Introduction
Melanoma, one of the most aggressive forms of skin
cancer, is derived from melanocytes and accounts for approximately
80% of skin cancer-related deaths worldwide (1). The incidence of melanoma has
increased considerably compared with that of other malignancies
around the world (2).
Approximately 200,000 new cases and 46,000 deaths due to melanoma
are reported worldwide (3).
Currently, the major therapies for patients with melanoma include
surgical resection, biotherapy, radiotherapy and chemotherapy
(4). Despite the advancements in
cancer therapy, melanoma is still considered to be therapy
resistant with a 5-year survival rate of less than 15% (5,6). The
poor prognosis of melanoma is mainly caused by the melanocytes
showing aberrant proliferation, resistance to apoptosis and highly
invasive potential and motility capacity, which are important
biological characteristics in the aggressive clinical course of the
metastatic disease (6,7). Therefore, further investigation on
the molecular mechanism underlying the formation and progression of
melanoma is needed to develop novel therapeutic targets for
melanoma treatment.
MicroRNA (miRNA/miR) is a type of non-coding,
single-strand, endogenous and short RNA molecules consisting of
19–22 nucleotides (8). miRNAs play
crucial roles in gene regulation through interaction with the
3′-untranslational regions (3′-UTRs) of their target genes in a
base-pairing manner, resulting in cleaving or repressing the
translation of their target mRNAs at the posttranscriptional level
(9). At present, over one thousand
miRNAs have been identified in the human genome, which may regulate
the expression of approximately 30% of all protein-coding genes
(10). Thus, miRNAs are found to
be involved in the regulation of diverse physiological and
pathological processes, such as cell proliferation, cycle,
apoptosis, differentiation, migration, invasion, metastasis,
motility and angiogenesis (11–13).
Many studies have reported that the aberrant expression of miRNAs
is associated with many disorders, particularly cancers such as
melanoma (14–16). A large number of miRNAs have been
identified as tumour suppressors or oncogenes in different types of
human malignancy, as a result of a change in the expression level
of their target genes (17–19).
These findings provide a strong basis for the importance of miRNAs
in the tumorigenesis and tumour development and emphasise the
implications of miRNAs as potential therapeutic targets for cancer
diagnosis, treatment and prognosis.
miR-675 is differentially expressed in several types
of human cancer and plays important roles in the pathogenesis of
several diseases (20–22), However, the expression levels and
the biological roles of miR-675 in melanoma remain unclear.
Therefore, the present study aimed to assess the expression of
miR-675 in melanoma, explore the effects of miR-675 on melanoma
cells and investigate the underlying molecular mechanisms that may
be involved in the actions of miR-675.
Materials and methods
Tissue samples and cell lines
This study was approved by the Ethics Committee of
Subei People's Hospital of Jiangsu province. Written informed
consent was provided by all of these patients participated in this
research. From May 2014 to February 2016, a total of 21 pairs of
melanoma tissues and corresponding adjacent normal tissues were
gathered from melanoma patients who suffered a surgical resection
at Subei People's Hospital of Jiangsu province. None patients had
been treated with chemotherapy, radiotherapy or other treatment
prior to surgery. All tissue samples were immediately frozen in
liquid nitrogen and stored in −80°C until further use.
Melanoma cell lines, including A375, A2058, HT144
and SK-MEL-28, were obtained from the American Type Culture
Collection (Manassas, VA, USA), and cultured in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% foetal bovine serum
(FBS), 100 mg/ml penicillin and 100 mg/ml streptomycin (all from
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Human
epidermal melanocytes (HEM) were acquired from ScienCell Research
Laboratories, Inc. (San Diego, CA, USA) and maintained in
melanocyte medium (ScienCell Research Laboratories, Inc.) according
to the manufacturer's protocol. All the cell lines were grown in a
humid incubator at 37°C with 5% CO2.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissue samples or cells
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. To detect the levels of
miR-675, complementary DNA (cDNA) was synthesized from total RNA
using TaqMan MicroRNA Reverse Transcription Kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. Quantitative PCR was performed using
TaqMan MicroRNA PCR Kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) on an ABI Prism 7500 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). To quantify
metadherin (MTDH) mRNA expression level, reverse transcription was
carried out with PrimeScript RT Reagent kit (Takara Biotechnology
Co., Ltd., Dalian, China). SYBR Premix Ex Taq™ (Takara
Biotechnology Co., Ltd.) was used to detect MTDH mRNA expression.
U6 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used
as normalization for miR-675 and MTDH mRNA expression,
respectively. Each experiment was performed in triplicate and
repeated three times. The relative levels of miRNA and mRNA were
analysed via the 2−ΔΔCq method (23).
Oligonucleotides, plasmids and cell
transfection
The miR-675 mimics and miRNA negative control mimics
(miR-NC) were chemically synthesized by GenePharma (Shanghai,
China). MTDH overexpression plasmid (pcDNA3.1-MTDH) and empty
pcDNA3.1 plasmid were acquired from GeneCopoeia (Guangzhou, China).
Cells were seeded into 6-well plates at a density of 5×105 cells
per well. Cell transfection was performed using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) as recommended by the
manufacturer's instructions. The medium was replaced with fresh
DMEM medium containing 10% FBS at 8 h following transfection.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
MTT assays were employed to evaluate the
proliferative ability of the melanoma cells. Transfected cells were
collected at 24 h following transfection, and plated into 96-well
plates at a density of 3,000 cells/well. At the indicated time (0,
24, 48, or 72 h incubation), MTT assay was performed according to
the manufacturer's instructions. Briefly, 20 µl of MTT (5 mg/ml;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) reagent was added
into each well. After incubation at 37°C with 5% CO2 for
4 h, the medium was removed followed by addition of 150 µl of DMSO
(Sigma-Aldrich; Merck KGaA). Finally, the absorbance was measured
using a microplate reader (Omega Bio-Tek, Inc., Norcross, GA, USA)
at a wavelength of 490 nm. Experiments were independently repeated
in triplicate.
