Introduction
Pneumonia is a common respiratory disease worldwide,
which is preventable and treatable (1); however, it is recognized as a leading
cause of mortality in children (2). It has been reported that between 1.1
and 1.4 million children succumb to pneumonia annually (3). Due to poor health care or incomplete
treatment, pediatric pneumonia often recurs, which can result in
numerous severe complications and a poor prognosis, thereby
affecting the child's development, even leading to mortality
(4,5). Although the prevalence of pediatric
mortality has been reduced due to recent surgical and medical
advances, pediatric pneumonia remains at a high risk of recurrence
and hospitalization (6,7). Therefore, it is essential to
investigate the underlying pathogenesis and to identify effective
therapeutic targets for the treatment of pediatric pneumonia.
MicroRNAs (miRNAs/miRs) are a group of small
non-coding RNAs, between 19 and 24 nucleotides long, which regulate
the expression of target genes by binding with complementary sites
on target 3′-untranslated region (3′UTR) (8). miRNAs have been reported to be
implicated in the majority of cellular processes, including cell
proliferation, apoptosis, migration, differentiation and
metabolism, as well as in innate immunity, inflammation and
infection (9,10). Previous studies have demonstrated
that miRNAs serve potential roles in some types of cancer, through
regulating gene expression levels and cancer
pathogenesis-associated pathways, including in colorectal cancer
(11), chronic lymphocytic
leukemia (12), breast cancer
(13), hepatocellular carcinoma
(14) and lung cancer (15).
In addition to the effects of miRNAs on cancer, much
attention has been paid to the potential roles of miRNAs in
pediatric lung development (16)
and respiratory diseases (17). A
previous study suggested that numerous miRNAs associated with
inflammation (miR-132, miR-181a, miR-221 and miR-222) may serve a
role in the pathogenesis of Streptococcus pneumoniae
meningitis (18). Recently,
miR-20a has been reported to be upregulated and participate in the
regulation of cell proliferation in lung cancer (19). However, it is unclear whether
miR-20a is associated with the pathogenesis of pediatric
pneumonia.
The present study evaluated the expression levels of
miR-20a in children with pneumonia and in a cell model of
lipopolysaccharide (LPS)-induced inflammatory lung injury (20). Subsequently, the role and
mechanisms of miR-20a in pediatric pneumonia-associated
inflammation were investigated.
Materials and methods
Patients
A total of 16 children (8 male and 8 female; mean
age ± standard deviation, 11.4±2.16 years; age range, 8–15) with
pneumonia were recruited to the present study. Pneumonia was
diagnosed according to clinical presentation, respiratory symptoms,
bacterial infection and chest X-ray. In addition, 16 gender-matched
children (mean age ± standard deviation, 11.3±2.24 years; age
range, 7–15) with fever were included as the control group; these
children were treated in the Emergency Department and had no
respiratory symptoms. These 32 patients were recruited from the
Jinan Maternity and Child Care Hospital (Jinan, China) between June
2012 and December 2013. None of the patients underwent any
treatment prior to blood collection. Venous blood samples (5 ml)
were collected from the patients and the present study was approved
by the Ethics Committee of the Jinan Maternity and Child Care
Hospital. Written informed consent was obtained from the legal
guardians of all participants.
Cell culture and treatment
The A549 human lung adenocarcinoma cell line was
purchased from the Shanghai Cell Bank of Chinese Academy of
Sciences (Shanghai, China). Cells were cultured in RPMI 1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum and 1%
penicillin/streptomycin (both Beijing Transgen Biotechnology Co.,
Ltd., Beijing, China) at 37°C in an incubator containing 5%
CO2. A total of 2 ml cells/well, at a concentration of
5×105 cells/ml, were cultured in a 6-well plate.
Subsequently, cells were separated into groups, and were treated
with LPS (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), miR-20a
mimic (sense 5′-UAAAGUGCUUAUAGUGCAGGUAG-3′ and antisense
5′-ACCUGCACUAUAAGCACUUUAUU-3′), mimic control (sense
5′-UGCUUAGUAUAAGAUGCAGGUAG-3′ and antisense
5′-ACGCUAUAUCUUAUAGCACACUU-3′) (both Shanghai GenePharma Co., Ltd.,
Shanghai, China) or the nuclear factor (NF)-κB inhibitor
pyrrolidine dithiocarbamate (PDTC; Sigma-Aldrich; Merck KGaA).
