Introduction
Salvia miltiorrhiza (SM), also known as
Danshen, is a member of the Labiatae family. It is a nontoxic tonic
herb used for improving microcirculation in traditional Chinese
medicine, and has been used in Asian countries to treat
cardiovascular diseases, including myocardial infarction, angina
pectoris and atherosclerosis, due to its multiple therapeutic
effects (1,2). Tanshinone IIA (Tan IIA) is a
bioactive compound extract from the dried roots of SM. Tan IIA has
been documented to possess antitumor activity in multiple types of
human cancer cell (3–7). The antitumor activity of Tan IIA is
primarily due to the inhibition of proliferation and the induction
of apoptosis (3,4). The induction of endoplasmic reticulum
stress has also been noted (5).
Tan IIA also decreases human cancer cell invasion and metastasis
(6). Thus, Tan IIA may be a
potential anti-cancer agent. A previous study reported that Tan IIA
inhibited the proliferation of non-small cell lung cancer A549
cells, potentially by decreasing the mitochondrial membrane
potential and causing apoptosis via to the induction of a higher
ratio of BCL2 associated X, apoptosis regulator/B-cell lymphoma 2
(7). However, the mechanisms
underlying the anti-cancer activity of Tan IIA remain to be further
elucidated.
The p53 tumor suppressor gene is involved in the
response to genotoxic stress exposure, including cell cycle arrest,
DNA repair, senescence or apoptosis (8). A previous study has demonstrated that
>50% of human cancers contain p53 mutations (9). P73 is a p53 family member that
generates two groups of isoforms, either containing a complete
transactivation domain (TAp73, also named P73α) or exhibiting a
truncated TA domain (10).
Inhibition of cell proliferation or induction of cell apoptosis is
induced by P73α in response to treatment with anti-cancer agents,
including doxorubicin (DOX) (11).
However, in contrast to p53, failure of tumor-formation in p73
knockout mice and observations of p73 overexpression in tumor
cells, do not support p73 as a classical tumor suppressor (12,13).
Thus, the functions of p73 remain to be further elucidated.
Murine double minute (MDM)2, as an antagonist of
p53/p73, is involved in downregulating p53/p73 activity (14). MDM2 inhibits p53 activity by
binding to the N-terminal domain of p53 and blocking p53-dependent
transcriptional activity, or by ubiquitination of p53 and targeting
it for proteasomal degradation (15). As with p53-MDM2 interaction,
binding of the P73 and MDM2 also results in the suppression of
p73-transcriptional activity (16). However, MDM2 does not ubiquitinate
p73 (17). MDM2 overexpression has
been observed in 7% of all human cancers, with higher frequencies
in soft tissue tumors, osteosarcomas and esophageal carcinomas
(18).
MDM4, also known as MDMX, is a homolog of MDM2.
Similar to MDM2, MDM4 also binds to and inhibits p53-and
p73-dependent transactivation (14). However, unlike MDM2, MDM4 does not
demonstrate appreciable ubiquitin ligase activity (14). MDM4 overexpression has been
reported in ~17% of mantle cell lymphomas, breast cancers, uterine
cancers, testicular cancers, stomach/small intestinal cancers,
colorectal cancers, lung cancers and malignant melanomas (19). However, the effects of MDM4 on
tumor properties, as well as its function during tumorigenesis,
remain unknown.
Although the mechanisms underlying MDM2 and MDM4
overexpression in human cancers are not known, overexpression of
the two proteins has been demonstrated to be associated with the
promotion of cancer and poor treatment outcome (20,21).
In the present study, the influence of Tan IIA exposure on MDM2 and
MDM4 expression and the inhibition of cell proliferation was
investigated in a p53-deficient cell model using the H1299 cell
line. The data demonstrated that Tan IIA downregulated expression
of MDM4, but not MDM2, and inhibited the viability of H1299 cells.
Herein, the cellular pathways involved in MDM4 downregulation,
which inhibited H1299 cell viability, were elucidated.
Materials and methods
Main reagents
Tan IIA (molecular formula,
C19H18O3; >96% high performance
liquid chromatography) was obtained from Herbasin (Shenyang) Co.,
Ltd. (Shenyang, China).
3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT),
dimethyl-sulfoxide (DMSO), actinomycin D, benzo(a)pyrene and DOX)
were obtained from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany).
RPMI-1640 and fetal bovine serum (FBS) were supplied by Gibco;
Thermo Fisher Scientific, Inc. (Waltham, MA, USA). The protein
isolation kit was purchased from Bio-Rad Laboratories, Inc.
(Hercules, CA, USA). MDM4 lentiviral activation particles (cat. no.
sc-417855-LAC) and control lentiviral activation particles (cat.
no. sc-437282), p73 small interfering RNA (siRNA; cat. no.
sc-36167), control siRNA-A (cat. no. sc-37007), and primary
antibodies of BCL2 binding component 3 (PUMA; cat. no. sc-374223),
anti-p73α (cat. no. sc-7238), anti-MDM2 (cat. no. sc-812),
anti-phorbol-12-myristate-13-acetate-induced protein 1 (Noxa; cat.
no. sc-56169) and anti-β-actin (cat. no. sc-8432) were all supplied
by Santa Cruz Biotechnology, Inc. (Dallas, TX, USA) and all diluted
in Tween buffered saline (1:200) for working solution. Anti-MDM4
(cat. no. ab16058) and anti-inhibitor of apoptosis 3 (IAP3; cat.
no. ab21278) antibodies were obtained from Abcam (Cambridge, UK).
Anti-Caspase 9 (cat. no. BS6444) and anti-Caspase 3 (cat. no.
BS1518) primary antibodies were supplied by Bioworld Technology,
Inc. (St. Louis Park, MN, USA). IRDye-conjugated rabbit anti-goat
(cat. no. 605-746-002), IRDye-conjugated rabbit anti-mouse (cat.
no. 610-446-C46) and IRDye-conjugated mouse anti-rabbit (cat. no.
18-4416-32) secondary antibodies were products of Rockland
immunochemicals, Inc. (Limerick, PA, USA). The concentrations of
all antibodies were determined according to the manufacturers'
protocol.
Cell culture and treatment
In the present study, the H1299 and 16HBE cells were
obtained from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China). The two cell lines
were grown in RPMI-1640 standard culture medium containing 10% FBS
(v/v), and cultured at 37°C in a humidified atmosphere consisting
of 5% CO2 and 95% air. Tan IIA was dissolved in pure
grade DMSO to create a 10 mM stock solution. Cells were treated
with Tan IIA, or 1.25 µg DOX or 32 µM benzo(a)pyrene at 37°C for 24
h. The mom-treatment group received nothing.
Western blotting
The 80 µM Tan IIA and 1.25 µg DOX treated and
untreated cells were harvested for protein extraction using whole
cell lysis buffer (1 M Tris-HCl, 5 M NaCl, 1% Nonidet P-40, 1%
sodium deoxycholate, 0.05% SDS, 1 mM phenylmethyl sulfonyl
fluoride), then centrifuged at 10,000 × g for 30 min at 4°C.
Supernatant was collected as whole cell lysate for western blotting
analysis. Protein concentrations of the whole cell lysate were
examined using a BCA Protein Assay kit (Beyotime Institute of
Biotechnology, Haimen, China), with FBS as a standard. The whole
cell lysates were boiled in SDS Laemmli sample buffer, then 20 µg
whole cell lysate/lane was separated by 12% SDS-polyacrylamide gel
and subsequently transferred to polyvinylidene fluoride membranes.