Transwell invasion assay
The invasiveness of melanoma cells was determined
using Transwell chambers (24-well plate, 8 µm pore; Corning
Incorporated, Corning, NY, USA) coated with Matrigel (BD
Bioscience, San Jose, CA, USA). Transfected cells were collected
after 48 h of incubation. 5×104 transfected cells in 200
µl FBS-free DMEM medium were seeded in the upper chambers. The
lower chambers were filled with DMEM with 20% FBS as a
chemoattractant. Following incubation at 37°C for 24 h, the cells
remaining on the upper chambers were removed using cotton swabs.
The invasive cells were fixed with 4% paraformaldehyde, stained
with 0.5% crystal violet and dried in air. A total of five randomly
selected fields were then examined under an inverted microscope
(IX73; Olympus Corporation, Tokyo, Japan) at a magnification of
200x. Each assay was performed in triplicate and repeated at least
three times.
Bioinformatics analysis
To predict the potential targets of miR-675,
bioinformatics analysis was performed using TargetScan (www.targetscan.org) and miRBase (http://www.mirbase.org/).
Luciferase reporter assay
MTDH was identified as a candidate target of miR-675
using bioinformatics analysis. A wild-type (Wt) MTDH 3′-UTR
containing the binding sequences of miR-675 (pGL3-MTDH-3′-UTR Wt)
and a mutant type (Mut) MTDH 3′-UTR lacking the binding sequences
of miR-675 (pGL3-MTDH-3′-UTR Mut) were chemically synthesized by
GenePharma. Cells were plated into 24-well plates at a density of
1.5×105 cells/well. After incubation overnight, cells
were cotransfected with miR-675 mimics or miR-NC, and
pGL3-MTDH-3′-UTR Wt or pGL3-MTDH-3′-UTR Mut, using Lipofectamine
2000 in accordance with the manufacturer's instructions. Luciferase
activities were measured 48 h after transfection using a
Dual-Luciferase® Reporter Assay System (Promega
Corporation, Madison, WI, USA), according to the manufacturer's
instructions. Renilla luciferase activity served as an internal
reference.
Western blotting analysis
Total protein was extracted from tissues or cells
using cold radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology, Haimen, China). The concentration of
total protein was examined using a bicinchoninic acid (BCA) Protein
Assay kit (Beyotime Institute of Biotechnology), according to the
manufacturer's instructions. Equal quantity of total protein was
separated by 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel
electrophoresis (PAGE) and transferred onto polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA).
Subsequently, the membranes were blocked with 5% skimmed milk in
TBS/0.1% Tween (TBST) at room temperature for 1 h and incubated
overnight at 4°C with primary antibodies: mouse anti-human
monoclonal MTDH (sc-517220; 1:1,000 dilution; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and mouse anti-human
monoclonal GAPDH (sc-66163; 1:1,000 dilution; Santa Cruz
Biotechnology, Inc.). Following three washes with TBST, the
membranes were incubated with corresponding horseradish
peroxidase-conjugated secondary antibodies (sc-2005; 1:1,000
dilution; Santa Cruz Biotechnology, Inc.) at room temperature for 1
h. Finally, the protein bands were visualized with enhanced
chemiluminescence (ECL) reagents (Bio-Rad Laboratories, Hercules,
CA, USA), and analyzed with ImageJ 1.49 (National Institutes of
Health, Bethesda, MD). GAPDH served as a loading control.
Statistical analysis
Data were expressed as mean ± standard deviation.
All statistical analyses were performed via Student's t-test or
one-way ANOVA with SPSS 18.0 software (SPSS, Inc., Chicago, IL,
USA). Student-Newman-Keuls test was used as a post hoc test
following ANOVA. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-675 expression is downregulated in
melanoma tissues and cell lines
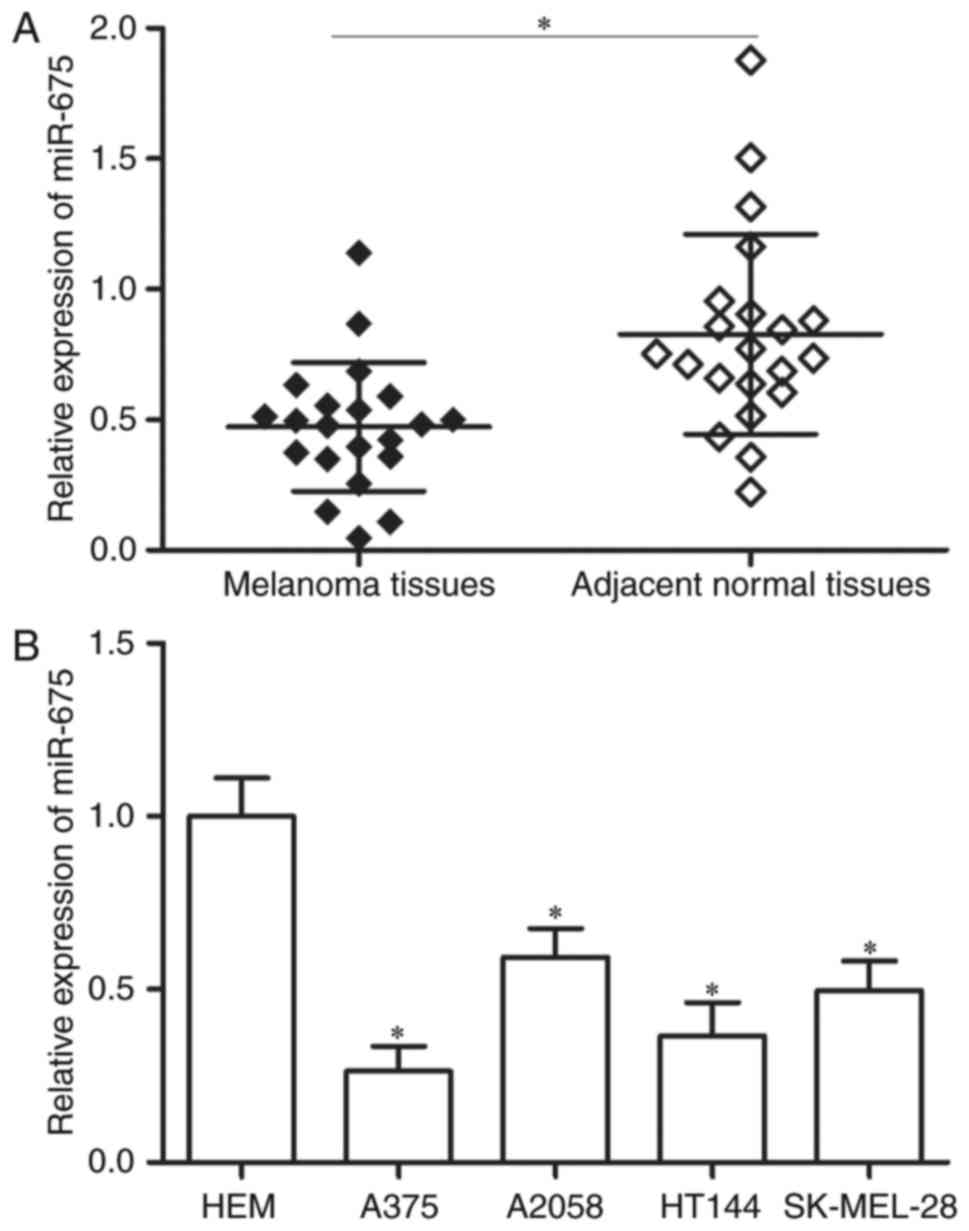
To investigate the roles of miR-675 in the
progression of melanoma, RT-qPCR was performed to measure miR-675
expression in 21 paired melanoma tissues and corresponding adjacent
normal tissues. As shown in Fig.
