensitivity of colorectal cancer
cells to 5-fluorouracil (40), and
another demonstrated that restoration of miR-197 expression
represses cell viability, colony formation and cell migration, and
induces apoptosis in multiple myeloma (24). However, Dai (34) et al and Wang (41) et al demonstrated that
miR-197 serves oncogenic roles in hepatocellular carcinoma by
promoting cell proliferation, migration and invasion in
vitro and in vivo. miR-197 knockdown was reported to
attenuate Wilms tumour cell proliferation and promote apoptosis
(37); whereas miR-197
overexpression was demonstrated to promote epithelial-mesenchymal
transition in pancreatic cancer (38). These conflicting findings indicated
that the roles of miR-197 are tissue specific in tumourigenesis and
tumour development. These studies have also suggested that miR-197
contributes to these types of human cancers and may serve as a
potential therapeutic target for their treatment.
Several miR-197 target genes have previously been
identified, including fusion 1 (23), GRB2-associated-binding protein in
glioblastoma (39), kangai 1/CD82
in hepatocellular carcinoma (34),
thymidylate synthase in colorectal cancer (40), induced myeloid leukaemia cell
differentiation protein MCL-1 in multiple myeloma (24), insulin-like growth factor-binding
protein 3 in Wilms tumour (37)
and p120 catenin in pancreatic cancer (38). In the present study, MTDH was
identified as a novel, direct and functional target of miR-197 in
gastric cancer. MTDH is located on chromosome 8q22 (42). MTDH is upregulated in numerous
human cancers, such as colorectal cancer (43), breast cancer (44), cervical cancer (45) and bladder cancer (46). MTDH serves important roles in
various biological processes in cancer occurrence and progression,
including cellular growth, apoptosis, metastasis and angiogenesis
(47,48).
In gastric cancer, MTDH expression levels are
increased, and it was significantly associated with differentiation
status, TNM staging, invasive depth and lymph node metastasis
(26). MTDH overexpression is
associated with poor survival in patients with gastric cancer.
Multivariate analyses have demonstrated that MTDH is an independent
prognostic factor for gastric cancer (27). The results of functional assays
revealed that MTDH is involved in gastric cancer proliferation,
cell cycle arrest, angiogenesis and metastasis (49–52).
The results of the current study demonstrated that miR-197
inhibited the proliferation and invasion of gastric cancer cells by
regulating the MTDH/PTEN/AKT signalling pathway. Therefore, the
miR-197/MTDH axis may provide a novel effective therapeutic target
for the disease.
In conclusion, miR-197 expression was downregulated
in gastric cancer and aberrantly expressed miR-197 may partially
influence gastric cancer cell proliferation and invasion by
directly targeting MTDH.
Acknowledgements
The present study was supported by the Shanghai
Baoshan District Medical Specialty Support Project (grant no.
BSZK_2014_B01).
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shen L, Shan YS, Hu HM, Price TJ, Sirohi
B, Yeh KH, Yang YH, Sano T, Yang HK, Zhang X, et al: Management of
gastric cancer in Asia: Resource-stratified guidelines. Lancet
Oncol. 14:e535–e547. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferro A, Peleteiro B, Malvezzi M, Bosetti
C, Bertuccio P, Levi F, Negri E, La Vecchia C and Lunet N:
Worldwide trends in gastric cancer mortality (1980–2011), with
predictions to 2015, and incidence by subtype. Eur J Cancer.
50:1330–1344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kato M and Asaka M: Recent knowledge of
the relationship between Helicobacter pylori and gastric cancer and
recent progress of gastroendoscopic diagnosis and treatment for
gastric cancer. Jpn J Clin Oncol. 40:828–837. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li L, Ying XJ, Sun TT, Yi K, Tian HL, Sun
R, Tian JH and Yang KH: Overview of methodological quality of
systematic reviews about gastric cancer risk and protective
factors. Asian Pac J Cancer Prev. 13:2069–2079. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Resende C, Thiel A, Machado JC and
Ristimäki A: Gastric cancer: Basic aspects. Helicobacter. 16 Suppl
1:S38–S44. 2011. View Article : Google Scholar
|
|
7
|
Thrumurthy SG, Chaudry MA, Chau I and
Allum W: Does surgery have a role in managing incurable gastric
cancer? Nat Rev Clin Oncol. 12:676–682. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ying SY, Chang DC and Lin SL: The microRNA
(miRNA): Overview of the RNA genes that modulate gene function. Mol
Biotechnol. 38:257–268. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guan A, Wang H, Li X, Xie H, Wang R, Zhu Y
and Li R: MiR-330-3p inhibits gastric cancer progression through
targeting MSI1. Am J Transl Res. 8:4802–4811. 2016.PubMed/NCBI
|
|
12
|
Wu SG, Huang YJ, Bao B, Wu LM, Dong J, Liu
XH, Li ZH, Wang XY, Wang L, Chen BJ and Chen W: miR-508-5p acts as
an anti-oncogene by targeting MESDC1 in hepatocellular carcinoma.