Following protein transfer, the membranes were blocked at 37°C for
30 min with Tris-buffered saline containing 0.1% Tween-20 and 5%
non-fat dry milk, then incubated with specific primary antibodies
which were dissolved in TBS overnight at 4°C and with the
corresponding IRDye-conjugated secondary antibodies (1:5,000
dilution for all) at room temperature for 1 h. The signals of
immunoreactive proteins were visualized using the Odyssey Infrared
Imaging System and Odyssey version 1.2 software (LI-COR
Biosciences, Lincoln, NE, USA). The relative densities of the
protein bands were analyzed using Quantity One software (version
4.62; Bio-Rad Laboratories, Inc.). The relative expression of
target proteins were normalized to the corresponding intensities of
β-actin. Three independent replicates were performed.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA of 32 µM Benzo(a)pyrene- or 80 µM Tan IIA
treated and untreated H1299 cells were extracted with TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. First-strand cDNA synthesis (45 cycles)
was performed on the 1,000-Series Thermal Cycling Platform with a
mixture of random monomers and oligo-dT primers according to the
manufacturer's protocol for the First Strand cDNA Synthesis kit
(K1612; Thermo Fisher Scientific, Inc.). cDNA amplification was
performed at 95°C for 10 sec and 56°C for 30 sec (45 cycles) on a
7900 Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.), using the Quanti-Fast SYBR Green RT-PCR kit
(Qiagen, Inc., Valencia, CA, USA), according to the manufacturer's
protocol. All specific primers for the target genes, as well as the
housekeeping gene, β-actin, were custom-designed and synthesized by
TaKaRa (Dalian, China; sequences not provided). Data was expressed
as n-fold change of untreated cells. Each sample mRNA was
normalized relative to β-actin using the 2−∆∆Cq method
(22).
Analysis of mRNA stability and protein
generation
The stability of mRNA was measured by using a
standard actinomycin D analysis. Cells were exposed to 5 µg/ml
actinomycin D with or without 80 µM Tan IIA for 0, 2 and 4 h, and
total RNA was isolated and analyzed as mentioned above. Protein
generation was measured using a standard protein-synthesis
inhibitor cycloheximide (CHX) as previously described (23). To arrest polyribosome migration,
the H1299 cells were treated with 35 µM CHX combined with or
without 80 µM Tan IIA for 0, 2 and 4 h, and were then lysed in
order to isolate cytoplasmic extracts in a hypotonic lysis buffer
containing 10 mM Tris, 1 mM EDTA, and 0.2% Triton X-100. Cell
lysates were added to a 15–45 % (w/v) sucrose gradient tube,
centrifuged at 4°C, 15,000 × g for 1 h, then the lysate was
collected from each gradient tube for western blot analysis.
Transfections
H1299 cells were plated into 6-well plates at a
density of 2.5×105 cells/well. Upon reaching 80%
confluence, cells were trypsinized and diluted in the RPMI-1640
medium. Transfection of MDM4 lentiviral activation particles (RNA)
and control lentiviral activation particles (non-specific RNA) into
H1299 cells was performed using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The p73 siRNA and control siRNA-A
transfection was performed using the Entranster-D transfection
reagent (Engreen Biosystem Ltd., Beijing, China), following the
manufacturer's protocol.
Cytotoxicity assay
The cells were seeded in 96-well plates (~3,000 per
well) for 24 h. Total of 80 µM Tan IIA dissolved in DMSO was added
to the cells. Following incubation for the 0, 8 and 24 h at 37°C,
the culture medium in each well was removed, cells were washed
twice with PBS, then incubated with fresh serum-free RPMI-1640
medium containing 10% MTT solution (v/v, concentration: 5 mg/ml) at
37°C for a further 4 h. To solubilize the formazan crystals in the
viable cells, the liquid in each well was replaced by 150 µl DMSO.
The absorbance of each well was measured at 570 nm using
Multi-Detection Microplate Readers (Synergy 2; BioTek Instruments,
Inc., Winooski, VA, USA). Data were expressed as a percentage of
the control measured in the absence of Tan IIA.
Cell apoptosis assay
An Annexin V-Fluorescein Isothiocyanate (FITC)
Apoptosis Detection kit [Multi Sciences (Lianke) Biotech Co., Ltd.,
Hangzhou, China] was used to measure the percentage of apoptotic
cells. Treated and untreated cells (~5×105 cells/ml)
were centrifuged at 1,000 × g for 5 min at room temperature
followed by washing twice with PBS, then the cells were
re-suspended in 0.5 ml pre-chilled binding buffer, 5 µl Annexin
V-FITC and 10 µl propidium iodide (PI) were added, and the cells
were incubated for 30 min at room temperature. Finally, the dead
cells were differentiated by FACScan flow cytometry using a Beckman
Coulter FC 500 flow cytometer (Beckman Coulter, Inc., Brea, CA,
USA) and analyzed using ModFit software (version 2.0.0; Verity
Software House, Inc., Topsham, ME, USA).
Statistical analysis
Data ware presented as the mean ± standard
deviation. Comparisons among different groups were analyzed using
one-way analysis of variance. If the variation was significant,
significant differences between the means of treated and un-treated
groups were analysed using Dunnett-t tests. P<0.05 was
considered to indicate a statistically significant difference. All
statistical analysis was performed using SPSS 22.0 statistical
software (IBM Corp., Armonk, NY, USA).
Results
Tan IIA suppresses MDM4 expression in
H1299 and 16HBE cells
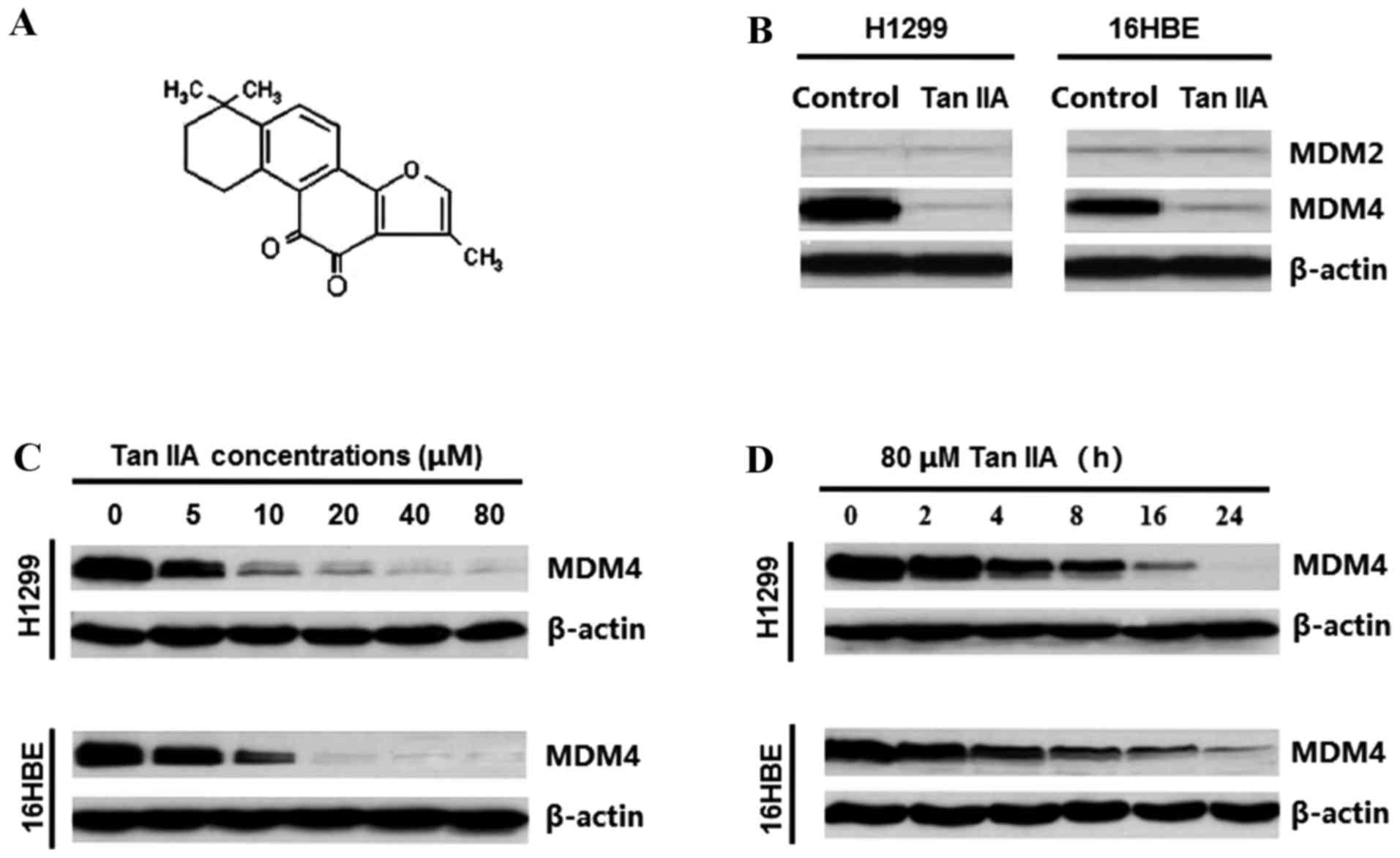
To investigate the effect of Tan IIA (Fig. 1A) on MDM2 and MDM4 expression in
H1299 and 16HBE cells, the two cell lines were treated with 40 µM
Tan IIA for 24 h. Tan IIA visibly downregulated MDM4, but not MDM2,
in the two cell lines studied (Fig.