Briefly, the cells were divided into the following four groups:
Control group (non-treated cells); LPS group, in which cells were
stimulated with LPS (1 µg/ml) for 12 h at 37°C to induce
inflammatory lung cell injury (20); LPS + miR-20a group, in which cells
overexpressing miR-20a were treated with LPS (1 µg/ml) for 12 h at
37°C; and LPS + miR-20a + PDTC group, in which cells overexpressing
miR-20a were treated with PDTC (100 mmol/l) for 30 min at 37°C and
then treated with LPS (1 µg/ml) for 12 h at 37°C (21). In addition, an LPS + mimic control
group, in which cells were transfected with mimic control and then
treated with LPS (1 µg/ml) for 12 h at 37°C, was generated. To
overexpress miR-20a, the miR-20a mimic (50 nM) was transfected into
A549 cells (1×105 cells) using 20 µl Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. Transfection with the mimic control was
the same as the transfection of miR-20a. Transfected cells were
harvested at 48 h post-transfection for subsequent experiments.
ELISA analysis
Blood samples were collected from the children and
were centrifuged at 1,500 × g for 10 min at 4°C. The supernatant
serum samples were then collected and stored at −80°C. The serum
concentrations of interleukin (IL)-6 (cat. no. S6050), tumor
necrosis factor (TNF)-α (cat. no. STA00C) and C-reactive protein
(CRP; cat. no. SCRP00) were determined using corresponding ELISA
kits (R&D Systems, Inc., Minneapolis, MN, USA) according to the
manufacturer's protocols. The absorbance was measured at 450 nm
using a microplate reader (Thermo Fisher Scientific, Inc.). The
concentrations of IL-6, TNF-α and CRP were calculated based on the
standard curve.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from blood samples using the
RNAprep pure Blood kit (Tiangen Biotech, Beijing, China) and total
RNA was extracted from treated cells using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, RNA was
measured using a UV spectrophotometer (BD Biosciences, San Diego,
CA, USA) at A260, and cDNA was generated from miRNAs
using stem-loop RT primers (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with the following cycling parameters: 30°C for
10 min, followed by 42°C for 30 min and 95°C for 5 min. Primer
sequences for miR-20a and U6 small nuclear (sn)RNA are presented in
Table I. SYBR®
Premix Ex Taq™ kit (Takara Biotechnology Co.,
Ltd., Dalian, China) was used to perform PCR amplification; the PCR
program was as follows: 94°C for 10 min, followed by 40 cycles at
94°C for 15 sec, 56°C for 30 sec and 72°C for 1 min, and finally
72°C for 10 min. U6 snRNA was used as an internal control. Relative
quantification and calculations were conducted using the
quantification cycle (Cq) method (2−ΔΔCq) (22).
 | Table I.Primer sequences for specific
genes. |
Table I.
Primer sequences for specific
genes.
| Gene | Primer | Sequence
(5′-3′) |
|---|
| miR-20a | Forward |
GCCGCGCTAAAGTGCTTATAGTG |
|
| Reverse |
CACCAGGGTCCGAGGT |
| U6 | Forward |
TGCGGGTGCTCGCTTCGGCAGC |
|
| Reverse |
CCAGTGCAGGGTCCGAGGT |
Western blotting
The cells were collected and the proteins were
extracted using RIPA buffer (Beyotime Institute of Biotechnology,
Shanghai, China) containing a cocktail of protease inhibitors
(Roche Diagnostics, Basel, Switzerland). Bicinchoninic acid protein
assay kit (Sangon Biotech Co., Ltd., Shanghai, China) was used to
determine protein concentration. Subsequently, equal amounts of
protein (30 µg/lane) were separated by 10% SDS-PAGE and were
transferred to a polyvinylidene fluoride membrane. After blocking
with 5% non-fat milk at room temperature for 2 h, the membrane was
incubated with rabbit anti-human inhibitor of NF-κB α (IκBα; cat.
no. SAB4501994; 1:500), phosphorylated (p)-NF-κB (cat. no.
SAB4301496; 1:500) polyclonal antibodies and β-actin monoclonal
antibody (cat. no. SAB2100037; 1:1,000) (all Sigma-Aldrich; Merck
KGaA) at 4°C overnight. β-actin was used as an internal control.
After washing with PBS, the membrane was incubated with horseradish
peroxidase-conjugated goat anti-rabbit secondary antibody (cat. no.
31460; 1:10,000; Thermo Fisher Scientific, Inc.) for 2 h at room
temperature. The membrane was washed again with PBS and color was
developed using a 3,3′-diaminobenzidine substrate (Sangon Biotech
Co., Ltd.). Image Pro Plus software (version 5.0; Media
Cybernetics, Inc., Rockville, MD, USA) was used to semi-quantify
the expression levels of the target proteins.
