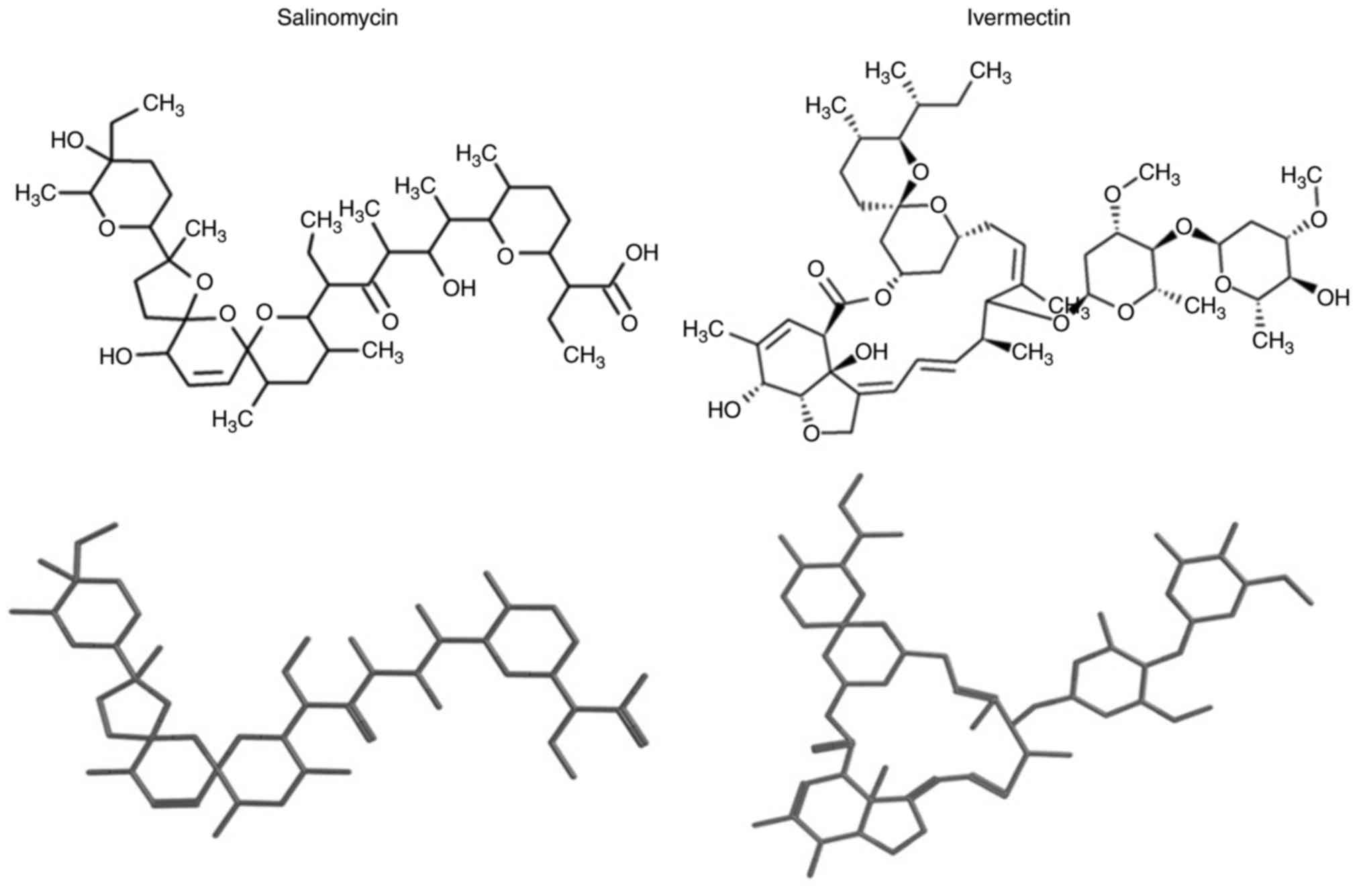
Introduction
Ivermectin, a polycyclic lactone pesticide produced
by bacterium Streptomyces avermitilis is a broad-spectrum
antiparasitic drug that has been used in human medicine since 1987
(1). It is the drug of choice in
treating onchocerciasis and strongyloidiasis, and as a therapeutic
option for mass population treatment campaigns for lymphatic
filariasis. As a microfilaricide, a single dose of ivermectin is
fast, effective and well tolerated, and causes little to no severe
inflammatory responses (2).
Commonly observed adverse effects from treatment with ivermectin
are mild and self-limiting and include: Skin reactions (such as
itching), musculoskeletal pain, fever, swelling of the face, joints
and limbs, headaches and dizziness, lymphadenopathy, eye reactions
and pain from parasitic nodules (3).