1B). In addition, Tan IIA downregulated MDM4 protein levels in
a dose-dependent manner in H1299 and 16HBE cells (Fig. 1B), even at low concentrations,
following treatment for 24 h. The inhibition of MDM4 by Tan IIA
occurred ~4 h post-exposure and was followed by a time-dependent
reduction during 8–24 h treatment (Fig. 1C).
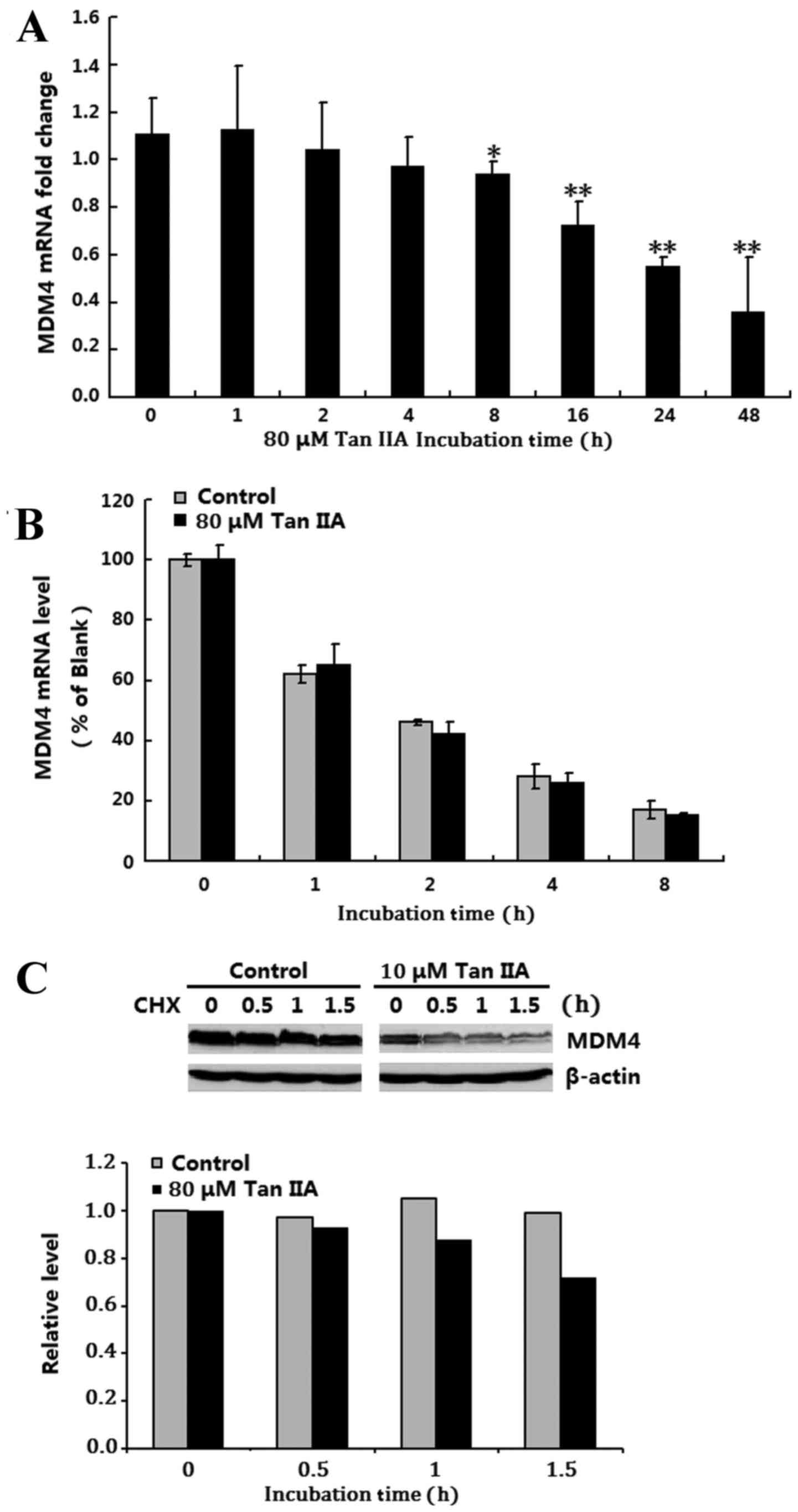
Tan IIA suppresses MDM4 expression
through the repression of MDM4 mRNA synthesis
To understand the mechanism by which Tan IIA
downregulates MDM4 expression, MDM4 mRNA expression levels were
examined in the p53 deficient H1299 cells following exposure to 80
µM Tan IIA for 1, 2, 4, 8, 16, 24 and 48 h using RT-qPCR. The
results demonstrated that Tan IIA significantly downregulated MDM4
mRNA expression 8–48 h post treatment compared with 0 h (P<0.05;
Fig. 2A). Next, the influence of
mRNA stability on the downregulation of MDM4 mRNA expression in Tan
IIA-treated H1299 cells was investigated using a pulse-chase assay
and RT-qPCR analysis. There was no difference in MDM4 mRNA
stability between the untreated control and the 80 µM Tan
IIA-treated group, which indicated that MDM4 mRNA stability was not
affected by Tan IIA treatment (Fig.
2B). The involvement of MDM4 protein stabilization in Tan
IIA-induced MDM4 inhibition in H1299 cells was then investigated
and a standard CHX pulse-chase assay and western blotting were
performed. There was no significant difference in MDM4 protein
half-life between the untreated control and the 80 µM Tan IIA
treated group (Fig. 2C). These
results suggested that Tan IIA downregulated MDM4 only through
inhibition of MDM4 mRNA synthesis.
