Introduction
Uveal melanoma (UM) is one of the most common
malignant tumors in the adult eye (1). The treatments for UM are
chemotherapy, radiotherapy and surgical resection (2). Despite the development of treatments,
UM exhibits a high mortality rate (3). A number of studies have investigated
the molecular mechanisms associated with the development and
progression of UM, with the aim of developing more effective
treatments for the disease (4).
Forkhead box O (FoxO) proteins are important for the
regulation of cell proliferation, differentiation, DNA damage
repair and autophagy (5,6). Forkhead box protein O3 (FoxO3a), a
transcription factor which is an important member of the FoxO
protein family, has been reported to be associated with numerous
diseases, including prostate cancer, breast cancer and leukemia
(7–9). The subcellular localization and
transcriptional activity of FoxO3a is regulated by a number of
post-translational modifications, including acetylation,
ubiquitination and, particularly, phosphorylation (10,11).
In response to growth and survival factors, including insulin like
growth factor I (IGF-1), Rac-α serine/threonine protein kinase
(Akt) is able to directly phosphorylate FoxO3a transcription
factors in three different phosphorylation sites and induce their
translocation from the nucleus to the cytoplasm, where FoxO3a
transcription factors may be degraded and inhibit cellular
regulatory functions (12). By
contrast, in conditions of oxidative stress, Akt inactivation leads
to FoxO3a protein dephosphorylation (activation), and FoxO3a may
promote the expression of target genes, including Bcl-2-like
protein 11 (Bim), cyclin-dependent kinase inhibitor 1 and
cyclin-dependent kinase inhibitor 1B (p27Kip1), and may
be associated with cell-cycle arrest and apoptosis (13–16).
Therefore, the FoxO3a family has been demonstrated to be associated
with various types of cancer. However, the effect of FoxO3a in the
development and formation of UM remains unclear.
Previous preliminary data (17) has demonstrated that the FoxO3a
transcription factor is associated with IGF-1-induced migration and
invasion of UM cells. Therefore, the present study investigated the
role of FoxO3a in the progression of UM. In the present study, UM
cells with FoxO3a overexpression or knockdown were used to
investigate the roles of FoxO3a in the development of UM. The
results of the present study demonstrated that FoxO3a inhibited
cell proliferation, induced apoptosis and led to G1 cell cycle
phase accumulation in UM cells. In addition, FoxO3a increased the
transcriptional activity and expression of Bim and
p27Kip1, and inhibited the expression of cyclin D1,
while FoxO3a knockdown exhibited the opposite effects. The data
from the present study indicated that FoxO3a serves an important
role in the cellular processes associated with UM, and that it may
be a potential target for further investigation into the treatment
of UM.
Materials and methods
Materials
All cell culture reagents and
Lipofectamine® 3000 reagent were purchased from
Invitrogen; Control N1-plasmid, FoxO3a-plasmid and small
interfering (si)FoxO3a-plasmid were from Promega Corporation
(Madison, WI, USA). MTT, dimethyl sulfoxide (DMSO), a Cell Cycle
kit, and Annexin V-fluorescein isothiocyanate (FITC)/propidium
iodide (PI) and bicinchoninic acid (BCA) assay kits were from
obtained from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany).
Antibodies against phosphorylated (p)-Akt (cat. no. 4060) and
FoxO3a (cat. no. 2497) were from Cell Signaling Technology, Inc.
(Danvers, MA, USA). Anti-Bim (cat. no. C04455H),
anti-p27Kip1 (cat. no. 12430), anti-Cyclin D1 (cat. no.
C00353H) and anti-β-actin (cat. no. C08432H) were from Signalway
Antibody LLC (College Park, MA, USA). The PrimeScript RT reagent
kit and SYBR Green Real-Time PCR kit were obtained from Roche
Diagnostics (Basel, Switzerland).
Cell culture and Lipofectamine
transfection
Human UM cells were purchased from Shanghai Bioleaf
Biotech Co., Ltd. (Shanghai, China). Cells were cultured in
Dulbecco's modified Eagle's medium supplemented with 10% fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and 1% penicillin-streptomycin, at 37°C in a
humidified atmosphere containing 5% CO2. When grown to
40% confluency, the UM cells were transfected with N1, FoxO3a and
siFoxO3a (5 µg) using 7.5 µl Lipofectamine® 3000
reagent, according to the manufacturer's protocol. A total of 6 h
following transfection, the culture medium was changed and cells
were incubated for 36 h. The transfected UM cells were used for
MTT, cell cycle and cellular apoptosis assays, western blotting and
the reverse transcription-quantitative polymerase chain reaction
(RT-qPCR).