Previous studies on the molecular pathogenesis of
cancer has improved our knowledge and increased interest in
repurposing non-cancer drugs to be used against this disease
(4). A study from 1996 reported
that ivermectin treatment in murine multidrug-resistant (MDR)-P388
and human MDR-CEM leukemia cells was a substrate and inhibitor of
P-glycoprotein-mediated multidrug resistance in cancer (5). Another study demonstrated that
ivermectin, at doses of 3–5 mg/kg, was able to suppress the growth
of human melanoma and a number of other cancer xenografts in mice
without adverse effects (6). In
2009, a high-throughput screen was performed to identify the
selective inhibitors of cancer stem-like cells (CSCs) and
demonstrated that salinomycin treatment reduced the proportion of
CSCs by >100-fold relative to paclitaxel, inhibited mammary
tumor growth in vivo and increased epithelial
differentiation of tumor cells, accompanied by a loss of expression
of breast CSC genes (7). As
salinomycin is an antiparasitic drug for veterinary use only,
similar compounds for human use were investigated that may also act
as selective inhibitors of CSC. The results of the present study
demonstrated that ivermectin exhibits a high degree of similarity
with salinomycin and preferentially inhibited the CSC subpopulation
in a breast cancer model.
Materials and methods
Computational searches
To systematically identify FDA approved drugs that
exhibit similar activity as salinomycin, a fast computational
search of the DrugBank database 4.0 of approved drugs was performed
(8). Using the principles of
similarity searching with two-dimensional fingerprints, the
chemical structure of salinomycin was used as a query to compute
the similarity of each of 1,623 compounds in DrugBank using the
Molecular ACCess System keys (9)
and the Tanimoto coefficient (10), as implemented in Molecular
Operating Environment (version 2010.10) (11). The selection of hit compounds was
based on similarity values and visual inspection.
Cell culture and drugs
The breast cancer cell line MDA-MB-231 was obtained
from the American Type Culture Collection (Manassas, VA, USA).
Cells were cultured in Dulbecco's modified Eagle's medium
(DMEM)/F12 medium supplemented with 10% of fetal bovine serum (FBS)
(both from Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) at 37°C in a humidified 5% CO2 atmosphere.
Ivermectin and paclitaxel were purchased from Sigma-Aldrich (Merck
KGaA, Darmstadt, Germany).
Cell viability assay
Cells were seeded (2×103 cells/well) into
96-well microplates (Corning Incorporated, Corning, NY, USA) into
0.1 ml of medium at 37°C for 24 h, and then treated with either
ivermectin (0.2, 0.4, 0.8, 1, 5 or 8 µM) or paclitaxel (0.001, 0.1,
10 or 1,000 nM) for 72 h at 37°C. The medium was replaced with
fresh medium containing drug or vehicle daily. Equal volumes of
ethanol and DMSO vehicles were used for ivermectin and paclitaxel,
respectively. Cell viability was evaluated by the colorimetric
assay CellTiter 96 AQueous One Solution Cell Proliferation assay
(Promega Corporation, Madison, WI, USA) according to the
manufacturer's protocol. Following 72 h treatment, the AQueous One
Solution reagent was added and incubated for an additional 4 h at
37°C. The optical density was measured at 490 nm, using a plate
reader and the effect of each treatment was expressed as a
percentage of cell viability relative to the untreated control
cells. Mean values and standard deviations were calculated;
experiments were performed in triplicate.
Clonogenic assay
MDA-MB-231 cells (4.5×105) were plated in
a 25 cm2 cell culture flask and incubated for 48 h to
allow the cells to proliferate to reach a cell density of 70%.
Cells were treated with different doses of ivermectin (0.2, 0.4,
0.8, 1, 5 and 8 µM) for 120 h. Control cells were treated with the
corresponding amounts of ethanol (the ivermectin vehicle). After
120 h, cells were collected and cultured in drug-free medium in a
dish (2 mm grid; 60×15 mm; Corning Incorporated) for 21 days.
Subsequently, colonies were fixed in methanol and acetic acid (3:1
v/v), and stained with 0.4% crystal violet (Sigma-Aldrich; Merck
KGaA). The number of colonies on the culture dish were counted with
a stereo microscope and quantified using ImageJ software version
1.50f (National Institutes of Health, Bethesda, MD, USA).