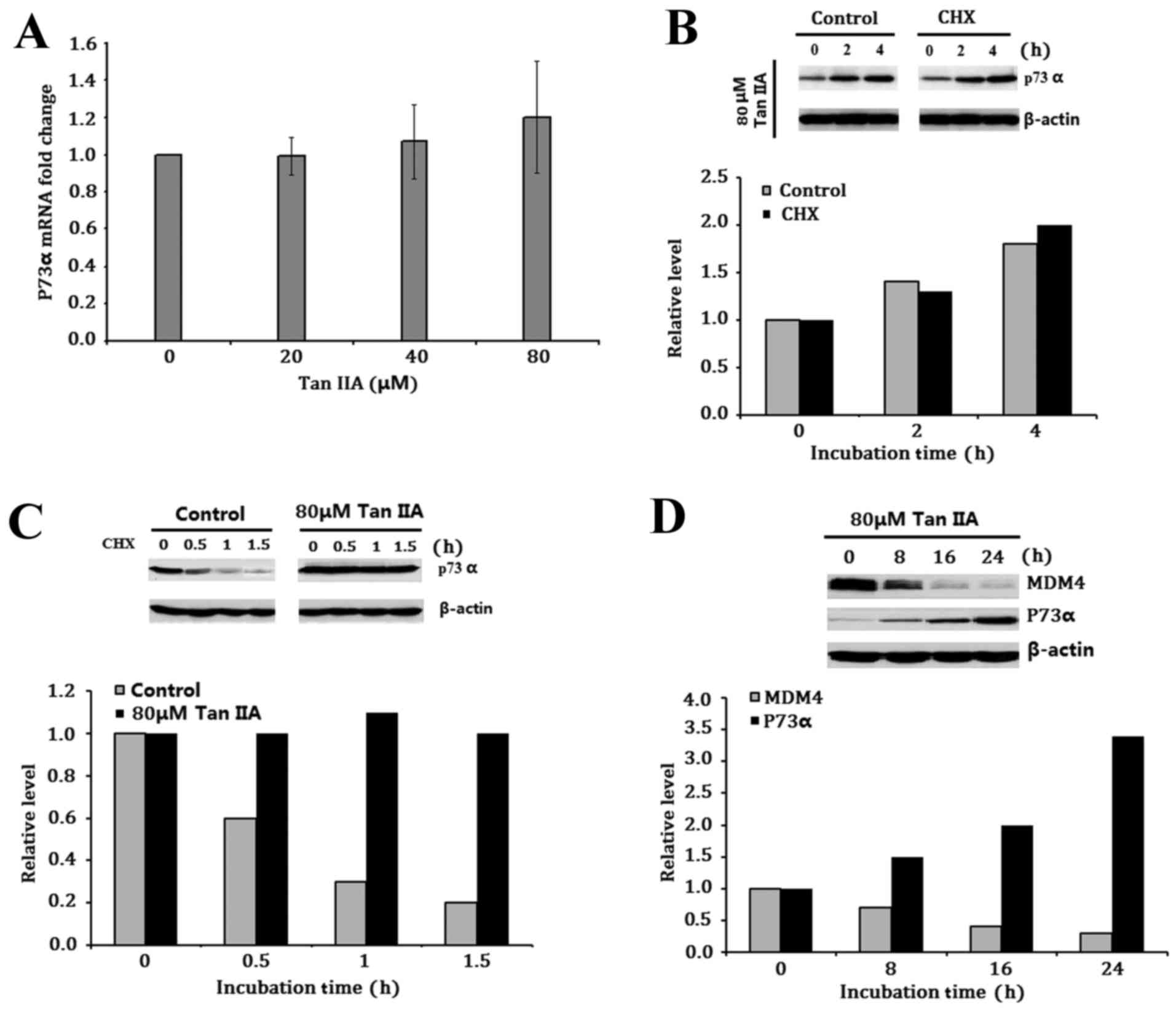
Tan IIA-induced MDM4 inhibition
upregulates P73α expression
The effect of Tan IIA-inhibited MDM4 expression on
P73α expression was then investigated. P73α mRNA levels were
measured using RT-qPCR. Treatment with 80 µM Tan IIA did not
demonstrate any directly inductive or inhibitory effects on p73
mRNA expression following treatment for 24 h (Fig. 3A). The involvement of Tan IIA in
modulation of P73α translation was then investigated, and the H1299
cells were exposed to 80 µM Tan IIA with or without the protein
synthesis inhibitor CHX. Tan IIA increased P73α protein expression
irrespective of CHX absence or presence, suggesting that Tan IIA
does not affect P73α translation (Fig.
3B). The turnover of P73α protein following Tan IIA treatment
was measured using a standard pulse-chase assay. The half-life of
P73α in the untreated control group was less than <0.5 h, while
it was significantly longer in 80 µM Tan IIA treated groups
(Fig. 3C), suggesting that the
observed upregulation of Tan IIA-induced P73α expression in H1299
cells is only due to post-translational modulation. In addition,
the relative levels of MDM4 and P73α were evaluated following
treatment of H1299 cells with 80 µM Tan IIA for 8, 16 and 24 h. Tan
IIA upregulated P73α levels following the downregulation of MDM4
levels (Fig. 3D).
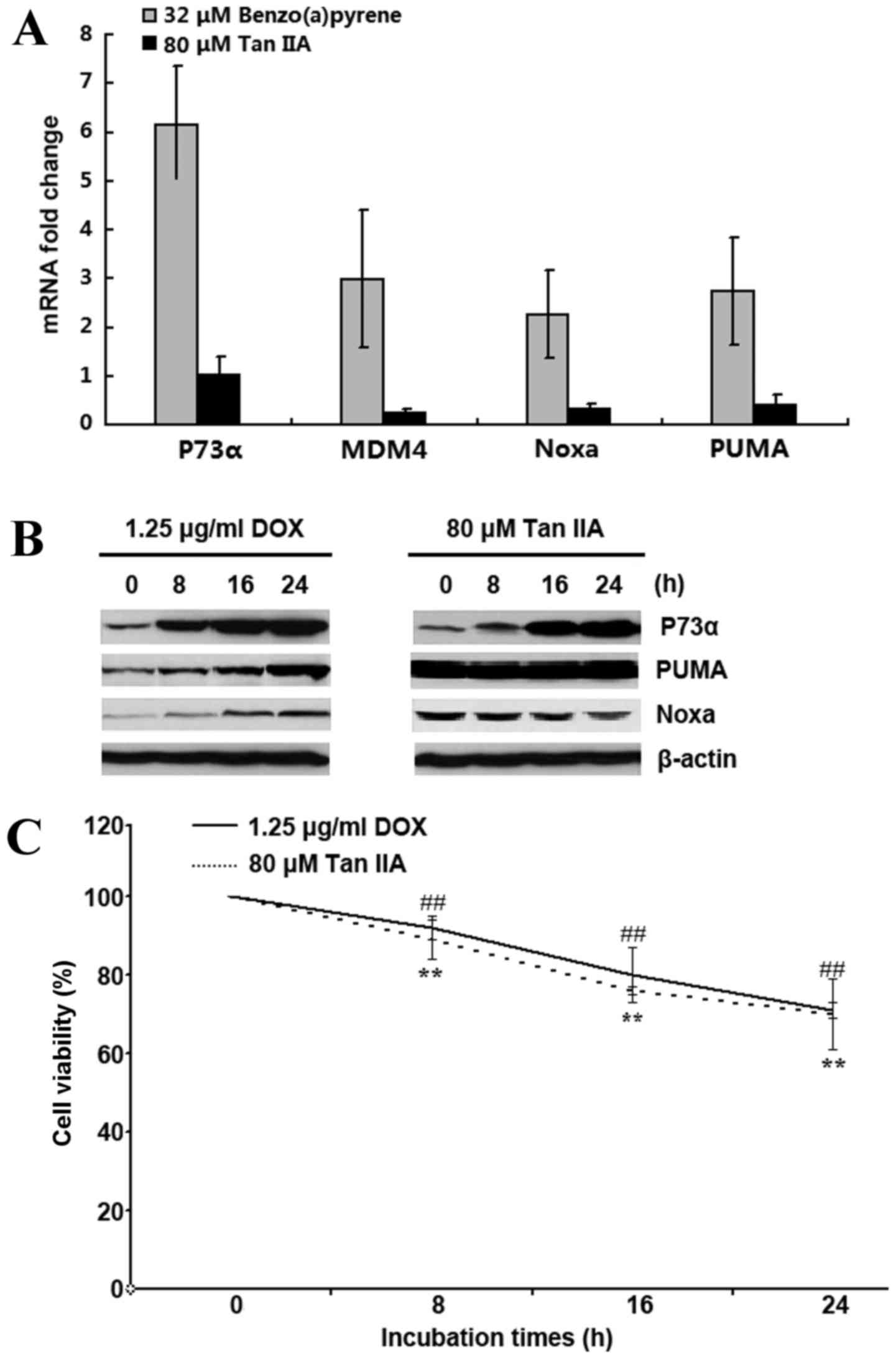
P73α-induced inhibition of cell
viability did not occur in the Tan IIA-treated H1299 cells
p73 has been observed to stimulate transcriptional
levels of PUMA and Noxa, which result in mitochondria-dependent
apoptosis (24,25). Thus, the expression levels of P73α,
MDM4, PUMA and Noxa were examined in H1299 cells following
treatment with 80 µM Tan IIA for 24 h using RT-qPCR. Compared with
benzo(a)pyrene that induces P73 mRNA (9), Tan IIA failed to upregulate and in
fact downregulated P73α, MDM4, PUMA and Noxa mRNA expression levels
(Fig. 4A). However, 80 µM Tan IIA
treatment for 8, 16 and 24 h visibly increased P73α protein
expression, similar to the effect of the 1.25 µg/ml DOX treated
group (Fig. 4B), but the increased
P73α levels failed to induce an increase in PUMA and Noxa protein
expression (Fig. 4B). Although
PUMA and Noxa were not upregulated, cell viability was inhibited in
the 80 µM Tan IIA-treated group, similar the 1.25 µg/ml DOX-treated
group (Fig. 4C).