Statistical analysis
All data are presented as the mean ± standard
deviation, and SPSS 16.0 statistics software (SPSS, Inc., Chicago,
IL, USA) was used to analyze the data. All data were initially
tested for Gaussian distribution. Subsequently, data were analyzed
by unpaired two-tailed t-test (for two groups) or one-way analysis
of variance followed by a Tukey post hoc test (for more than two
groups). P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-20a expression in patients with
pneumonia and in LPS-induced cells
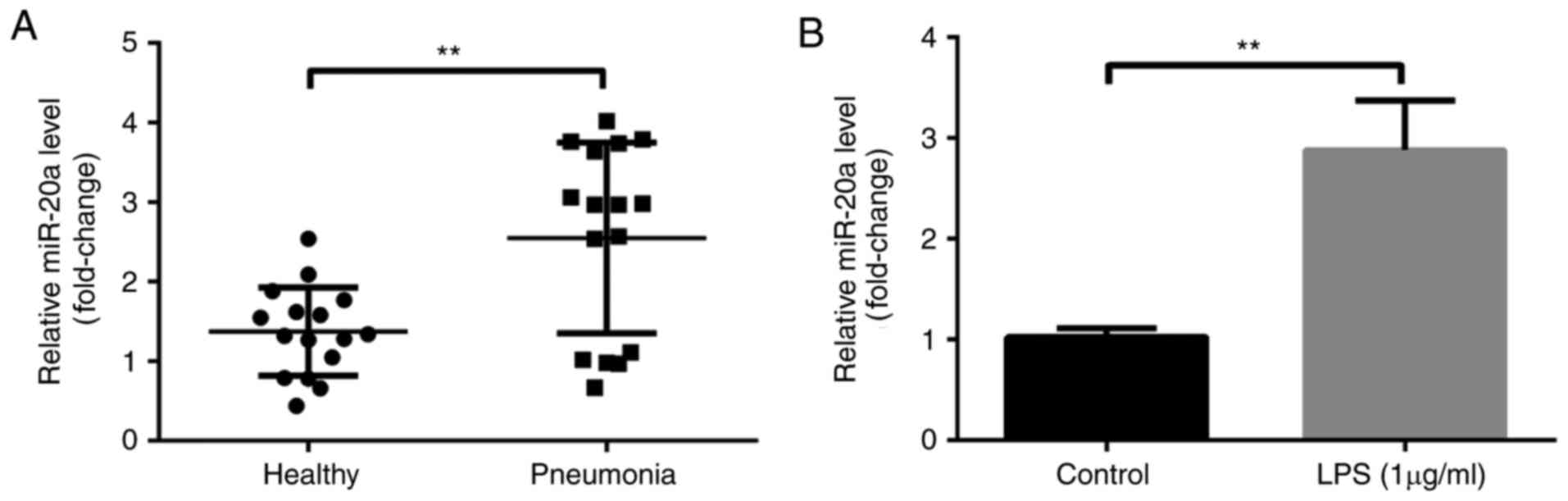
To explore the expression levels of miR-20a in
patients with pneumonia, an RT-qPCR analysis was performed; the
results demonstrated that miR-20a expression was higher in patients
with pneumonia compared with in healthy controls (P<0.01;
Fig. 1A). In addition, LPS was
used to induce inflammatory injury in A549 cells. Compared with in
the control group, the expression levels of miR-20a were increased
in the LPS group (P<0.01; Fig.
1B).
Effects of miR-20a overexpression on
inflammation in pneumonia
The present study investigated the association
between miR-20a and the inflammatory reaction. ELISA analysis
revealed that the serum concentrations of inflammatory factors,
including IL-6, TNF-α and CRP, were higher in patients with
pneumonia compared with in healthy controls (P<0.05; Fig. 2A). The results of an RT-qPCR
analysis indicated that the expression levels of miR-20a were
significantly upregulated following transfection of cells with
miR-20a mimic compared with in the mimic control group (P<0.001;
Fig. 2B), confirming that miR-20a
was successfully overexpressed. Subsequent experiments demonstrated
that the concentrations of inflammatory factors, including IL-6,
TNF-α and CRP, were increased in the LPS group compared with in the
control group (P<0.05; Fig.
2C-E). In addition, overexpression of miR-20a further increased
the concentrations of IL-6, TNF-α and CRP compared with the LPS
group (P<0.05). The levels of inflammatory cytokines in the LPS
+ mimic control group were not significantly different compared
with the LPS group (data not shown).