1A, miR-675 was significantly downregulated in melanoma tissues
compared with that in corresponding adjacent normal tissues
(P<0.05). In addition, the analysis of miR-675 expression in
four human melanoma cell lines (A375, A2058, HT144 and SK-MEL-28)
and the human epidermal melanocytes (HEMs) indicated that
expression levels of miR-675 were lower in melanoma cell lines
(Fig. 1B; P<0.05). These
results suggested that miR-675 may serve important roles in
melanoma formation and progression.
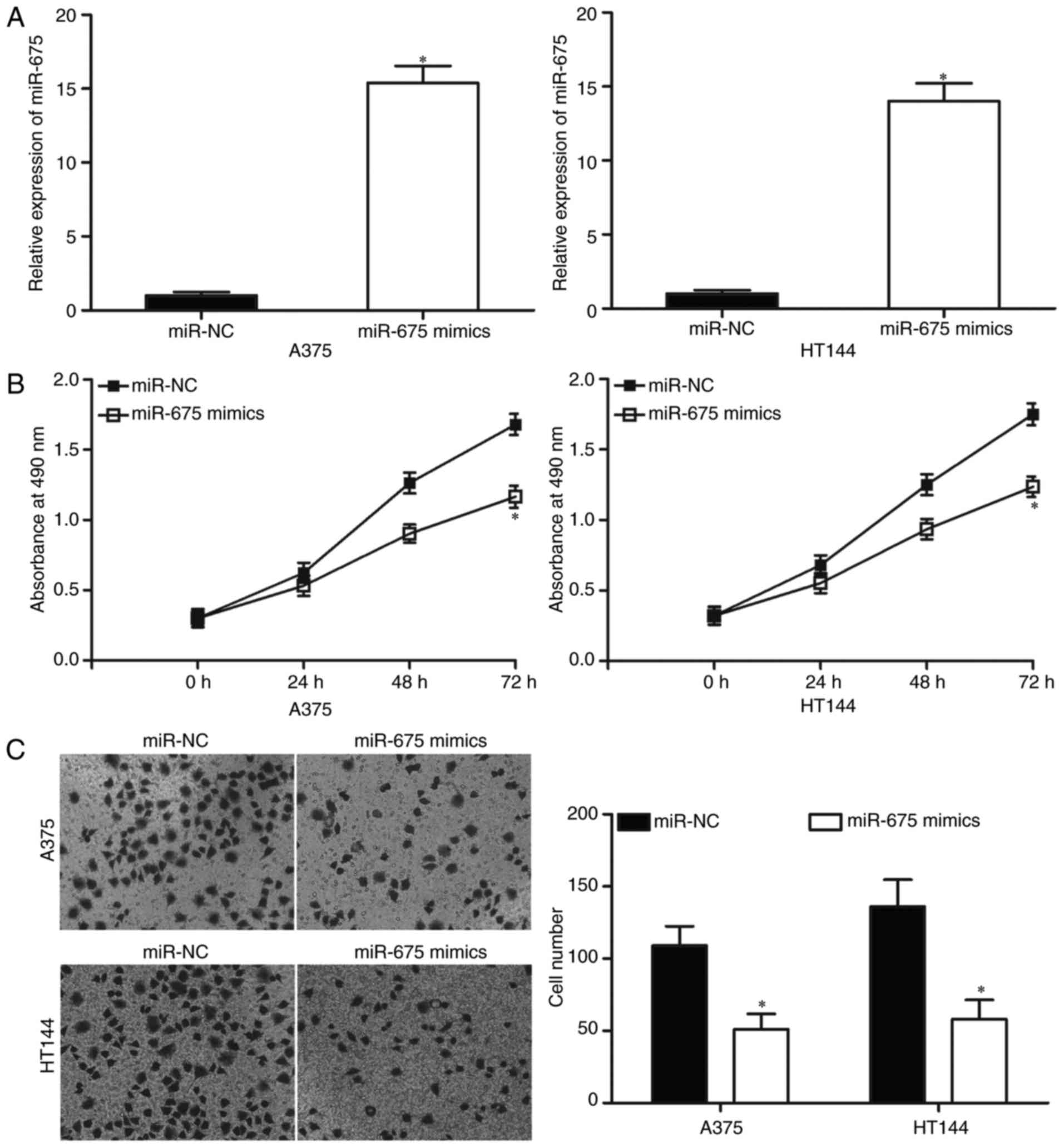
miR-675 inhibits melanoma cell
proliferation and invasion in vitro
miR-675 is lowly expressed in melanoma and thus
might serve as a tumour suppressor. Therefore, we determined
whether the deregulation of miR-675 in malignant melanoma cells
affects biological behaviours of melanoma. A375 and HT144 cells,
which exhibited relatively low miR-675 expression, were chosen for
subsequent functional assays and transfected with miR-675 mimics or
miR-NC. The data from RT-qPCR analysis confirmed that miR-675 was
markedly upregulated in A375 and HT144 cells after transfection
with miR-675 mimics (Fig. 2A;
P<0.05).