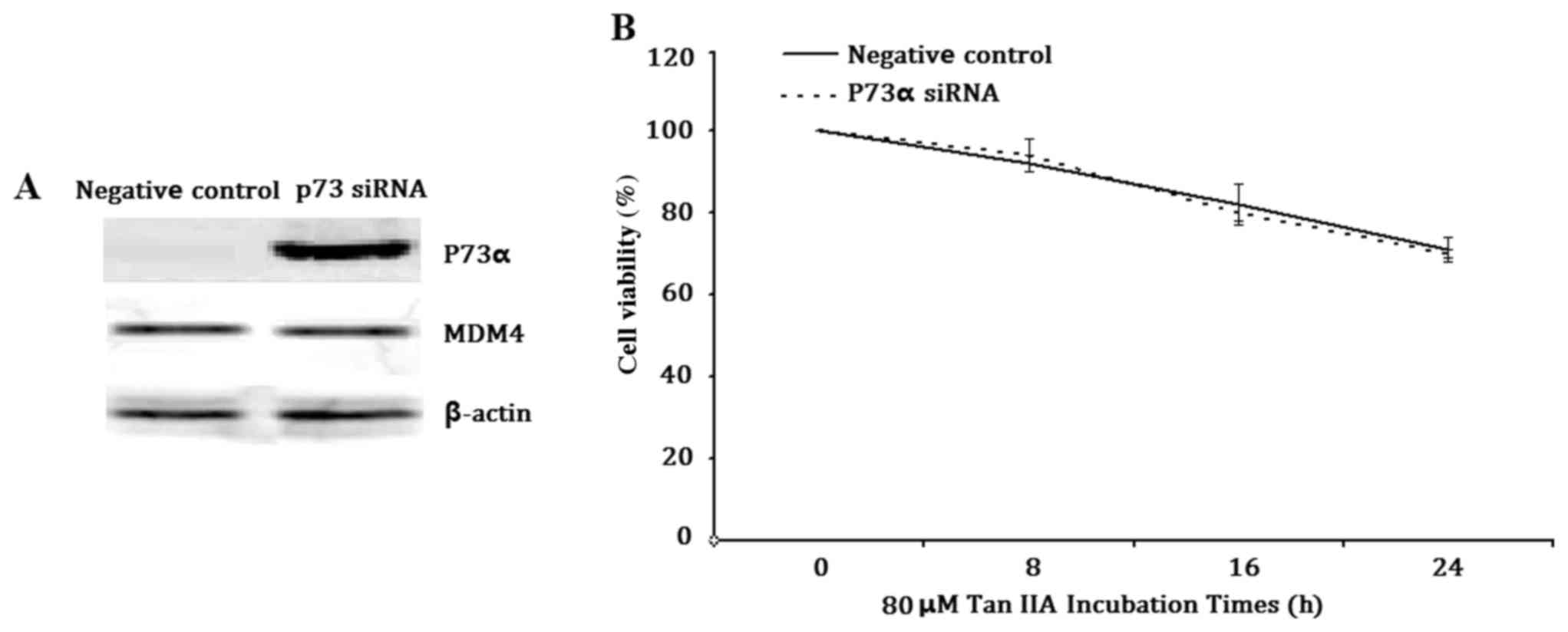
Knockdown of P73α does not affect Tan
IIA-inhibited H1299 cell viability
To further confirm whether P73α exerted a tumor
suppressor function in this cell model, P73α was inhibited using
siRNA (Fig. 5A). Notably, there
was no difference in cell viability between the P73α siRNA group
and negative control group following treatment of the cells with 80
µM Tan IIA for 8, 16 or 24 h (Fig.
5B). These results suggested that P73α does not regulate
transcriptional activation in response to Tan IIA treatment in this
cell model.
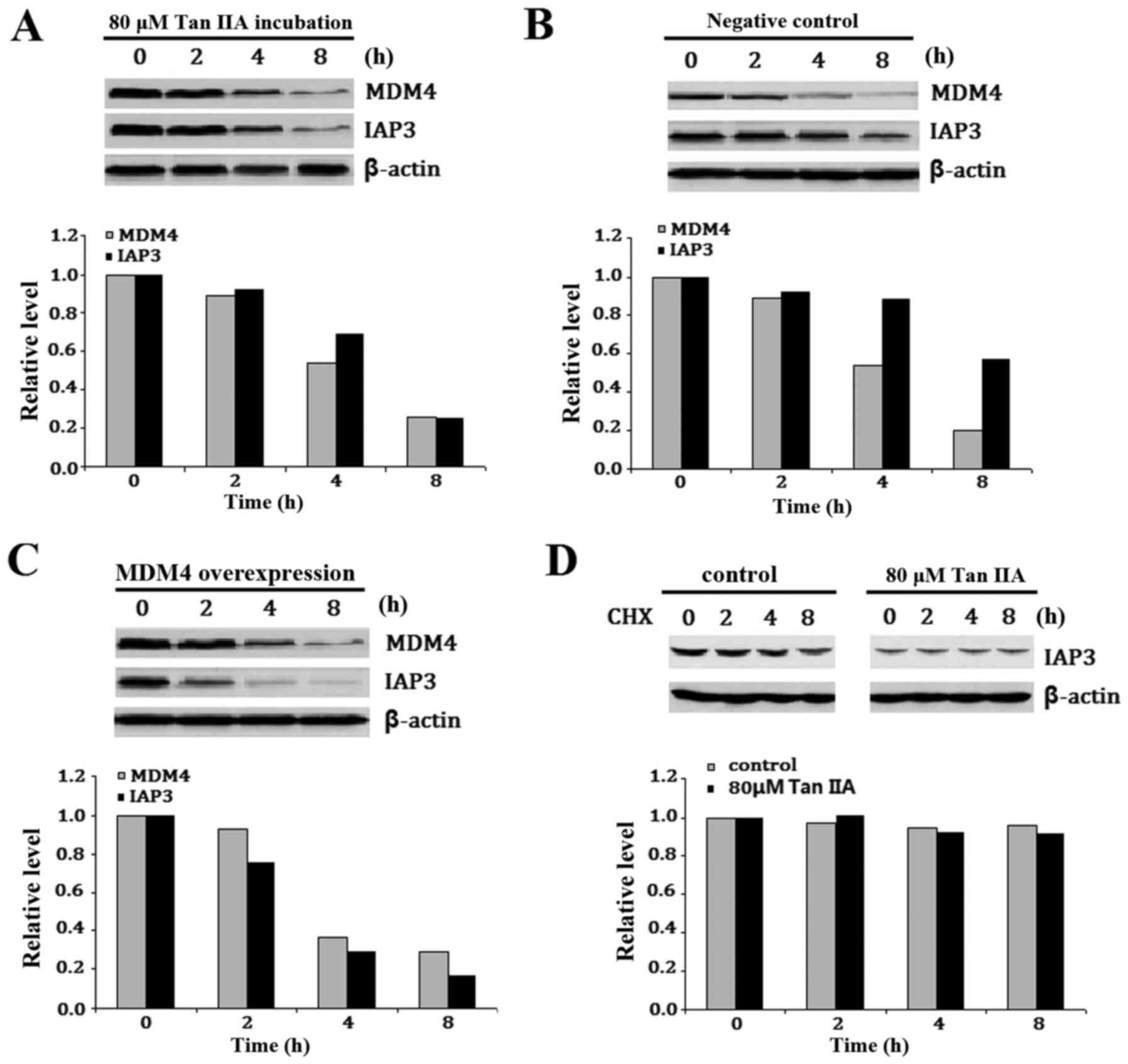
Tan IIA inhibits IAP3 in H1299 cells
overexpressing MDM4
IAP3 is a known translational target of MDM2
(26) and thus it was hypothesized
that Tan IIA-induced downregulation of MDM4 may also affect IAP3
expression. To investigate this, IAP3 protein expression was
examined following treatment of H1299 cells with or without 80 µM
Tan IIA for 2, 4 and 8 h. IAP3 expression was visibly reduced in
the Tan IIA-treated H1299 cells following MDM4 downregulation
(Fig. 6A). To further confirm the
involvement of Tan IIA-inhibited MDM4 in the reduction of IAP3,
H1299 cells were transfected with MDM4 lentiviral activation
particles, which express wild-type MDM4, and were termed
MDM4-overexpressing H1299 cells. Empty vector control lentiviral
activation particles were also transfected and termed the negative
control. The cells were then treated with or without 80 µM Tan IIA
for 2, 4 and 8 h. A visible reduction of IAP3 expression was
detected in the MDM4-overexpressing H1299 cells compared with the
negative control H1299 cells (Fig. 6B
and C). In addition, no appreciable difference was detected in
IAP protein stability between H1299 cells treated with or without
Tan IIA (Fig. 6D). These results
indicated that Tan IIA suppressed IAP3 expression in H1299 cells