Cell viability assay
Cells (1–2×105/well) were seeded into
96-well plates, followed by overnight incubation, and were
transfected with the different plasmids. The transfected UM cells
were selected using puromycin to generate stable cell lines.
Briefly, prior to puromycin selection, the working concentration of
puromycin for UM cells was determined using a series of
concentrations ranging from 1 to 10 µg/ml. The lowest concentration
that induced cell death in 100% of non-transfection UM cells in 3–4
days from the start of puromycin selection was chosen as the
optimal concentration for the subsequent experiments. Following 48
h after transfection, UM cells were trypsinized, split into 10-cm
culture dishes (200–500 cells/dish) and cultured in DMEM containing
10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) and 3 µg/ml
puromycin (Sigma-Aldrich; Merck KGaA. During the selection period,
the medium containing puromycin was exchanged every 2 days for ~3
weeks. Puromycin resistant cell colonies were manually picked,
subcloned and passaged. UM cells at passages 3–5 were used for
these experiments. The media was removed and replaced with fresh
medium containing MTT (1 mg/ml), and the cells were incubated at
37°C for 4 h. The formazan crystals were dissolved by adding 100 µl
DMSO and the absorbance was measured at 570 nm using a Bio-Rad 680
plate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
experiment was performed in triplicate.
Cell cycle assay
The transfected UM cells (2–3×104) were
collected, washed twice with ice-cold PBS and fixed with 70%
ethanol overnight at 4°C. The cells were incubated with 20 µg/ml
RNase and stained with 20 µg/ml PI for 30 min at 4°C in the dark.
Cell cycle distribution was analyzed by flow cytometry (EPICS XL
Flow Cytometer; Beckman Coulter, Inc., Brea, CA, USA) using Flowjo
software (version 7.6.1; Tree Star, Inc., Ashland, OR, USA). All
experiments were repeated 3 times.
Cellular apoptosis assay
UM cells (5×105 per well) were seeded
into 6-well plates. Following transfection for 36 h, the adherent
and floating cells were harvested and washed twice with ice-cold
PBS, and resuspended with Annexin V binding buffer. The cell
supernatant was stained at room temperature with 5 µl Annexin
V-FITC and 15 µl PI. The number of apoptotic cells was analyzed
using flow cytometry (as described in the Cell cycle assay
section). Each experiment was repeated three times.
Western blotting
A total of 36 h post-transfection, UM cells were
collected and lysed with radioimmunoprecipitation assay lysis
buffer [20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1%
Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM b-glycerophosphate,
1 mM Na3VO4, 1 mg/ml leupeptin and 1 mM
PMSF]. Protein concentration was determined using a BCA Protein
Assay kit (Sigma-Aldrich; Merck KGaA). Total proteins (30
µg/sample) were separated by 10% SDS-PAGE and transferred onto a
polyvinylidene fluoride membrane. The membrane was subsequently
blocked in 5% non-fat milk and incubated overnight at 4°C with
primary antibodies (anti-p-Akt, FoxO3a, cyclin D1,
p27Kip1, Bim and β-actin; 1:1,000). Following washing,
blots were further incubated with the corresponding horseradish
peroxidase-conjugated secondary antibody (cat. no. SA00001-1;
ProteinTech Group, Inc., Chicago, IL, USA; 1:10,000) at room
temperature for 1 h. and visualized with a chemiluminescence system
(ChemiDoc Touch; Bio-Rad Laboratories, Inc.). Image J v 1.48
software (National Institutes of Health, Bethesda, MA, USA) was
used to quantify optical density.
RT-qPCR analysis
Total RNA from transfected UM cells was isolated
using TRIzol reagent (cat. no. QXT94240; Sigma; Merck KGaA) and
cDNA was synthesized from total RNA (500 ng) using the Roche First
Stand cDNA Synthesis kit (Roche Diagnostics). In order to detect
the relative gene expression levels, a SYBR Green Real-Time PCR kit
was used. The thermocycling conditions used for PCR were as
follows: 95°C for 3 min, followed by 40 cycles of 95°C for 30 sec,
Ta (data available on request) for 30 sec and 72°C for 30 sec. The
data were analyzed using the comparative 2−ΔΔCq method
(18). The primers were as
follows: 60S ribosomal protein PL19 forward,
5′-GAGACAAAGTGGGAGCCAGCGA-3′ and reverse,
5′-ACCCTCCAGGAAGCGAGAATGC-3′; FoxO3a forward,
5′-CTCCCTACGCCAGTCTCCCAT-3′ and reverse,
5′-TGAGTCCGAAGTGAGCAGGTCC-3′; Bim forward,
5′-ATAAGCTAAAGAGGCTGAAAGAG-3′ and reverse,
5′-GAATGAAATGAGTCCCCAAAAC-3′; p27Kip1 forward,
5′-AAAAGCAACAGAAACCTATCCTCAC-3′ and reverse,
5′-ATTCAAAACTCCCAAGCACCTC-3′; and Cyclin D1 forward, 5′- CCC TCG
GTG TCC TAC TTC AAA TGT-3′ and reverse,
5′-GGAAGCGGTCCAGGTAGTTCAT-3′.