Neoplasma. 64:40–47. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu H, Duan P, Zhu H and Rao D: miR-613
inhibits bladder cancer proliferation and migration through
targeting SphK1. Am J Transl Res. 9:1213–1221. 2017.PubMed/NCBI
|
|
14
|
Xu J, Jiang N, Shi H, Zhao S, Yao S and
Shen H: (Corrigendum) miR-28-5p promotes the development and
progression of ovarian cancer through inhibition of N4BP1. Int J
Oncol. 50:22362017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu Y, Tang Z, Jiang B, Chen J and Fu Z:
miR-198 functions as a tumor suppressor in breast cancer by
targeting CUB domain-containing protein 1. Oncol Lett.
13:1753–1760. 2017.PubMed/NCBI
|
|
16
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen P, Zhao H, Huang J, Yan X, Zhang Y
and Gao Y: MicroRNA-17-5p promotes gastric cancer proliferation,
migration and invasion by directly targeting early growth response
2. Am J Cancer Res. 6:2010–2020. 2016.PubMed/NCBI
|
|
19
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Babashah S and Soleimani M: The oncogenic
and tumour suppressive roles of microRNAs in cancer and apoptosis.
Eur J Cancer. 47:1127–1137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng G, Wang H, Zhang X, Yang Y, Wang L,
Du L, Li W, Li J, Qu A, Liu Y and Wang C: Identification and
validation of reference genes for qPCR detection of serum microRNAs
in colorectal adenocarcinoma patients. PLoS One. 8:e830252013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xin J, Zhang XK, Xin DY, Li XF, Sun DK, Ma
YY and Tian LQ: FUS1 acts as a tumor-suppressor gene by
upregulating miR-197 in human glioblastoma. Oncol Rep. 34:868–876.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang Y, Li F, Saha MN, Abdi J, Qiu L and
Chang H: miR-137 and miR-197 induce apoptosis and suppress
tumorigenicity by targeting MCL-1 in multiple myeloma. Clin Cancer
Res. 21:2399–2411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCt Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dong L, Qin S, Li Y, Zhao L, Dong S, Wang
Y, Zhang C and Han S: High expression of astrocyte elevated gene-1
is associated with clinical staging, metastasis, and unfavorable
prognosis in gastric carcinoma. Tumour Biol. 36:2169–2178. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jian-bo X, Hui W, Yu-long H, Chang-hua Z,
Long-juan Z, Shi-rong C and Wen-hua Z: Astrocyte-elevated gene-1
overexpression is associated with poor prognosis in gastric cancer.
Med Oncol. 28:455–462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du C, Yi X, Liu W, Han T, Liu Z, Ding Z,
Zheng Z, Piao Y, Yuan J, Han Y, et al: MTDH mediates trastuzumab
resistance in HER2 positive breast cancer by decreasing PTEN
expression through an NFκB-dependent pathway. BMC Cancer.
14:8692014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu C, Kong X, Wang H, Zhang N, Kong X,
Ding X, Li X and Yang Q: MTDH mediates estrogen-independent growth
and tamoxifen resistance by down-regulating PTEN in MCF-7 breast
cancer cells. Cell Physiol Biochem. 33:1557–1567. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shrestha S, Hsu SD, Huang WY, Huang HY,
Chen W, Weng SL and Huang HD: A systematic review of microRNA
expression profiling studies in human gastric cancer. Cancer Med.