overexpressing MDM4 at the transcriptional and translational
level.
Tan IIA sensitizes H1299 cells
overexpressing MDM4 to DOX-induced apoptosis
To evaluate the effect of Tan IIA on H1299 cells
overexpressing MDM4, the cells were exposed to 5–80 µM Tan IIA for
24 h. Treatment with Tan IIA at a concentration of 20–80 µM
significantly decreased cell viability in a dose-dependent manner
in MDM4-overexpressing and negative control H1299 cells, and the
inhibitory effect was stronger in MDM4-overexpressing H1299 cells
(Fig. 7A). In addition, the
potential synergistic effect of Tan IIA in DOX-induced apoptosis
was assessed by treating the H1299 cells overexpressing MDM4 with
80 µM Tan IIA and/or 1.25 µg/ml DOX for 24 h. Tan IIA and DOX
significantly induced apoptosis in H1299 cells overexpressing MDM4,
and when the same concentrations were given in combination,
significantly more cells underwent apoptosis (Fig. 7B). These results suggested that Tan
IIA may work in synergy with DOX, and may be able to reverse
DOX-resistance in p53-deficient tumor cells.
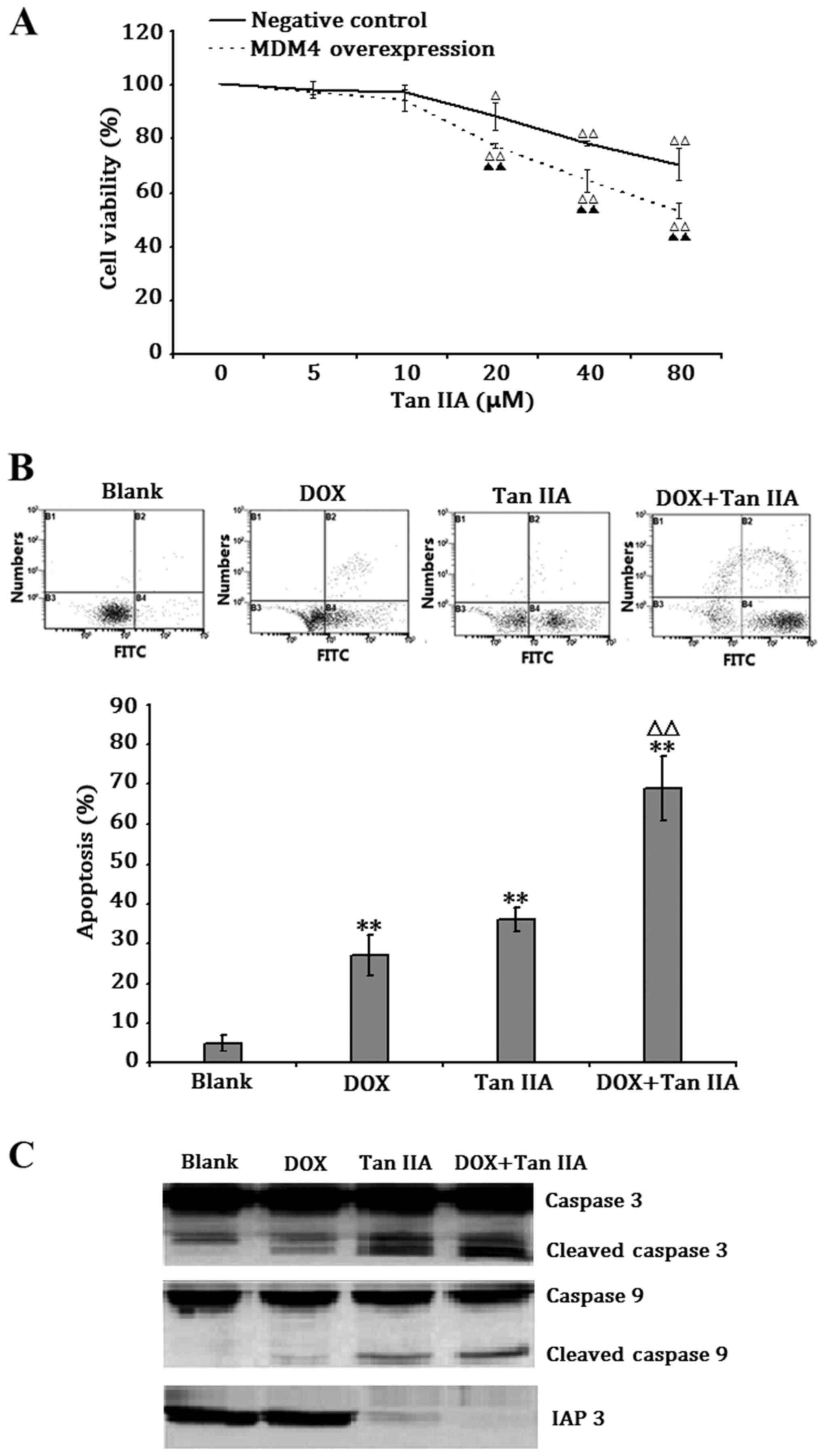 | Figure 7.Tan IIA sensitized H1299 cells
overexpressing MDM4 to DOX-induced apoptosis. (A) Transfected H1299
cell viability following exposure to the indicated concentrations
of Tan IIA for 24 h. (B) Apoptosis of H1299 cells overexpressing
MDM4 following exposure to 80 µM Tan IIA or 1.25 µg/ml DOX, alone
or in combination, for 24 h. (C) IAP3, caspase 3, cleaved caspase
3, caspase 9 and cleaved caspase 9 protein levels in H1299 cells
following exposure to 80 µM Tan IIA or 1.25 µg/ml DOX, alone or in
combination, for 24 h. Data represent the mean ± standard deviation
of apoptosis from three independent experiments. **P<0.01 vs.
unexposed group (Blank). ΔP<0.05 vs. DOX-treated
group, ΔΔP<0.01 vs. vs. untransfected H1299 cells.