An MTT assay was conducted to explore effect of
miR-675 overexpression on melanoma cell proliferation. Restoration
expression of miR-675 suppressed the proliferation of A375 and
HT144 compared with the miR-NC group (Fig. 2B; P<0.05), indicating that
miR-675 may serve a suppressive role in the proliferation of
melanoma cells. To further characterise the effect of miR-675 on
cell invasion ability, Transwell invasion assays were performed. As
depicted in Fig. 2C, the
upregulation of miR-675 decreased the invasion capacities of A375
and HT144 cells, relative to the miR-NC group (P<0.05).
Collectively, these data suggested that miR-675 may play
tumour-suppressing roles in melanoma.
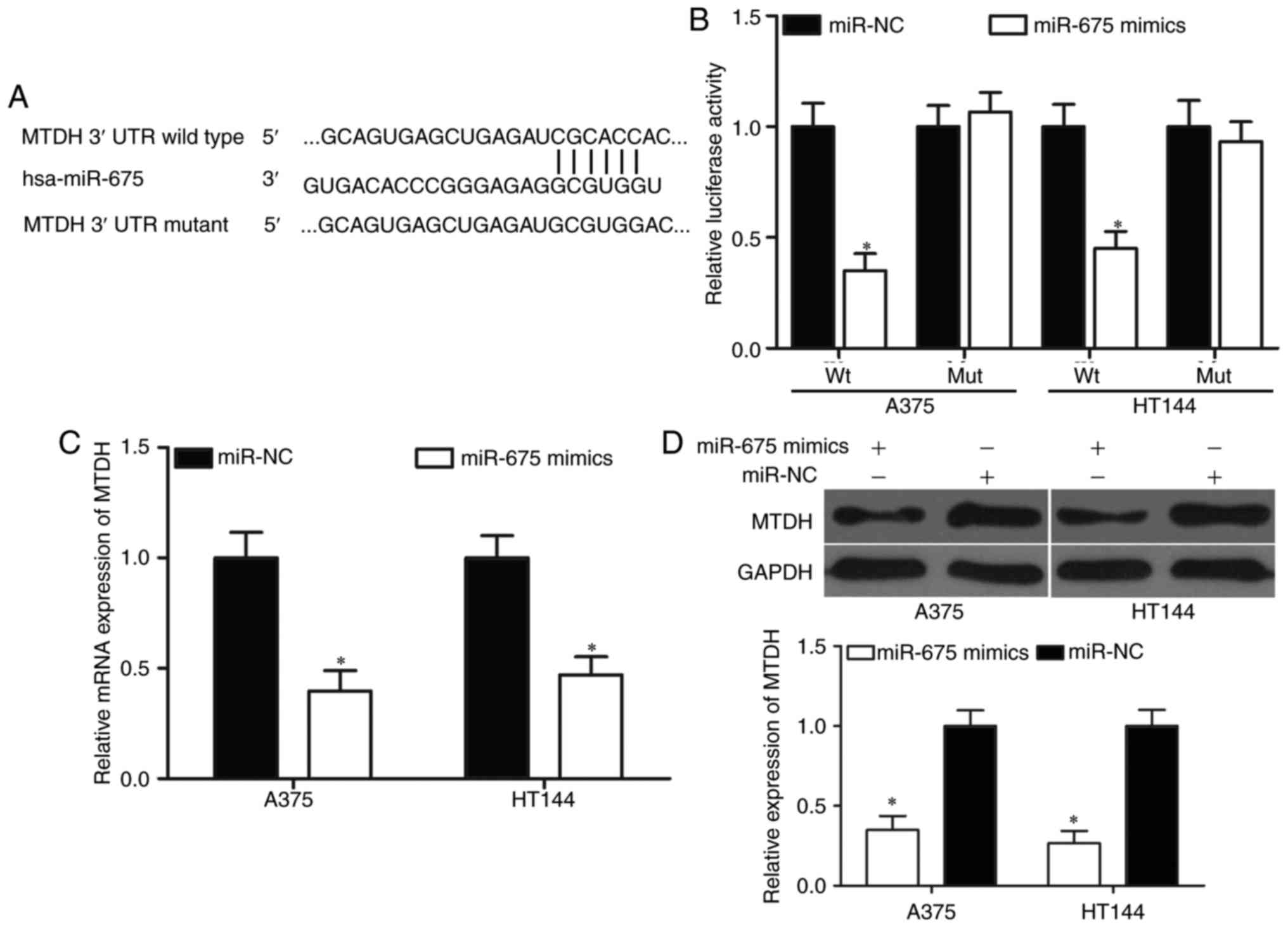
MTDH is a direct target of miR-675 in
melanoma
To investigate the underlying mechanism responsible
for miR-675-mediated tumour-suppressing roles in melanoma,
bioinformatics analysis was performed to predict the potential
targets of miR-675. Among these potential target genes, MTDH
(Fig. 3A) caught our attention
because it is upregulated in melanoma and participated in the
regulation of biological behaviours of melanoma (24–26).
Luciferase reporter assays were used to verify a direct interaction
between miR-675 and the 3′-UTR of MTDH. miR-675 mimics or miR-NC
was transfected into A375 and HT144 cells, together with
pGL3-MTDH-3′-UTR Wt or pGL3-MTDH-3′-UTR Mut. As shown in Fig. 3B, miR-675 overexpression decreased
the luciferase activities of the MTDH with a wild-type 3′-UTR
vector (P<0.05) but not the mutant vector, implying that miR-675
directly binds the 3′-UTR of MTDH. To further confirm whether
miR-675 has regulation effect on MTDH endogenous in melanoma, we
detected MTDH mRNA and protein expression in A375 and HT144 cells
after transfection with miR-675 mimics or miR-NC. The results
revealed that MTDH mRNA (Fig. 3C;
P<0.05) and protein (Fig. 3D;
P<0.05) expressions were significantly suppressed by miR-675
overexpression in A375 and HT144 cells. Overall, these results
suggested that MTDH is a direct target gene of miR-675 in
melanoma.