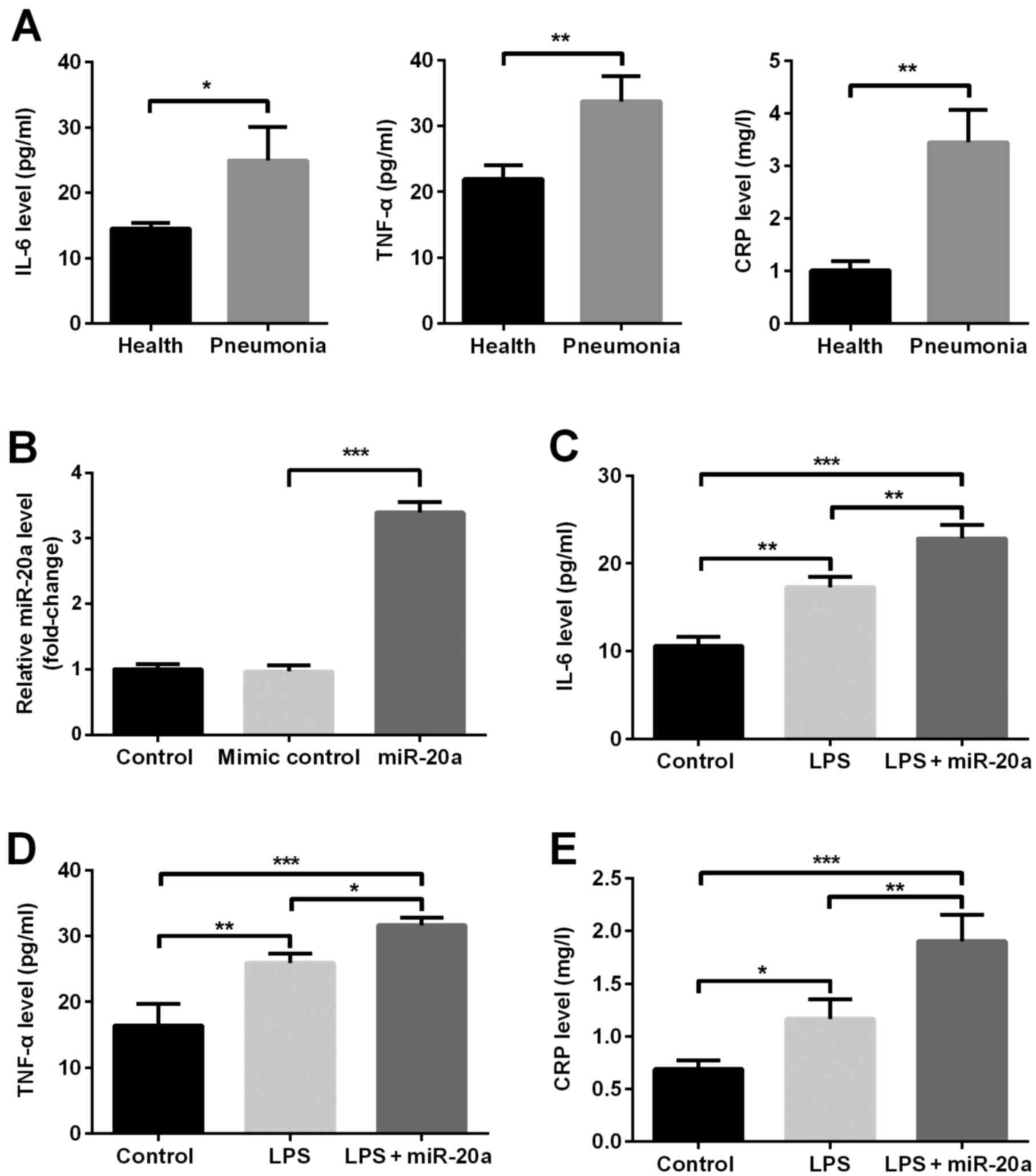 | Figure 2.miR-20a promotes the expression of
inflammatory factors, including IL-6, TNF-α and CRP, in A549 cells
with LPS-induced inflammatory injury. (A) Concentrations of IL-6,
TNF-α and CRP in children with pneumonia and in healthy children
(n=16/group). (B) Expression levels of miR-20a in transfected A549
cells; and the concentrations of (C) IL-6, (D) TNF-α and (E) CRP in
A549 cells. In addition, an LPS + mimic control group was
generated; the results from this group were not significantly
different compared with the LPS group; therefore, data are not
shown. Data are presented as the mean ± standard deviation.