3:878–888. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang TY, Liu SG, Zhao BS, Qi B, Qin XG and
Yao WJ: Implications of microRNA-197 downregulated expression in
esophageal cancer with poor prognosis. Genet Mol Res. 13:5574–5581.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dai W, Wang C, Wang F, Wang Y, Shen M,
Chen K, Cheng P, Zhang Y, Yang J, Zhu R, et al: Anti-miR-197
inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem
Biophys Res Commun. 446:541–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mavridis K, Gueugnon F, Petit-Courty A,
Courty Y, Barascu A, Guyetant S and Scorilas A: The oncomiR miR-197
is a novel prognostic indicator for non-small cell lung cancer
patients. Br J Cancer. 112:1527–1535. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shaker O, Maher M, Nassar Y, Morcos G and
Gad Z: Role of microRNAs −29b-2, −155, −197 and −205 as diagnostic
biomarkers in serum of breast cancer females. Gene. 560:77–82.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu J, Liu G, Zhao Z, Jia W and Xia H:
MicroRNA-197 mediates the overgrowth and anti-apoptotic effects by
downregulating insulin-like growth factor-binding protein-3 during
nephroblastoma tumorigenesis. Fetal Pediatr Pathol. 35:287–298.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hamada S, Satoh K, Miura S, Hirota M,
Kanno A, Masamune A, Kikuta K, Kume K, Unno J, Egawa S, et al:
miR-197 induces epithelial-mesenchymal transition in pancreatic
cancer cells by targeting p120 catenin. J Cell Physiol.
228:1255–1263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tian LQ, Liu EQ, Zhu XD, Wang XG, Li J and
Xu GM: MicroRNA-197 inhibits cell proliferation by targeting GAB2
in glioblastoma. Mol Med Rep. 13:4279–4288. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sun Z, Zhou N, Han Q, Zhao L, Bai C, Chen
Y, Zhou J and Zhao RC: MicroRNA-197 influences 5-fluorouracil
resistance via thymidylate synthase in colorectal cancer. Clin
Transl Oncol. 17:876–883. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang H, Su X, Yang M, Chen T, Hou J, Li N
and Cao X: Reciprocal control of miR-197 and IL-6/STAT3 pathway
reveals miR-197 as potential therapeutic target for hepatocellular
carcinoma. Oncoimmunology. 4:e10314402015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wan L and Kang Y: Pleiotropic roles of
AEG-1/MTDH/LYRIC in breast cancer. Adv Cancer Res. 120:113–134.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gnosa S, Shen YM, Wang CJ, Zhang H,
Stratmann J, Arbman G and Sun XF: Expression of AEG-1 mRNA and
protein in colorectal cancer patients and colon cancer cell lines.
J Transl Med. 10:1092012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li J, Zhang N, Song LB, Liao WT, Jiang LL,
Gong LY, Wu J, Yuan J, Zhang HZ, Zeng MS and Li M: Astrocyte
elevated gene-1 is a novel prognostic marker for breast cancer
progression and overall patient survival. Clin Cancer Res.
14:3319–3326. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu JQ, Zhou Q, Zhu H, Zheng FY and Chen
ZW: Overexpression of astrocyte elevated gene-1 (AEG-1) in cervical
cancer and its correlation with angiogenesis. Asian Pac J Cancer
Prev. 16:2277–2281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang G, Zhang L, Lin S, Li L, Liu M, Chen
H, Cao M, Liu D, Huang YR and Bo J: AEG-1 is associated with tumor
progression in nonmuscle-invasive bladder cancer. Med Oncol.
31:9862014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hu G, Wei Y and Kang Y: The multifaceted
role of MTDH/AEG-1 in cancer progression. Clin Cancer Res.
15:5615–5620. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Emdad L, Lee SG, Su ZZ, Jeon HY, Boukerche
H, Sarkar D and Fisher PB: Astrocyte elevated gene-1 (AEG-1)
functions as an oncogene and regulates angiogenesis. Proc Natl Acad
Sci USA. 106:21300–21305. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang CF, Xia YH, Zheng QF, Li ZJ, Guo XH,
Zhou HC, Zhang LL, Dong LP, Han Y, Liu ZE, et al: Effect of
silencing AEG-1 with small interfering RNA on the proliferation and
cell cycle of gastric carcinoma SGC-7901 cells. Zhonghua Zhong Liu
Za Zhi. 35:22–27. 2013.(Article In Chinese). PubMed/NCBI
|
|
50
|
Tang Y, Liu X, Su B, Zhang Z, Zeng X, Lei
Y, Shan J, Wu Y, Tang H and Su Q: MicroRNA-22 acts as a metastasis
suppressor by targeting metadherin in gastric cancer. Mol Med Rep.
11:454–460. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shen X, Si Y, Yang Z, Wang Q, Yuan J and
Zhang X: MicroRNA-542-3p suppresses cell growth of gastric cancer
cells via targeting oncogene astrocyte-elevated gene-1. Med Oncol.
32:3612015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li S, Guo X, Ma X, Tang C, Ke Z and Huang
W: Expression of astrocyte elevated gene-1 closely correlates with
the angiogenesis of gastric cancer. Oncol Lett. 7:1447–1454.
2014.PubMed/NCBI
|