Statistical analysis
The results of the present study were analyzed using
SPSS software (version 19.0; IBM Corp., Armonk, NY, USA). All data
are expressed as the mean ± standard error, and were evaluated
using one-way analysis of variance followed by Tukey's post hoc
test for multiple comparisons or a two-sided t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
FoxO3a activity affects UM cell
viability
In order to observe whether or not FoxO3a activity
is associated with UM cell growth, the N1 control vector, FoxO3a
and siFoxO3a were transfected into UM cells. Following the
establishment of stable cell lines, cell number equivalent was
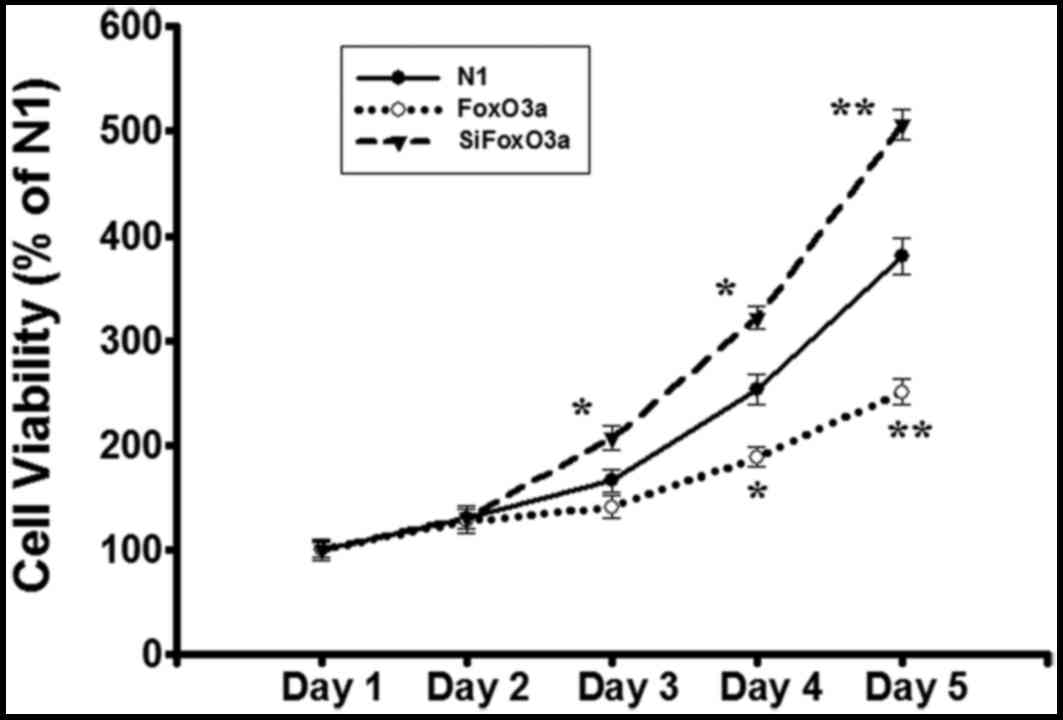
detected using the MTT assay. Compared with N1 group, cell
viability was decreased in FoxO3a group, while the viability of
siFoxO3a group was markedly increased (Fig. 1). These data indicated that the
FoxO3a activity inhibits UM cell growth.