*P<0.05; **P<0.01; ***P<0.001. CRP, C-reactive protein;
IL-6, interleukin-6; LPS, lipopolysaccharide; miR, microRNA; TNF-α,
tumor necrosis factor-α. |
Effects of miR-20a overexpression on
the NF-κB signaling pathway in LPS-induced cells
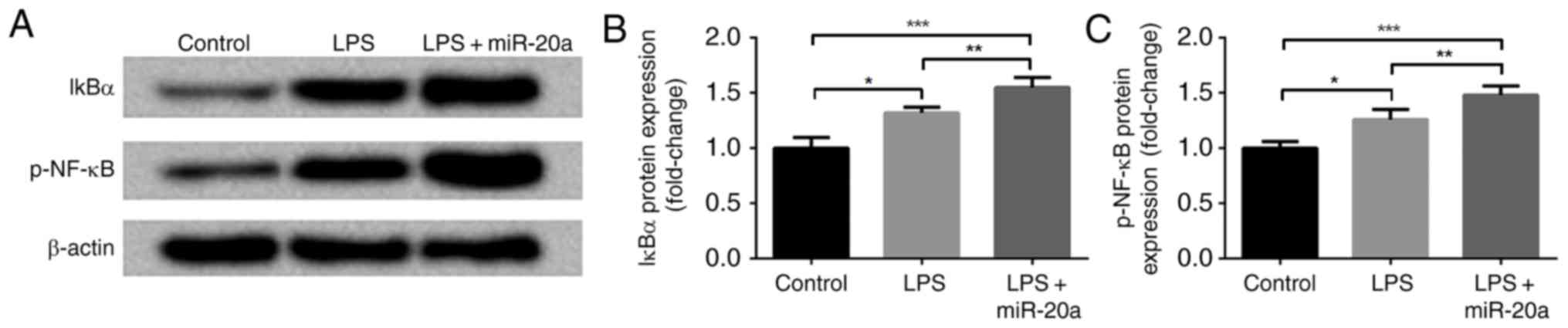
To further investigate the mechanism underlying the
effects of miR-20a on the induction of inflammation, NF-κB
signaling pathway-associated proteins, including IκBα and p-NF-κB,
were measured by western blotting. As shown in Fig. 3A-C, the protein expression levels
of IκBα and p-NF-κB were increased in the LPS group compared with
in the control group (P<0.05). The expression of total NF-κB was
consistent with that of β-actin (data not shown). In addition, the
expression levels of IκBα and p-NF-κB were further increased in the
LPS + miR-20a group compared with in the LPS group (P<0.05). The
expression levels of IκBα and p-NF-κB in LPS + mimic control group
were not significantly different compared with the LPS group (data
not shown).
Effects of the NF-κB inhibitor PDTC on
miR-20a-induced inflammation in LPS-treated cells
To evaluate the role of the NF-κB signaling pathway
in miR-20a-induced inflammation, the NF-κB inhibitor PDTC was used
to suppress the NF-κB signaling pathway. ELISA was subsequently
used to detect the expression levels of inflammatory factors. The
results demonstrated that following treatment with the NF-κB
inhibitor PDTC, the expression levels of IL-6, TNF-α and CRP were
markedly decreased compared with in the LPS + miR-20a group
(P<0.05; Fig. 4). The levels of
inflammatory cytokines in the LPS + mimic control group were not
significantly different compared with the LPS group (data not
shown).
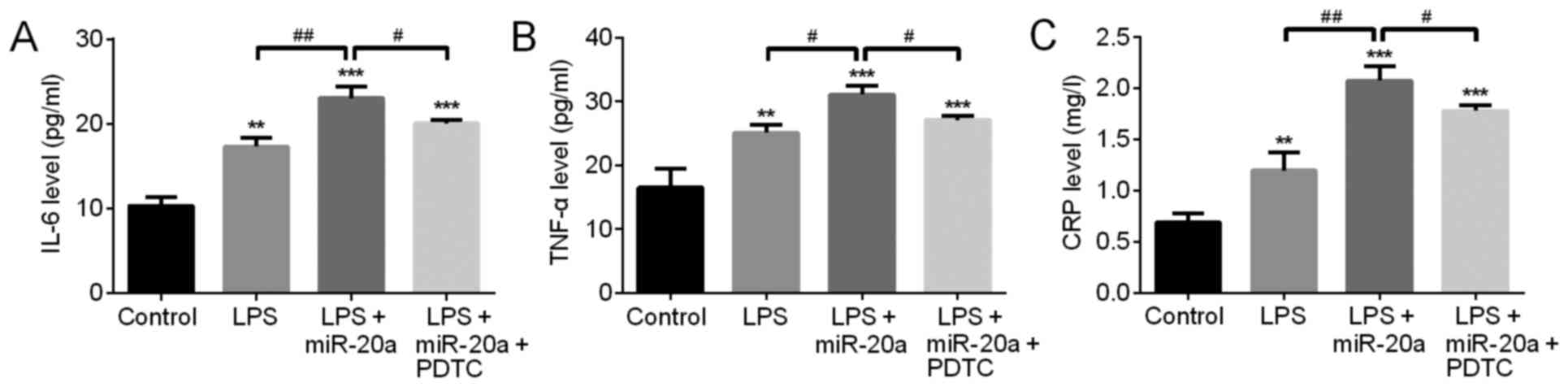 | Figure 4.NF-κB inhibitor PDTC inhibits the
proinflammatory role of miR-20a in A549 cells with LPS-induced
inflammatory injury. The expression levels of (A) IL-6, (B) TNF-α
and (C) CRP were detected by ELISA analysis. In addition, an LPS +
mimic control group was generated; the results from this group were
not significantly different compared with the LPS group; therefore,
data are not shown. Data are presented as the mean ± standard
deviation. **P<0.01, ***P<0.001 vs. control group.