Tan IIA, tanshinone IIA; MDM4, murine double minute 4; DOX,
doxorubicin; IAP3, inhibitor of apoptosis 3. |
Discussion
Tan IIA has been widely studied due to its
anticancer properties. Tan IIA has previously been reported to
inhibit cell proliferation and to induce apoptosis through caspase-
and mitochondria-dependent pathways (27). In the present study, the potential
effects of Tan IIA on the expression of two p53/p73 negative
regulators, MDM2 and MDM4, were investigated in p53-deficient H1299
cells. Tan IIA suppressed MDM4 expression, which resulted in the
accumulation of P73α and inhibition of H1299 cell viability.
However, the pro-apoptotic function of P73, which induces PUMA and
Noxa, were not activated. Tan IIA was also demonstrated to suppress
IAP3 expression, which in turn activated Caspase 3 and Caspase 9,
as well as induction of cell apoptosis.
Inhibitor-of-apoptosis proteins (IAPs) are the only
known endogenous proteins that act to suppress apoptosis via
inhibition of initiator and effector caspases (28). Among the IAPs, only IAP3 is a
direct inhibitor of caspases, and is the most effective caspase
inhibitor compared with the other IAPs (29). IAP3 protein has been demonstrated
to bind to and inhibit the activated forms of initiator and
effector caspases, which are the enzymes that induce
mitochondrial-dependent apoptosis (30). IAP3 expression levels are
positively correlated with disease progression and, furthermore,
its overexpression is associated with tumorigenesis (31,32).
As well as contributing to cancer development, IAP3 has also been
reported to contribute to chemotherapy resistance, and inhibiting
this protein was demonstrated to efficiently sensitize cells to
apoptosis in response to anti-cancer agents (33,34).
MDM2 binds to the IAP3 internal ribosome entry site (IRES) RNA
through its Really Interesting New Gene (RING) finger domain and
upregulates IRES-regulated IAP3 translation, which results in
overexpression of IAP3 and resistance to anticancer therapy
(26). Since MDM4 also has a RING
finger domain through which it is able to interact with MDM2
(35), Tan IIA-induced MDM4
inhibition may affect IAP3 expression. To investigate this, the
present study examined whether Tan IIA-induced suppression of MDM4
results in downregulation of IAP3 expression. Tan IIA was
demonstrated to suppress IAP3 expression at the transcriptional and
translational level, and the inhibition of IAP3 induced by Tan IIA
was greater in H1299 cells overexpressing MDM4 than in the negative
control H1299 cells.
The inhibition of viability in the H1299 cell model
was mainly due to Tan IIA-induced downregulation of IAP3, and this
was further confirmed by observation that there was no induction of
P73α function and activation of caspase 3 and caspase 9 in these
cells. The results demonstrated that, although the suppression of
MDM4 induced by Tan IIA upregulated P73α expression in the H1299
cells, the two P73 transcriptional targets, PUMA and Noxa, were not
activated. Although the reason why PUMA and Noxa were not activated
by Tan IIA-induced upregulation of P73α was not examined, it is
possible to hypothesize that the stability of PUMA and Noxa mRNA
was decreased by Tan IIA exposure, even though their promoters may
have been activated by the upregulation of P73α. The interactions
among these proteins require further elucidation. As a Food and
Drug Administration-approved chemotherapeutic drug, DOX is used to
treat a wide range of types of cancer (36,37).
However, clinical use of DOX is limited by its severe side effects,
including kidney injury (38).
When DOX is given in combination with other antitumor agents,
including curcumin or tripeptide, its therapeutic effect is
elevated while side effects are relieved, and combination therapy
has become the first choice for DOX-based chemotherapy (36,39).
The results of the present study further confirmed the results of
previous reports (3–7); demonstrating that Tan IIA-treated
H1299 cells overexpressing MDM4 were more sensitive to DOX-induced
apoptosis, supporting the potential of Tan IIA as an agent for
sensitization of cells to the anti-cancer effect of DOX. This
facilitates the avoidance or reduction of the side effects of Tan
IIA as it can be effectively used at a lower dose.
In conclusion, the present study demonstrated that
Tan IIA exposure resulted in the inhibition of p53 deficient H1299
cell viability through the MDM4-IAP3-caspase signaling pathway, and
increased sensitivity of H1299 cells to DOX-induced apoptosis.
Thus, it may be possible to use Tan IIA alone or combination with
other anti-cancer agents to treat p53-deficient and/or
MDM4-overexpressing cancer cells. Further in vivo studies
are required to confirm the contribution of Tan IIA to tumor
therapy.
Glossary
Abbreviations
Abbreviations:
|
Tan IIA
|
Tanshinone IIA
|
|
SM
|
Salvia miltiorrhiza
|
|
MDM4
|
murine double minute 4
|
|
DMSO
|
dimethyl sulfoxide
|
|
CHX
|
cycloheximide
|
|
IAP3
|
inhibitor-of-apoptosis protein 3
|
|
DOX
|
doxorubicin
|
References
|
1
|
Yang W, Ju JH, Jeon MJ, Han X and Shin I:
Danshen (Salvia miltiorrhiza) extract inhibits proliferation of
breast cancer cells via modulation of Akt activity and p27 level.
Phytother Res. 24:198–204. 2010.PubMed/NCBI
|
|
2
|
Franek KJ, Zhou Z, Zhang WD and Chen WY:
In vitro studies of baicalin alone or in combination with Salvia
miltiorrhiza extract as a potential anti-cancer agent. Int J Oncol.
26:217–224. 2005.PubMed/NCBI
|
|
3
|
Munagala R, Aqil F, Jeyabalan J and Gupta
RC: Tanshinone IIA inhibits viral oncogene expression leading to
apoptosis and inhibition of cervical cancer. Cancer Lett.
356:536–546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang JF, Feng JG, Han J, Zhang BB and Mao
WM: The molecular mechanisms of Tanshinone IIA on the apoptosis and
arrest of human esophageal carcinoma cells. Biomed Res Int.
2014:5827302014.PubMed/NCBI
|
|
5
|
Hwang SL, Yang JH, Jeong YT, Kim YD, Li X,
Lu Y, Chang YC, Son KH and Chang HW: Tanshinone IIA improves
endoplasmic reticulum stress-induced insulin resistance through
AMP-activated protein kinase. Biochem Biophys Res Commun.
430:1246–1252. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shan YF, Shen X, Xie YK, Chen JC, Shi HQ,
Yu ZP, Song QT, Zhou MT and Zhang QY: Inhibitory effects of
tanshinone II-A on invasion and metastasis of human colon carcinoma
cells. Acta Pharmacol Sin. 11:1537–1542. 2009. View Article : Google Scholar
|
|
7
|
Chiu TL and Su CC: Tanshinone IIA induces
apoptosis in human lung cancer A549 cells through the induction of
reactive oxygen species and decreasing the mitochondrial membrane
potential. Int J Mol Med. 25:231–236. 2010.PubMed/NCBI
|
|
8
|
Reincke S, Govbakh L, Wilhelm B, Jin H,
Bogdanova N, Bremer M, Karstens JH and Dörk T: Mutation analysis of
the MDM4 gene in German breast cancer patients. BMC Cancer.