FoxO3a activity affects UM cell cycle
distribution
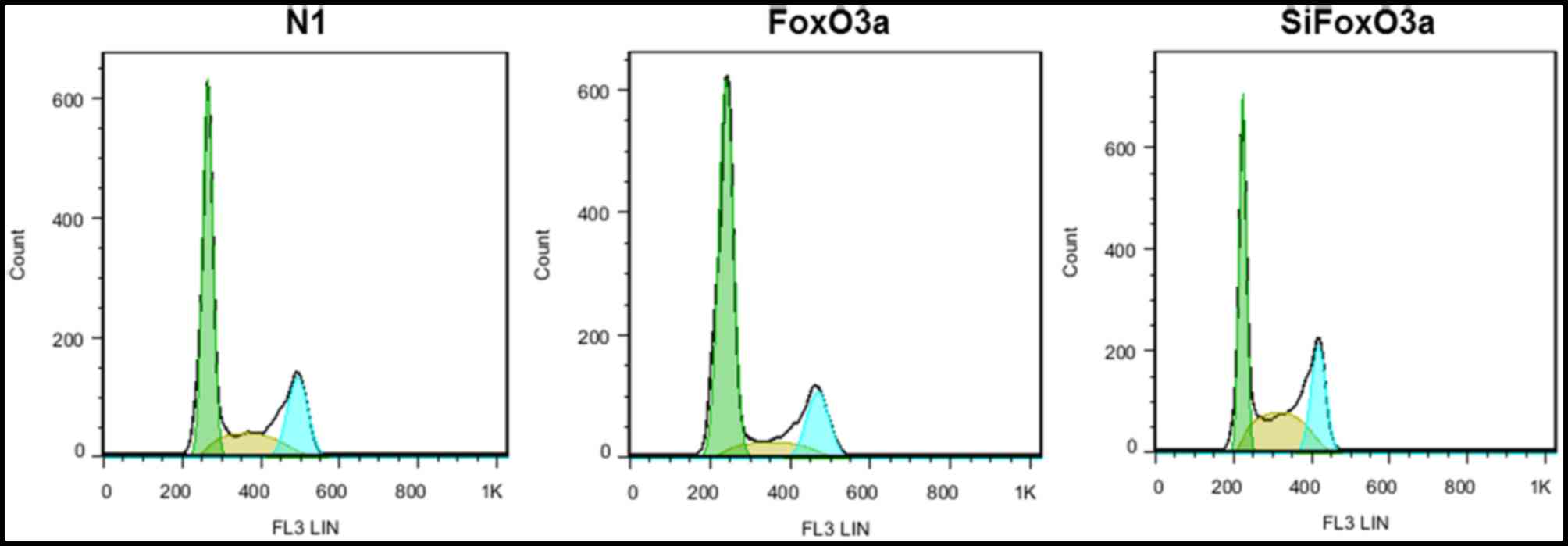
The present study investigated the association
between FoxO3a activity and cell cycle distribution. UM cells were
transfected with N1, FoxO3a and siFoxO3a, and the cell cycle
distribution of UM cells was determined using flow cytometry. For
the FoxO3a group, the proportion of UM cells at the G0/G1 phase
increased, while cells at the S and G2/M phases decreased. By
contrast, in the siFoxO3a group of UM cells, the cells were
arrested in G0/G1 phase, and the number of cells at the S and G2/M
phases decreased (Fig. 2).
FoxO3a activity promotes UM cell
apoptosis
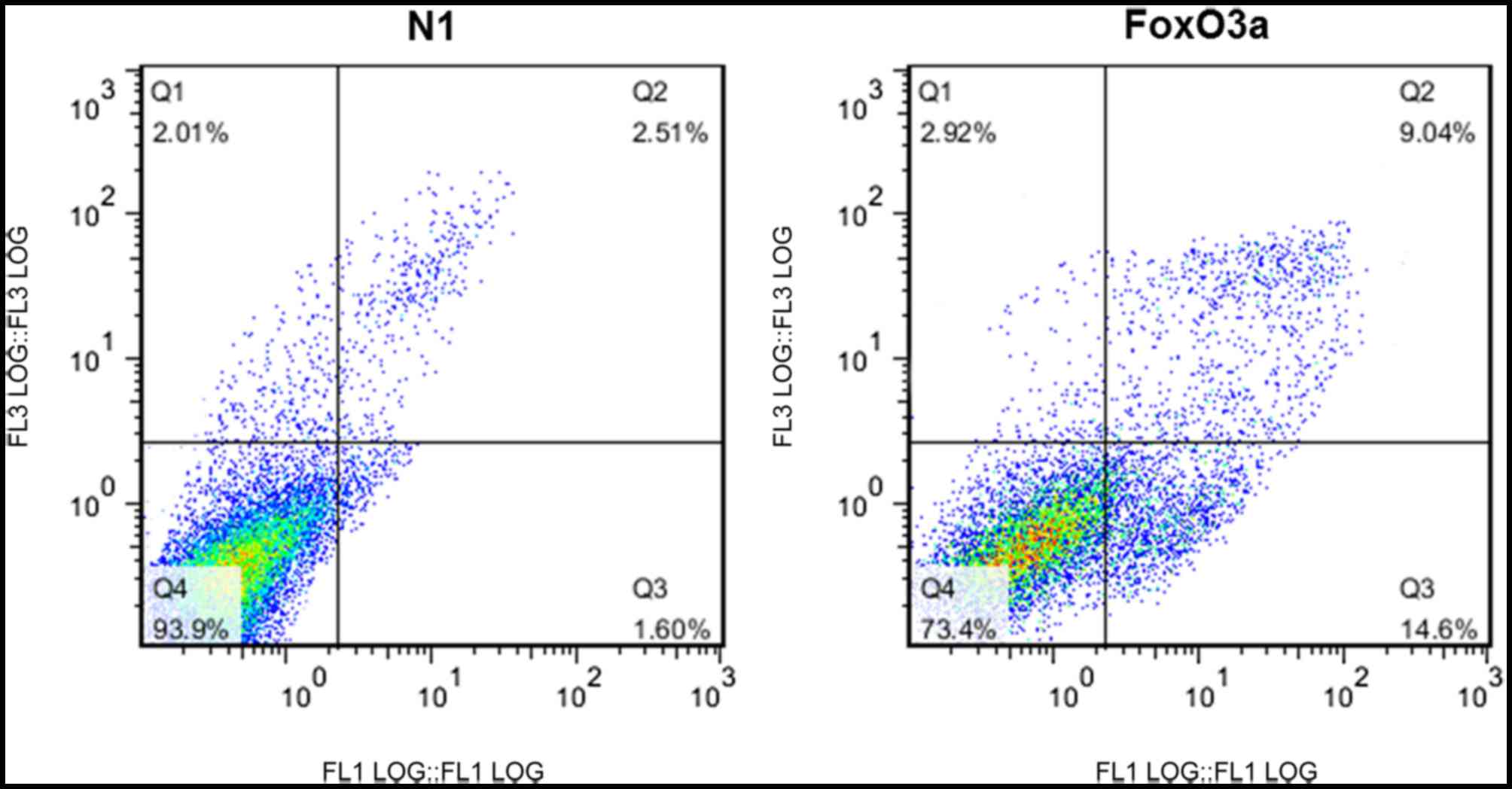
In order to examine the effect of FoxO3a on
apoptosis in UM cells, the cells were transfected with N1 or FoxO3a