#P<0.05, ##P<0.01 vs. LPS + miR-20a
group. CRP, C-reactive protein; IL-6, interleukin-6; LPS,
lipopolysaccharide; miR, microRNA; PDTC, pyrrolidine
dithiocarbamate; TNF-α, tumor necrosis factor-α. |
Discussion
miRNAs have been reported to have a role in
inflammatory diseases (23). The
results of the present study demonstrated that the expression
levels of miR-20a were increased in children with pneumonia and in
lung cells with LPS-induced inflammatory injury. Furthermore, the
results indicated that miR-20a overexpression may promote the
expression of inflammatory factors, including IL-6, TNF-α and CRP.
miR-20a overexpression also increased the expression levels of
NF-κB signaling pathway-associated proteins, including IκBα and
p-NF-κB, whereas treatment with the NF-κB inhibitor PDTC was able
to inhibit the enhancing effects of miR-20a on the inflammatory
reaction.
It has previously been demonstrated that miRNAs
exert marked regulatory effects on inflammation, and various miRNAs
have been identified and considered to participate in the
pathological mechanisms of inflammatory diseases (24). Bazzoni et al (25) reported that in neutrophils and
macrophages, miR-9 overexpression could be induced by LPS and
negatively regulated NF-κB signaling pathway-dependent inflammatory
responses. miR-21 has also been identified to target tumor
suppressor genes and negatively regulate the LPS-stimulated
Toll-like receptor (TLR)4 pathway, which may result in activation
of NF-κB and production of IL-10 (26). Downregulated miR-125b has been
indicated to promote the expression of TNF-α in LPS-induced
activated macrophages (27).
Furthermore, miR-155 has been reported to suppress the inflammatory
reaction through regulating the expression of myeloid
differentiation primary response protein 88, which serves an
important role in the TLR pathway (28). Taganov et al (29) revealed that miR-146 may be
significantly upregulated in human monocytic cells stimulated by
LPS, and miR-146 was able to inhibit the expression of target
genes, including IL-1 receptor-associated kinase 1 and TNF
receptor-associated factor 6, and negatively regulate TLR
signaling. It was also demonstrated that miR-146a was upregulated
in monocytes stimulated by inflammatory cytokines, including TNF-α
and IL-1β (29). Following
activation of the TLR pathway, miR-147 has been reported to be
upregulated and to inhibit the excessive inflammatory responses in
murine lung macrophages (30).
The present study focused on the role of miR-20a in
pediatric pneumonia. miR-20a is a member of the miR-17-92 cluster,
which is involved in tumorigenesis (31). Previous studies have demonstrated
that miR-20a is associated with cell proliferation (32) and cell cycle progression (33) by regulating the expression of
transcription factor E2F1. Fan et al (34) also reported that miR-20a
overexpression was able to promote cell proliferation and invasion
by inhibiting the expression of targeted amyloid precursor protein
in ovarian cancer cells. Furthermore, it has been demonstrated that
miR-20 serves an important role in inflammatory responses. Philippe
et al (35) reported that
miR-20 could effectively inhibit the production of inflammatory
cytokines, including IL-6, C-X-C motif chemokine ligand 10, IL-1β
and TNF-α, in LPS-activated fibroblast-like synoviocytes.
Conversely, miR-20a was reported to promote macrophage inflammatory
responses through modulating the expression of signal-regulatory
protein α both in vitro and in vivo (36). The various effects of miR-20 on the
inflammatory response may be associated with the different target
genes of miR-20 in various cell types. In the present study, a high
expression of miR-20a was detected in pediatric pneumonia, and the
results revealed that miR-20a overexpression increased the
expression levels of IL-6, TNF-α and CRP in lung cells with
LPS-induced inflammatory injury. These findings indicated that
miR-20a may be involved in the inflammatory response in pediatric
pneumonia.
The present study also investigated the mechanism
underlying the effects of miR-20a on inflammation in pediatric
pneumonia. The NF-κB signaling pathway is considered closely
associated with inflammation; the activation of NF-κB induces the
secretion of proinflammatory cytokines, including IL-1 and TNF-α,
chemokines and adhesion molecules. Previous studies have
demonstrated that NF-κB is activated in numerous inflammatory
disorders, including arthritis (37), glomerulonephritis (38), gastritis (39) and asthma (40). The interaction between miRNAs
(miR-21, miR-146, miR-155, miR-181b and miR-301a) and NF-κB
activation has been demonstrated in human disease (41). Bhaumik et al (42) demonstrated that NF-κB activity and
the inflammatory pathway can be negatively regulated by miR-146a/b
in breast cancer cells. Notably, the results of the present study
preliminarily confirmed that the NF-κB signaling pathway was
activated by miR-20a overexpression, and the proinflammatory role
of miR-20a could be inhibited following treatment of LPS-induced
cells with an NF-κB inhibitor. These results suggested that miR-20a
may promote inflammation by activating the NF-κB signaling
pathway.