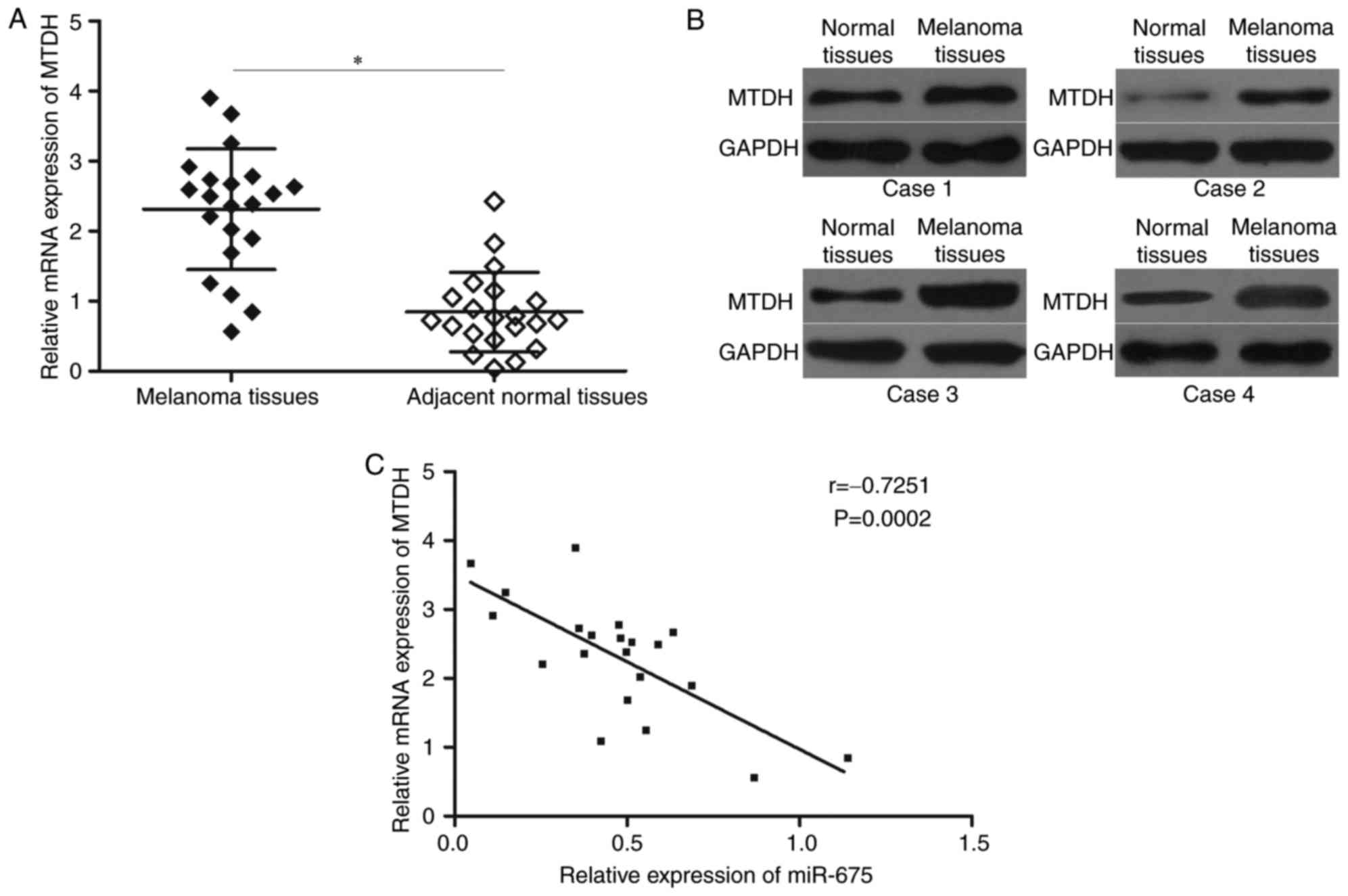
MTDH is upregulated in melanoma
tissues and negatively correlated with miR-675 expression
To further explore the relationship between miR-675
and MTDH, we measured MTDH mRNA expression in melanoma tissues and
corresponding adjacent normal tissues. The data from RT-qPCR
analysis revealed that that levels of MTDH mRNA were significantly
upregulated in melanoma tissues compared with those in adjacent
normal tissues (Fig. 4A;
P<0.05). In addition, Western blotting analysis indicated that
MTDH protein was increased in melanoma tissues than adjacent normal
tissues (Fig. 4B). Furthermore,
Spearman's correlation analysis demonstrated an inverse association
between MTDH mRNA and miR-675 expression levels in melanoma tissues
(Fig. 4C; r=−0.7251; P=0.0002).
These results suggested that increased levels of MTDH in melanoma,
at least in part, may be attributed to a downregulation in
miR-675.