and double-stained with Annexin V-FITC/PI. Cellular apoptosis was
analyzed using flow cytometry. The apoptosis levels in UM cells
transfected with FoxO3a were significantly increased compared with
those in cells transfected with N1 (Fig. 3).
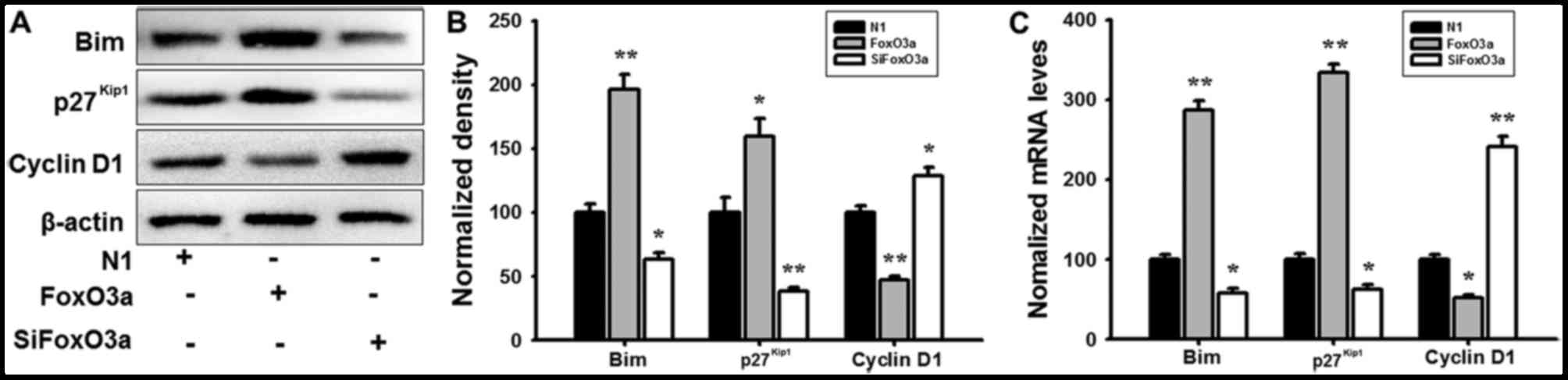
FoxO3a regulates the activity of its
downstream targets
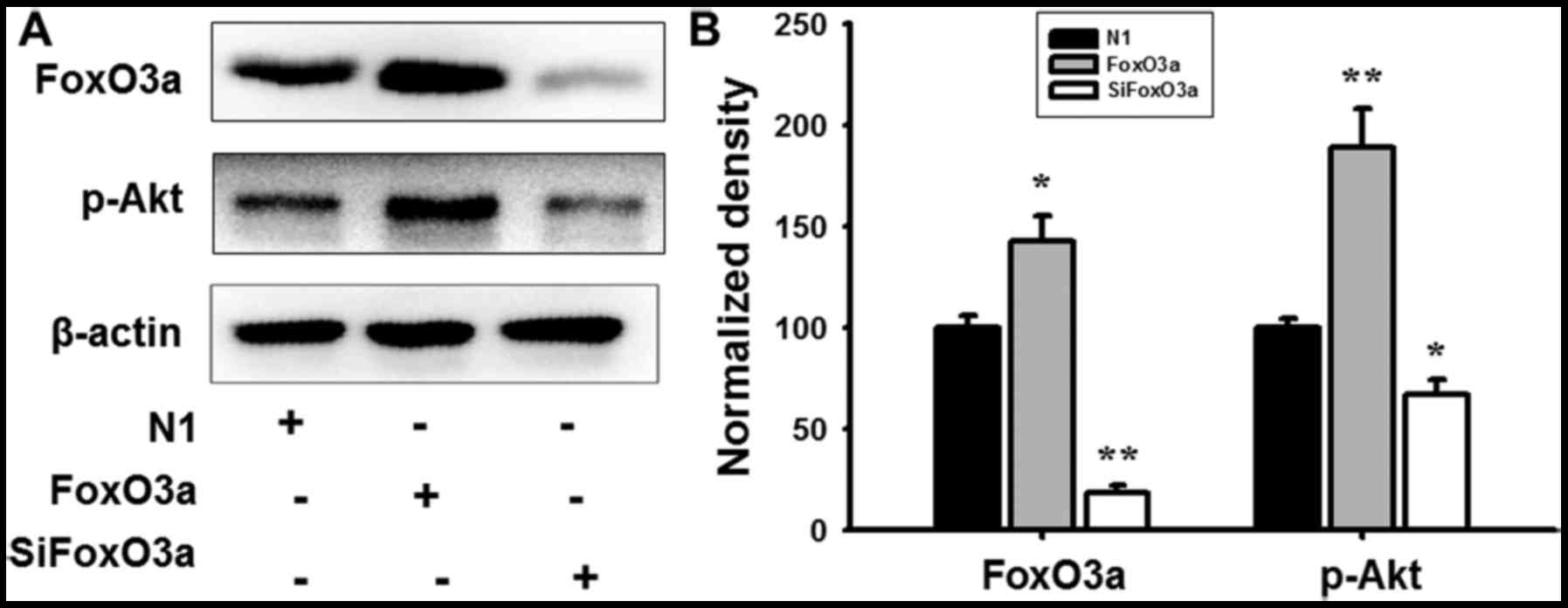
In order to further investigate the regulatory
mechanism of FoxO3a in UM cells, mRNA expression and protein levels
were detected using RT-qPCR analysis and western blotting. FoxO3a
overexpression promoted the phosphorylation of Akt (Fig. 4), and markedly increased the mRNA
and protein expression of Bim and p27Kip1; however, the
cyclin D1 transcription and expression levels were decreased in UM
cells compared with N1 (Fig. 5).
When cells were transfected with siFoxO3a, the opposite cellular
effects were observed (Figs. 4 and
5). The results of the present
study demonstrated that FoxO3a may regulate the activity of Bim and
p27Kip1.
Discussion
FoxO transcription factors belong to the large
forkhead box family of proteins, consisting of FoxO1, FoxO3a, FoxO4
and FoxO6 in mammals (19). The
FoxO transcription factors are well-known regulators of genes which
are involved in important cellular processes, including cell cycle
arrest, apoptosis, DNA repair and resistance to oxidative stress
(20). FoxO3a is an important FoxO
transcription factor and is associated with the formation and
development of tumors. Previous studies have demonstrated a
negative association between FoxO3a, and cancer grade and spread.
In ovarian cancer, liver cancer, breast cancer and prostate cancer,
FoxO3a expression is decreased (8,21–23).