8:522008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang Y, Rao K, Yang G, Chen X, Wang Q,
Liu A, Zheng H and Yuan J: Benzo(a)pyrene induces p73 mRNA
expression and necrosis in human lung adenocarcinoma H1299 cells.
Environ Toxicol. 27:202–210. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alexandrova EM and Moll UM: Role of p53
family members p73 and p63 in human hematological malignancies.
Leuk Lymphoma. 53:2116–2129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fedorova NE, Emelianova SS, Vinogradskaya
GR, Chichev EV, Murzakova AV, Kirichenko AA, Verbenko VN and Kushch
AA: Effect of Anti-Cancer Drug Doxorubicine on Cytomegalovirus
Infected Human Fibroblasts. Tsitologiia. 57:260–268. 2015.(In
Russian). PubMed/NCBI
|
|
12
|
Zaika AI, Kovalev S, Marchenko ND and Moll
UM: Overexpression of the wild type p73 gene in breast cancer
tissues and cell lines. Cancer Res. 59:3257–3263. 1999.PubMed/NCBI
|
|
13
|
Stiewe T and Putzer BM: p73 in apoptosis.
Apoptosis. 6:447–452. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Momand J, Villegas A and Belyi VA: The
evolution of MDM2 family genes. Gene. 486:23–30. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pei D, Zhang Y and Zheng J: Regulation of
p53: A collaboration between Mdm2 and Mdmx. Oncotarget. 3:228–235.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Michael D and Oren M: The p53 and Mdm2
families in cancer. Curr Opin Genet Dev. 12:53–59. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeng X, Chen L, Jost CA, Maya R, Keller D,
Wang X, Kaelin WG Jr, Oren M, Chen J and Lu H: MDM2 suppresses p73
function without promoting p73 degradation. Mol Cell Biol.
19:3257–3266. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Katayama A, Ogino T, Bandoh N, Takahara M,
Kishibe K, Nonaka S and Harabuchi Y: Overexpression of small
ubiquitin-related modifier-1 and sumoylated Mdm2 in oral squamous
cell carcinoma: Possible involvement in tumor proliferation and
prognosis. Int J Oncol. 31:517–524. 2007.PubMed/NCBI
|
|
19
|
Laurie NA, Donovan SL, Shih CS, Zhang J,
Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al:
Inactivation of the p53 pathway in retinoblastoma. Nature.
444:61–66. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gustafsson B, Axelsson B, Gustafsson B,
Christensson B and Winiarski J: MDM2 and p53 in childhood acute
lymphoblastic leukemia: Higher expression in childhood leukemias
with poor prognosis compared to long-term survivors. Pediatr
Hematol Oncol. 18:497–508. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gustafsson B, Christenson B, Hjalmar V and
Winiarski J: Cellular expression of MDM2 and p53 in childhood
leukemias with poor prognosis. Med Pediatr Oncol. 34:117–124. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Croons V, Martinet W, De Herman AG and
Meyer GR: Differential effect of the protein synthesis inhibitors
puromycin and cycloheximide on vascular smooth muscle cell
viability. J Pharmacol Exp Ther. 325:824–832. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Melino G, Bernassola F, Ranalli M, Yee K,
Zong WX, Corazzari M, Knight RA, Green DR, Thompson C and Vousden
KH: p73 Induces apoptosis via PUMA transactivation and Bax
mitochondrial translocation. J Biol Chem. 27:279: 8076–8083.
2004.
|
|
25
|
Grande L, Bretones G, Rosa-Garrido M,
Garrido-Martin EM, Hernandez T, Fraile S, Botella L, de Alava E,
Vidal A, Garcia del Muro X, et al: Transcription factors Sp1 and
p73 control the expression of the proapoptotic protein NOXA in the
response of testicular embryonal carcinoma cells to cisplatin. J
Biol Chem. 287:26495–26505. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gu L, Zhu N, Zhang H, Durden DL, Feng Y
and Zhou M: Regulation of XIAP translation and induction by MDM2
following irradiation. Cancer Cell. 15:363–375. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang J, Wang J, Jiang JY, Liu SD, Fu K
and Liu HY: Tanshinone IIA induces cytochrome c-mediated caspase
cascade apoptosis in A549 human lung cancer cells via the JNK
pathway. Int J Oncol. 45:683–690. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chow KU, Nowak D, Boehrer S, Ruthardt M,
Knau A, Hoelzer D, Mitrou PS and Weidmann E: Synergistic effects of
chemotherapeutic drugs in lymphoma cells are associated with
down-regulation of inhibitor of apoptosis proteins (IAPs),
prostate-apoptosis-response-gene 4 (Par-4), death-associated
protein (Daxx) and with enforced caspase activation. Biochem
Pharmacol. 66:711–724. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
LaCasse EC, Mahoney DJ, Cheung HH,
Plenchette S, Baird S and Korneluk RG: IAP-targeted therapies for
cancer. Oncogene. 27:6252–6275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eckelman BP, Salvesen GS and Scott FL:
Human inhibitor of apoptosis proteins: Why XIAP is the black sheep
of the family. EMBO Rep. 7:988–994. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang R, Li B, Wang X, Lin F, Gao P, Cheng
SY and Zhang HZ: Inhibiting XIAP expression by RNAi to inhibit
proliferation and enhance radiosensitivity in laryngeal cancer cell
line. Auris Nasus Larynx. 36:332–339. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qiao L, Li GH, Dai Y, Wang J, Li Z, Zou B,
Gu Q, Ma J, Pang R, Lan HY and Wong BC: Gene expression profile in
colon cancer cells with respect to XIAP expression status. Int J
Colorectal Dis. 24:245–260. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Paschall AV, Zimmerman MA, Torres CM, Yang
D, Chen MR, Li X, Bieberich E, Bai A, Bielawski J, Bielawska A and
Liuet K: Ceramide targets xIAP and cIAP1 to sensitize metastatic
colon and breast cancer cells to apoptosis induction to suppress
tumor progression. BMC Cancer. 14:242014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Golovine K, Makhov P, Uzzo RG, Kutikov A,
Kaplan DJ, Fox E and Kolenko VM: Cadmium down-regulates expression
of XIAP at the post-transcriptional level in prostate cancer cells
through an NF-kappaB-independent, proteasome-mediated mechanism.
Mol Cancer. 9:1832010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang L, He G, Zhang P, Wang X, Jiang M and
Yu L: Interplay between MDM2, MDMX, Pirh2 and COP1: The negative
regulators of p53. Mol Biol Rep. 38:229–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhu ZF, Chen LJ, Lu R, Jia J, Liang Y, Xu
Q, Zhou CL, Wang L, Wang S and Yao Z: Tripeptide tyroserleutide
plus doxorubicin: Therapeutic synergy and side effect attenuation.
BMC Cancer. 8:3422008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Carvalho C, Santos RX, Cardoso S, Correia
S, Oliveira PJ, Santos MS and Moreira PI: Doxorubicin: The good,
the bad and the ugly effect. Curr Med Chem. 16:3267–3285. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zeidán Q, Strauss M, Porras N and Anselmi
G: Differential long-term subcellular responses in heart and liver
to adriamycin stress. Exogenous L-carnitine cardiac and hepatic
protection. J Submicrosc Cytol Pathol. 34:315–321. 2002.PubMed/NCBI
|
|
39
|
Wang J, Ma W and Tu P: Synergistically
Improved Anti-tumor Efficacy by Co-delivery Doxorubicin and
Curcumin Polymeric Micelles. Macromol Biosci. 15:1252–1261. 2015.
View Article : Google Scholar : PubMed/NCBI
|