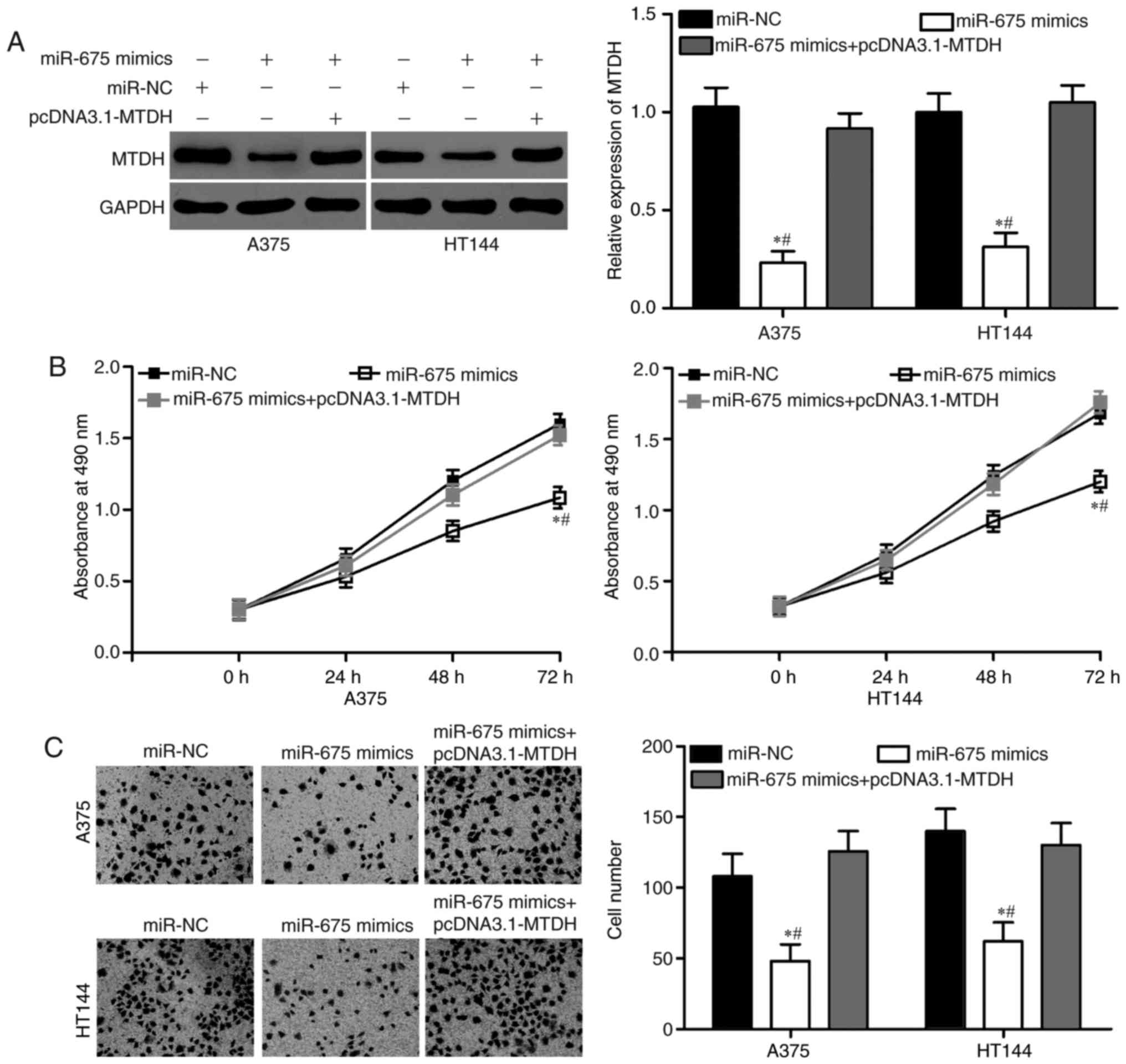
Restoration of MTDH expression
reverses the antitumor effects of miR-675 in melanoma
To further determine whether MTDH is a functional
target of miR-675 in melanoma, we performed a rescue experiment
involving the transfection of MTDH overexpression plasmids into
miR-675-expressing A375 and HT144 cells. As shown in Fig. 5A, the decreased expression of MTDH
protein was markedly restored by the transfection of pcDNA3.1-MTDH
in A375 and HT144 cells (P<0.05). Functional experiments
demonstrated that the upregulation of MTDH could effectively
reverse the tumour-suppressing effects on cell proliferation
(Fig. 5B; P<0.05) and invasion
(Fig. 5C; P<0.05) induced by
miR-675 overexpression in A375 and HT144 cells. These results
suggested that miR-675 exerted its suppressive effects in melanoma,
at least partially, by repressing MTDH.
Discussion
The dysregulation of miRNAs is involved in the
development and progression of melanoma (27–29).
Therefore, miRNAs may be investigated as novel diagnostic or
prognostic biomarkers and promising therapeutic targets in the
treatment of patients with melanoma. In the present study, the
expression of miR-675 was significantly downregulated in melanoma
tissues and cell lines. In addition, we illustrated that miR-675
overexpression inhibited cell proliferation and invasion of
melanoma in vitro. Importantly, this is the first study to
identify MTDH as a direct target gene of miR-675 in melanoma. MTDH
is highly expressed in melanoma tissues and negatively correlated
with miR-675 expression level. The upregulation of MTDH rescued the
antitumor effects of miR-675 in melanoma. Thus, miR-675 may play
tumour-suppressing roles in melanoma by targeting MTDH in
pathophysiologic process of melanoma.
An increasing number of studies reported that
miR-675 is upregulated and plays an oncogenic role in the
development and progression of multiple kinds of human cancer. For
example, miR-675 was upregulated in breast cancer tissues and
correlated with tumour grade (20). miR-675 overexpression increased the
aggressive phenotype of breast cancer cells including promoted cell
proliferation and migration in vitro and increased tumour
growth and metastasis in vivo (21). miR-675 was also identified as an
oncogene in bladder cancer (22),
head and neck squamous cell carcinoma (30) and hepatocellular carcinoma
(31,32). However, miR-675 was found to be
lowly expressed in non-small cell lung cancer. Low miR-675
expression was correlated with lymph node metastasis and TNM stage
(33). Resumption expression of
miR-675 suppressed non-small cell lung cancer growth, colony
formation and metastasis in vitro and decreased the
tumorigenicity graft growth of nude mice in vivo (33). The downregulation of miR-675 was
also observed in pancreatic cancer (34), adrenocortical adenomas (35), prostate cancer (36) and glioma (37). These findings suggested that
miR-675 expression and roles exhibits tissue specificity and may be
a prognostic marker and promising molecular therapeutic target in
these specific types of cancer.