In addition, the nuclear transport and abnormal cytoplasmic
distribution of FoxO3a is associated with poor prognosis of breast
cancer (9). Upregulating FoxO3a
expression inhibits the proliferation of breast cancer cells
(24,25). Previous studies have demonstrated
that IGF-1 is able to activate the phosphatidylinositol 3-kinase
(PI3K)/Akt signaling pathway in UM cells, and the activated
PI3K/Akt pathway has been demonstrated to promote the
phosphorylation of FoxO3a (26).
FoxO3a phosphorylation induces its translocation into the cytoplasm
from the nucleus and inhibits its function. Therefore, the present
study investigated whether FoxO3a activation may be a target for
intervention and treatment for the development of UM, and whether
there exists a negative feedback mechanism in the activation
process.
The PI3K/Akt signaling pathway is an important
pathway for tumor cell proliferation, differentiation and
apoptosis, and FoxO3a is the principal downstream target of the
kinase activity of Akt (27). Akt
is able to phosphorylate FoxO3a and promote its interaction with
14-3-3 protein, which leads to nuclear exclusion and inhibits
FoxO3a activity (28,29). When the PI3K/Akt pathway is
inhibited by siRNA or the pharmacological antagonist LY294002, the
phosphorylation of FoxO3a is decreased, and FoxO3a
dephosphorylation is increased, in breast cells (30). FoxO3a dephosphorylation induces its
accumulation in the nucleus and an increase in its activity; FoxO3a
activity has been demonstrated to upregulate the expression of
p27Kip1 and to inhibit cell proliferation (30). Due to the aforementioned previous
results, FoxO3a overexpression or knockdown plasmids were
constructed in the present study, in order to investigate the tumor
suppressive effect and associated molecular mechanisms of FoxO3a in
UM cells. The results demonstrated that FoxO3a overexpression
inhibited cell proliferation, promoted cellular apoptosis and led
to the accumulation of cells at the G1 cell cycle phase. Western
blot analysis demonstrated that FoxO3a overexpression promoted Akt
phosphorylation, increased the transcriptional activity and protein
expression of Bim and p27Kip1, and inhibited cyclin D1
transcription and expression. The results of the present study
demonstrated that the effects of FoxO3a on the cell cycle and
apoptosis may be regulated by the transcription and expression of
Bim, p27Kip1 and cyclin D1.
In conclusion, the results of the present study
demonstrated that FoxO3a activity affects the proliferation,
apoptosis and cell cycle of UM cells, altering cell fate and
impacting on tumor development. Therefore, FoxO3a may serve a role
in the formation and development of UM, and it may represent a
candidate target for the treatment of UM.
Acknowledgements
The present study was supported by the Guangdong
Provincial Project of Science and Technology (grant no.
2011B050200005), the National Natural Science Foundation of China
(grant no. 31371088), the University of Macau (grant nos.
SRG2015-00004-FHS and MYRG2016-00052-FHS), and the Science and
Technology Development Fund of Macau (grant no. FDCT
021/2015/A1).
References
|
1
|
Hu DN, Yu GP, McCormick SA, Schneider S
and Finger PT: Population-based incidence of uveal melanoma in
various races and ethnic groups. Am J Ophthalmol. 140:612–617.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Damato B and Heimann H: Personalized
treatment of uveal melanoma. Eye (Lond). 27:172–179. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu YC, Tsai CC, Lee FL, Lee SM and Kao
SC: Mortality from uveal melanoma treated by enucleation -a 16-year
survey in Taiwan. Acta Ophthalmol. 91:e583–e584. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shoushtari AN and Carvajal RD: Treatment
of uveal melanoma. Cancer Treat Res. 167:281–293. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang X, Tang N, Hadden T and Rishi A:
Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta.
1813:1978–1986. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang H, Quirion R, Little PJ, Cheng Y,
Feng ZP, Sun HS, Xu JP and Zheng WH: Forkhead box O transcription
factors as possible mediators in the development of major
depression. Neuropharmacology. 99:527–537. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu H: Targeting forkhead box
transcription factors FOXM1 and FOXO in leukemia (Review). Oncol
Rep. 32:1327–1334. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shukla S, Shukla M, MacLennan GT, Fu P and
Gupta S: Deregulation of FOXO3A during prostate cancer progression.
Int J Oncol. 34:1613–1620. 2009.PubMed/NCBI
|
|
9
|
Fonseca EAI, Oliveira MA, Eichler R,
Akamine EH, Carvalho MHC, Barbosa AM, Dekker RFH, Khaper N and
Fortes ZB: Antitumor effect of metformin in breast cancer is
associated with AMPK and FOXO3a activation. Cancer Res. 73:2013.