In conclusion, the present study confirmed that
miR-20a was upregulated in children with pediatric pneumonia and in
human lung adenocarcinoma A549 cells with LPS-induced inflammatory
injury. In addition, overexpression of miR-20a was able to promote
inflammation, which may be associated with the NF-κB signaling
pathway in pediatric pneumonia. However, further studies are
required to investigate the target molecules of miR-20a in
pediatric pneumonia.
References
|
1
|
He C, Kang L, Miao L, Li Q, Liang J, Li X,
Wang Y and Zhu J: Pneumonia mortality among children under 5 in
China from 1996 to 2013: An analysis from national surveillance
system. PLoS One. 10:e01336202015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wardlaw T, Salama P, Johansson EW and
Mason E: Pneumonia: The leading killer of children. Lancet.
368:1048–1050. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu L, Johnson HL, Cousens S, Perin J,
Scott S, Lawn JE, Rudan I, Campbell H, Cibulskis R, Li M, et al:
Global, regional, and national causes of child mortality: An
updated systematic analysis for 2010 with time trends since 2000.
Lancet. 379:2151–2161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bhutta ZA, Das JK, Walker N, Rizvi A,
Campbell H, Rudan I and Black RE: Lancet Diarrhoea and Pneumonia
Interventions Study Group: Interventions to address deaths from
childhood pneumonia and diarrhoea equitably: What works and at what
cost? Lancet. 381:1417–1429. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Leyenaar JK, Lagu T, Shieh MS, Pekow PS
and Lindenauer PK: Management and outcomes of pneumonia among
children with complex chronic conditions. Pediatr Infect Dis J.
33:907–911. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Plioplys AV and Kasnicka I: Nebulized
tobramycin: Prevention of pneumonias in patients with severe
cerebral palsy. J Pediatr Rehabil Med. 4:155–158. 2011.PubMed/NCBI
|
|
7
|
Loddenkemper T, Syed TU, Ramgopal S,
Gulati D, Thanaviratananich S, Kothare SV, Alshekhlee A and
Koubeissi MZ: Risk factors associated with death in in-hospital
pediatric convulsive status epilepticus. PLoS One. 7:e474742012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jovanovic M and Hengartner MO: miRNAs and
apoptosis: RNAs to die for. Oncogene. 25:6176–6187. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Michael MZ, O'Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
12
|
Calin GA, Liu CG, Sevignani C, Ferracin M,
Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al:
MicroRNA profiling reveals distinct signatures in B cell chronic
lymphocytic leukemias. Proc Natl Acad Sci USA. 101:11755–11760.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Khoshgoo N, Kholdebarin R, Iwasiow BM and
Keijzer R: MicroRNAs and lung development. Pediatr Pulmonol.
48:317–323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Booton R and Lindsay MA: Emerging role of
MicroRNAs and long noncoding RNAs in respiratory disease. Chest.
146:193–204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Omran A, Jagoo M, Ashhab MU, He F, Kong H,
Peng J and Yin F: MicroRNAs expression changes in acute
Streptococcus pneumoniae meningitis. Translational Neurosci.
5:131–136. 2014. View Article : Google Scholar
|
|
19
|
Hayashita Y, Osada H, Tatematsu Y, Yamada
H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y and
Takahashi T: A polycistronic microRNA cluster, miR-17-92, is
overexpressed in human lung cancers and enhances cell
proliferation. Cancer Res. 65:9628–9632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qi W, Li H, Cai XH, Gu JQ, Meng J, Xie HQ,
Zhang JL, Chen J, Jin XG, Tang Q, et al: Lipoxin A4 activates
alveolar epithelial sodium channel gamma via the
microRNA-21/PTEN/AKT pathway in lipopolysaccharide-induced
inflammatory lung injury. Lab Invest. 95:1258–1268. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Romacho T, Azcutia V, Vazquez-Bella M,
Matesanz N, Cercas E, Nevado J, Carraro R, Rodríguez-Mañas L,
Sánchez-Ferrer CF and Peiró C: Extracellular PBEF/NAMPT/visfatin
activates pro-inflammatory signalling in human vascular smooth
muscle cells through nicotinamide phosphoribosyltransferase
activity. Diabetologia. 52:2455–2463. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Plank M, Maltby S, Mattes J and Foster P:
Targeting translational control as a novel way to treat
inflammatory disease: The emerging role of microRNAs. Clin Exp
Allergy. 43:981–999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
O'Connell RM, Rao DS and Baltimore D:
microRNA regulation of inflammatory responses. Annu Rev Immunol.