The identification of the miRNA target genes is
important for understanding its roles in carcinogenesis and
progression (38). Multiple
targets of miR-675 have been identified, including c-Cbl(21),
Cbl-b(21) in breast cancer, ZEB1(34) in pancreatic cancer,
GPR55(33) in non-small cell lung cancer, TGFBI(36) in prostate
cancer, CDK6(37) in glioma, p53(22) in bladder cancer, AKT(32) and
Cdc25A(31) in hepatocellular carcinoma. In the current study, MTDH,
also known as astrocyte elevated gene-1, was identified as a direct
target of miR-675 in melanoma. It was first discovered in human
foetal astrocytes in 2002 (39)
and was found to be overexpressed in several types of human cancer,
such as colorectal cancer (40),
breast cancer (41), cervical
cancer (42), gastric cancer
(43) and bladder cancer (44). Increasing evidence indicated that
MTDH is participated in the regulation of tumorigenesis and tumour
development by regulating cell proliferation, cell cycle,
apoptosis, migration, invasion, metastasis,
epithelial-to-mesenchymal transition and angiogenesis (45–47).
In melanoma, MTDH expression level increased in tumour tissues and
is significantly associated with metastatic rate (24,48).
Functional study indicated that MTDH underexpression inhibited the
proliferation, migration and invasion and induced cell cycle arrest
and apoptosis in melanoma. Additionally, in vivo experiments
revealed that the downregulation of MTDH reduced the growth of
melanoma xenografts in nude mice (26). Combined with the present findings,
miR-675/MTDH axis may present a potentially effective therapeutic
strategy for the treatments of patients with melanoma.
In conclusion, the results of the present study
provide evidence suggesting that miR-675 may play a
tumour-suppressing role in melanoma, partly by targeting MTDH. The
miR-675/MTDH pathway may provide novel insights into the
pathogenesis of melanoma and could be investigated as a novel
potential therapeutic target for the treatment of this disease.
Future work is needed to explore whether the potential of miR-675
may be fully realised in melanoma.
References
|
1
|
Miller AJ and Mihm MC Jr: Melanoma. N Engl
J Med. 355:51–65. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eggermont AM, Suciu S, Rutkowski P, Kruit
WH, Punt CJ, Dummer R, Salès F, Keilholz U, de Schaetzen G and
Testori A; EORTC Melanoma Group, : Long term follow up of the EORTC
18952 trial of adjuvant therapy in resected stage IIB-III cutaneous
melanoma patients comparing intermediate doses of
interferon-alpha-2b (IFN) with observation: Ulceration of primary
is key determinant for IFN-sensitivity. Eur J Cancer. 55:111–121.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pacheco I, Buzea C and Tron V: Towards new
therapeutic approaches for malignant melanoma. Expert Rev Mol Med.
13:e332011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fang W, Fan Y, Fa Z, Xu J, Yu H, Li P and
Gu J: microRNA-625 inhibits tumorigenicity by suppressing
proliferation, migration and invasion in malignant melanoma.
Oncotarget. 8:13253–13263. 2017.PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Adams BD, Parsons C, Walker L, Zhang WC
and Slack FJ: Targeting noncoding RNAs in disease. J Clin Invest.
127:761–771. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li S, Xu X, Xu X, Hu Z, Wu J, Zhu Y, Chen
H, Mao Y, Lin Y, Luo J, et al: MicroRNA-490-5p inhibits
proliferation of bladder cancer by targeting c-Fos. Biochem Biophys
Res Commun. 441:976–981. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Wu W, Xu S, Zhang J, Zhang J, Yu
Q, Jiao Y, Wang Y, Lu A, You Y, et al: MicroRNA-105 inhibits human
glioma cell malignancy by directly targeting SUZ12. Tumour Biol.
39:10104283177057662017.PubMed/NCBI
|
|
16
|
Asahchop EL, Akinwumi SM, Branton WG,
Fujiwara E, Gill MJ and Power C: Plasma microRNA profiling predicts
HIV-associated neurocognitive disorder. AIDS. 30:2021–2031. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang W, Zhang H, Tang M, Liu L, Zhou Z,
Zhang S and Wang L: MicroRNA-592 targets IGF-1R to suppress
cellular proliferation, migration and invasion in hepatocellular
carcinoma. Oncol Lett. 13:3522–3528. 2017.PubMed/NCBI
|
|
18
|
Zhang P, Kong F, Deng X, Yu Y, Hou C,
Liang T and Zhu L: MicroRNA-326 suppresses the proliferation,
migration and invasion of cervical cancer cells by targeting ELK1.
Oncol Lett. 13:2949–2956. 2017.PubMed/NCBI
|
|
19
|
Liang Z, Wang X, Xu X, Xie B, Ji A, Meng
S, Li S, Zhu Y, Wu J, Hu Z, et al: MicroRNA-608 inhibits
proliferation of bladder cancer via AKT/FOXO3a signaling pathway.
Mol Cancer. 16:962017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhai LL, Wang P, Zhou LY, Yin JY, Tang Q,
Zhang TJ, Wang YX, Yang DQ, Lin J and Deng ZQ: Over-expression of
miR-675 in formalin-fixed paraffin-embedded (FFPE) tissues of
breast cancer patients. Int J Clin Exp Med. 8:11195–11201.
2015.PubMed/NCBI
|
|
21
|
Vennin C, Spruyt N, Dahmani F, Julien S,
Bertucci F, Finetti P, Chassat T, Bourette RP, Le Bourhis X and
Adriaenssens E: H19 non coding RNA-derived miR-675 enhances
tumorigenesis and metastasis of breast cancer cells by
downregulating c-Cbl and Cbl-b. Oncotarget. 6:29209–29223. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu C, Chen Z, Fang J, Xu A, Zhang W and
Wang Z: H19-derived miR-675contributes to bladder cancer cell
proliferation by regulating p53 activation. Tumour Biol.