View Article : Google Scholar
|
|
10
|
Obsil T and Obsilova V: Structure/function
relationships underlying regulation of FOXO transcription factors.
Oncogene. 27:2263–2275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vogt PK, Jiang H and Aoki M: Triple layer
control: Phosphorylation, acetylation and ubiquitination of FOXO
proteins. Cell Cycle. 4:908–913. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang H, Zhou X, Huang J, Mu N, Guo Z, Wen
Q, Wang R, Chen S, Feng ZP and Zheng W: The role of Akt/FoxO3a in
the protective effect of venlafaxine against corticosterone-induced
cell death in PC12 cells. Psychopharmacology (Berl). 228:129–141.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Biggs WH, Meisenhelder J, Hunter T,
Cavenee WK and Arden KC: Protein kinase B/Akt-mediated
phosphorylation promotes nuclear exclusion of the winged helix
transcription factor FKHR1. P Natl Acad Sci USA. 96:7421–7426.
1999. View Article : Google Scholar
|
|
14
|
Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo
P, Hu LS, Anderson MJ, Arden KC, Blenis J and Greenberg ME: Akt
promotes cell survival by phosphorylating and inhibiting a forkhead
transcription factor. Cell. 96:857–868. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang ED, Nuñez G, Barr FG and Guan KL:
Negative regulation of the forkhead transcription factor FKHR by
Akt. J Biol Chem. 274:16741–16746. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cui M, Huang Y, Zhao Y and Zheng J:
Transcription factor FOXO3a mediates apoptosis in HIV-1-infected
macrophages. J Immunol. 180:898–906. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan F, Liao R, Farhan M, Wang T, Chen J,
Wang Z, Little PJ and Zheng W: Elucidating the role of the FoxO3a
transcription factor in the IGF-1-induced migration and invasion of
uveal melanoma cancer cells. Biomed Pharmacother. 84:1538–1550.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Paik JH, Kollipara R, Chu G, Ji H, Xiao Y,
Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al: FoxOs are
lineage-restricted redundant tumor suppressors and regulate
endothelial cell homeostasis. Cell. 128:309–323. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Burgering BM and Kops GJ: Cell cycle and
death control: Long live Forkheads. Trends Biochem Sci. 27:352–360.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ning Y, Luo C, Ren K, Quan M and Cao J:
FOXO3a-mediated suppression of the self-renewal capacity of
sphere-forming cells derived from the ovarian cancer SKOV3 cell
line by 7-difluoromethoxy1-5,4′-di-n-octyl genistein. Mol Med Rep.
9:1982–1988. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He LY, Wei X, Du L, Liu L, Xu F, Min J, Li
C, Tao DD, Chen Q, Hu JB and Gong JP: Remarkably reduced expression
of FoxO3a in metaplastic colorectum, primary colorectal cancer and
liver metastasis. J Huazhong Univ Sci Technolog Med Sci.
33:205–211. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin CH, Chang CY, Lee KR, Lin HJ, Chen TH
and Wan L: Flavones inhibit breast cancer proliferation through the
Akt/FOXO3a signaling pathway. BMC Cancer. 15:9582015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang JY, Zong CS, Xia W, Yamaguchi H, Ding
Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, et al: ERK promotes
tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation.
Nat Cell Biol. 10:138–148. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang WS, Dolloff NG and El-Deiry WS: ERK
and MDM2 prey on FOXO3a. Nat Cell Biol. 10:125–126. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang H, Zhang Q, Zhang L, Little PJ, Xie
X, Meng Q, Ren Y, Zhou L, Gao G, Quirion R and Zheng W:
Insulin-like growth factor-1 induces the phosphorylation of PRAS40
via the PI3K/Akt signaling pathway in PC12 cells. Neurosci Lett.
516:105–109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang K, Moran A, Holst J and Wang Q:
PI3K/Akt signalling regulates leucine transport in prostate cancer.
Bju Int. 116:34–35. 2015.
|
|
28
|
Matsuzaki H, Daitoku H, Hatta M, Tanaka K
and Fukamizu A: Insulin-induced phosphorylation of FKHR (Foxo1)
targets to proteasomal degradation. Proc Natl Acad Sci USA. 100:pp.
11285–11290. 2003; View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wen Q, Duan X, Liao R, Little P, Gao G,
Jiang H, Lalit S, Quirion R and Zheng W: Characterization of
intracellular translocation of Forkhead transcription factor O
(FoxO) members induced by NGF in PC12 cells. Neurosci Lett.
498:31–36. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|