30:295–312. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bazzoni F, Rossato M, Fabbri M, Gaudiosi
D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA and
Locati M: Induction and regulatory function of miR-9 in human
monocytes and neutrophils exposed to proinflammatory signals. Proc
Natl Acad Sci USA. 106:5282–5287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sheedy FJ, Palsson-McDermott E, Hennessy
EJ, Martin C, O'Leary JJ, Ruan Q, Johnson DS, Chen Y and O'Neill
LA: Negative regulation of TLR4 via targeting of the
proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat
Immunol. 11:141–147. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tili E, Michaille JJ, Cimino A, Costinean
S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA and
Croce CM: Modulation of miR-155 and miR-125b levels following
lipopolysaccharide/TNF-alpha stimulation and their possible roles
in regulating the response to endotoxin shock. J Immunol.
179:5082–5089. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang B, Xiao B, Liu Z, Li N, Zhu ED, Li
BS, Xie QH, Zhuang Y, Zou QM and Mao XH: Identification of MyD88 as
a novel target of miR-155, involved in negative regulation of
Helicobacter pylori-induced inflammation. FEBS Lett.
584:1481–1486. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Taganov KD, Boldin MP, Chang KJ and
Baltimore D: NF-κB-dependent induction of microRNA miR-146, an
inhibitor targeted to signaling proteins of innate immune
responses. Proc Natl Acad Sci USA. 103:12481–12486. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu G, Friggeri A, Yang Y, Park YJ,
Tsuruta Y and Abraham E: miR-147, a microRNA that is induced upon
Toll-like receptor stimulation, regulates murine macrophage
inflammatory responses. Proc Natl Acad Sci USA. 106:15819–15824.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Olive V, Jiang I and He L: mir-17-92, a
cluster of miRNAs in the midst of the cancer network. Int J Biochem
Cell Biol. 42:1348–1354. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
O'Donnell KA, Wentzel EA, Zeller KI, Dang
CV and Mendell JT: c-Myc-regulated microRNAs modulate E2F1
expression. Nature. 435:839–843. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pickering MT, Stadler BM and Kowalik TF:
miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate
cell cycle progression. Oncogene. 28:140–145. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fan X, Liu Y, Jiang J, Ma Z, Wu H, Liu T,
Liu M, Li X and Tang H: miR-20a promotes proliferation and invasion
by targeting APP in human ovarian cancer cells. Acta Biochim
Biophys Sin (Shanghai). 42:318–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Philippe L, Alsaleh G, Pichot A, Ostermann
E, Zuber G, Frisch B, Sibilia J, Pfeffer S, Bahram S, Wachsmann D
and Georgel P: MiR-20a regulates ASK1 expression and TLR4-dependent
cytokine release in rheumatoid fibroblast-like synoviocytes. Ann
Rheum Dis. 72:1071–1079. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhu D, Pan C, Li L, Bian Z, Lv Z, Shi L,
Zhang J, Li D, Gu H, Zhang CY, et al: MicroRNA-17/20a/106a modulate
macrophage inflammatory responses through targeting
signal-regulatory protein α. J Allergy Clin Immunol. 132:426–436.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Han Z, Boyle DL, Manning AM and Firestein
GS: AP-1 and NF-kB regulation in rheumatoid arthritis and murine
collagen-induced arthritis. Autoimmunity. 28:197–208. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sakurai H, Hisada Y, Ueno M, Sugiura M,
Kawashima K and Sugita T: Activation of transcription factor
NF-κappa B in experimental glomerulonephritis in rats. Biochim
Biophys Acta. 1316:132–138. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
van Den Brink GR, ten Kate FJ, Ponsioen
CY, Rive MM, Tytgat GN, van Deventer SJ and Peppelenbosch MP:
Expression and activation of NF-κappa B in the antrum of the human
stomach. J Immunol. 164:3353–3359. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hart LA, Krishnan VL, Adcock IM, Barnes PJ
and Chung KF: Activation and localization of transcription factor,
nuclear factor-κappaB, in asthma. Am J Respir Crit Care Med.
158:1585–1592. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma X, Buscaglia Becker LE, Barker JR and
Li Y: MicroRNAs in NF-κB signaling. J Mol Cell Biol. 3:159–166.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bhaumik D, Scott GK, Schokrpur S, Patil
CK, Orjalo AV, Rodier F, Lithgow GJ and Campisi J: MicroRNAs
miR-146a/b negatively modulate the senescence-associated
inflammatory mediators IL-6 and IL-8. Aging (Albany NY). 1:402–401.
2009. View Article : Google Scholar : PubMed/NCBI
|