37:263–270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kang DC, Su ZZ, Sarkar D, Emdad L, Volsky
DJ and Fisher PB: Cloning and characterization of HIV-1-inducible
astrocyte elevated gene-1, AEG-1. Gene. 353:8–15. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang Y and Li LP: Progress of cancer
research on astrocyte elevated gene-1/Metadherin (Review). Oncol
Lett. 8:493–501. 2014.PubMed/NCBI
|
|
26
|
Zhang Y, Peng G, Wang Y, Cui L, Wu W, Wang
L, Liu C and Han X: Silencing of astrocyte elevated gene-1 inhibits
proliferation and migration of melanoma cells and induces
apoptosis. Clin Exp Pharmacol Physiol. 44:815–826. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bustos MA, Ono S, Marzese DM, Oyama T,
Iida Y, Cheung G, Nelson N, Hsu SC, Yu Q and Hoon DSB: miR-200a
regulates CDK4/6 Inhibitor effect by targeting CDK6 in metastatic
melanoma. J Invest Dermatol. 137:1955–1964. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He J, Tian N, Yang Y, Jin L, Feng X, Hua
J, Lin S, Wang B, Li H and Wang J: miR-185 enhances the inhibition
of proliferation and migration induced by ionizing radiation in
melanoma. Oncol Lett. 13:2442–2448. 2017.PubMed/NCBI
|
|
29
|
Zeng HF, Yan S and Wu SF: MicroRNA-153-3p
suppress cell proliferation and invasion by targeting SNAI1 in
melanoma. Biochem Biophys Res Commun. 487:140–145. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guan GF, Zhang DJ, Wen LJ, Xin D, Liu Y,
Yu DJ, Su K, Zhu L, Guo YY and Wang K: Overexpression of lncRNA
H19/miR-675 promotes tumorigenesis in head and neck squamous cell
carcinoma. Int J Med Sci. 13:914–922. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu YQ, Weng J, Li SQ, Li B and Lv J:
miR-675 promotes the growth of hepatocellular carcinoma cells
through the Cdc25A pathway. Asian Pac J Cancer Prev. 17:3881–3885.
2016.PubMed/NCBI
|
|
32
|
Lv J, Ma L, Chen XL, Huang XH and Wang Q:
Downregulation of LncRNAH19 and miR-675 promotes migration and
invasion of human hepatocellular carcinoma cells through
AKT/GSK-3β/Cdc25A signaling pathway. J Huazhong Univ Sci Technolog
Med Sci. 34:363–369. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He D, Wang J, Zhang C, Shan B, Deng X, Li
B, Zhou Y, Chen W, Hong J, Gao Y, et al: Down-regulation of
miR-675-5p contributes to tumor progression and development by
targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol
Cancer. 14:732015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Zhang Y, Wei H, Zhang X, Wu Y,
Gong A, Xia Y, Wang W and Xu M: The mir-675-5p regulates the
progression and development of pancreatic cancer via the
UBQLN1-ZEB1-mir200 axis. Oncotarget. 8:24978–24987. 2017.PubMed/NCBI
|
|
35
|
Schmitz KJ, Helwig J, Bertram S, Sheu SY,
Suttorp AC, Seggewiss J, Willscher E, Walz MK, Worm K and Schmid
KW: Differential expression of microRNA-675, microRNA-139-3p and
microRNA-335 in benign and malignant adrenocortical tumours. J Clin
Pathol. 64:529–535. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng
R, Wang Y, Huang J, Xu M, Yan J and Yu J: lncRNA H19/miR-675 axis
represses prostate cancer metastasis by targeting TGFBI. FEBS J.
281:3766–3775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li C, Lei B, Huang S, Zheng M, Liu Z, Li Z
and Deng Y: H19 derived microRNA-675 regulates cell proliferation
and migration through CDK6 in glioma. Am J Transl Res. 7:1747–1764.
2015.PubMed/NCBI
|
|
38
|
Yao J, Zhang P, Li J and Xu W:
MicroRNA-215 acts as a tumor suppressor in breast cancer by
targeting AKT serine/threonine kinase 1. Oncol Lett. 14:1097–1104.
2017.PubMed/NCBI
|
|
39
|
Su ZZ, Kang DC, Chen Y, Pekarskaya O, Chao
W, Volsky DJ and Fisher PB: Identification and cloning of human
astrocyte genes displaying elevated expression after infection with
HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid
subtraction hybridization, RaSH. Oncogene. 21:3592–3602. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gnosa S, Shen YM, Wang CJ, Zhang H,
Stratmann J, Arbman G and Sun XF: Expression of AEG-1 mRNA and
protein in colorectal cancer patients and colon cancer cell lines.
J Transl Med. 10:1092012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li J, Zhang N, Song LB, Liao WT, Jiang LL,
Gong LY, Wu J, Yuan J, Zhang HZ, Zeng MS and Li M: Astrocyte
elevated gene-1 is a novel prognostic marker for breast cancer
progression and overall patient survival. Clin Cancer Res.
14:3319–3326. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu JQ, Zhou Q, Zhu H, Zheng FY and Chen
ZW: Overexpression of astrocyte elevated gene-1 (AEG-1) in cervical
cancer and its correlation with angiogenesis. Asian Pac J Cancer
Prev. 16:2277–2281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dong L, Qin S, Li Y, Zhao L, Dong S, Wang
Y, Zhang C and Han S: High expression of astrocyte elevated gene-1
is associated with clinical staging, metastasis and unfavorable
prognosis in gastric carcinoma. Tumour Biol. 36:2169–2178. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang G, Zhang L, Lin S, Li L, Liu M, Chen
H, Cao M, Liu D, Huang YR and Bo J: AEG-1 is associated with tumor
progression in nonmuscle-invasive bladder cancer. Med Oncol.
31:9862014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hu G, Wei Y and Kang Y: The multifaceted
role of MTDH/AEG-1 in cancer progression. Clin Cancer Res.
15:5615–5620. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Emdad L, Lee SG, Su ZZ, Jeon HY, Boukerche
H, Sarkar D and Fisher PB: Astrocyte elevated gene-1 (AEG-1)
functions as an oncogene and regulates angiogenesis. Proc Natl Acad
Sci USA. 106:pp. 21300–21305. 2009; View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Du Y, Jiang B, Song S, Pei G, Ni X, Wu J,
Wang S, Wang Z and Yu J: Metadherin regulates actin cytoskeletal
remodeling and enhances human gastric cancer metastasis via
epithelial-mesenchymal transition. Int J Oncol. 51:63–74. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zou B and Wang S: Expression and
prognostic value of MTDH, Bcl-2 and E-cadherin in sinonasal
malignant mucosal melanoma. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke
Za Zhi. 50:1009–1014. 2015.(In Chinese). PubMed/NCBI
|