Spheroid culture
MDA-MB-231 cells (4.5×105) were grown in
a 25 cm2 cell culture flask and incubated in DMEM/F12
medium, 10% fetal bovine serum, penicillin (100 U/ml) and
streptomycin (1.0 mg/ml) at 37°C in humidified atmosphere of 5%
CO2 for 48 h, to allow the cells to proliferate to a
confluence of ~80%. Cells were collected and sub-cultured
(1×103 cells) in 24-well ultra-low attachment multiwell
plates (Corning Costar; Corning Incorporated) for 15 days with 1 ml
MammoCult Basal Medium plus supplements (STEMCELL Technologies,
Inc., Vancouver, BC, Canada), 0.48 µg/ml hydrocortisone and 4 µg/ml
heparin, at 37°C in a humidified atmosphere of 5% CO2
and 95% air. The medium was changed every 2 days. Following 15 days
incubation, the spheroids (confluence, ~60–70%) were dispersed and
stained with 0.4% trypan blue to assess the cell viability and
counted with a TC10 Automated Cell Counter (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Cells (1×103) were seeded into
a 96-well plate and exposed to ivermectin (0.25, 0.5, 1, 2, 4 and 8
µM) or paclitaxel (0.1, 0.1, 1, 10 and 100 nM) for 72 h, or their
respective vehicles, ethanol or DMSO.
Sorting the cluster of differentiation
(CD)44+/CD24− subpopulation of MDA-MB-231
cells by flow cytometry
MDA-MB-231 cells (5×105) were seeded in
75 cm2 culture flasks and incubated in complete medium
for 48 h to achieve a confluence of ~90%. Cells were washed once
with PBS and then harvested with 0.05% trypsin + 0.025% EDTA.
Detached cells were washed with blocking reagent (1% FBS in PBS)
and resuspended in 1 ml wash buffer (1×106 cells/100
µl), containing the fluorophore-conjugated monoclonal antibodies
against CD44 (fluorescein isothiocyanate-conjugated; catalog no.
555742; BD Biosciences, Franklin Lakes, NJ, USA) and against human
CD24 (phycoerythrin-conjugated; catalog no. 311106; BioLegend,
Inc., San Diego, CA, USA) or their respective isotype controls
(FITC-IgG2bκ, catalog no. 555742; PE-IgG2aκ, catalog no. 555574; BD
Biosciences). Antibodies were used at the manufacturer's
recommended concentrations (dilution: 20 µg/1×106 cells
in 100 µl) and incubated at 4°C in the dark for 40 min. Labeled
cells were washed in the wash buffer twice. The stem cell marker
profiling analysis was performed using a BD FACScanto II (BD
Biosciences). Live cell sorting was performed using a BD FACSAria
(BD Biosciences). The percentage of cells in different marker
populations was evaluated using BD FACSDiva version 6.1.3 (BD
Biosciences). The sorted cells were resuspended three times in
DMEM-F12-10% FBS with 10,000 U/ml penicillin, 10 mg/ml streptomycin
and 0.025 mg/ml amphotericin B. Cells were counted using the trypan
blue method and seeded (1×103 cells/well) in a 96-well
plate with an ultra-low attachment surface (Corning-Costar; Corning
Inc.) in Human MammoCult™ Proliferation supplement
(STEMCELL Technologies, Inc.), 0.48 µg/ml hydrocortisone and 4
µg/ml heparin, for 24 h at 37°C in humidified atmosphere of 5%
CO2. Cells were treated with ivermectin (0.25, 0.5, 1,
2, 4 or 8 µM) or paclitaxel (0.1, 0.1, 1,10 or 100 nM) or their
respective vehicle controls for 72 h. Finally, cells were stained
with 0.4% trypan blue to assess cell viability and counted with
TC10 Automated Cell Counter (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) as aforementioned.
Expression of stemness genes by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Unsorted, whole population MDA-MB-231 cells were
treated with ivermectin for 72 h and total RNA was isolated when
cells reached ~70% confluence, using TRIzol reagent (Gibco; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
RNA purity and integrity were assessed by spectrophotometric
analysis using a NanoDrop 2000c (Thermo Fisher Scientific, Inc.)
and denaturing 2% agarose gel; bands were visualized using a
MiniBIS Pro D-Transilluminator (DNR Bio-Imaging Systems Ltd., Neve
Yamin, Israel).
A total of 1 µg total RNA was used for cDNA
synthesis with the GeneAmp RNA PCR Core kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.). cDNA was used with iQ SYBR Green
SuperMix (Bio-Rad Laboratories, Inc.), according to the
manufacturer's protocol. qPCR reactions were run in triplicate
using an ABI Prism 7000 (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The qPCR cycling conditions were as follows: 10
min at 95°C; 40 cycles of 30 sec at 95°C, 30 sec at 60°C and 30 sec
at 72°C. Data were analyzed using the 2−ΔΔCq method
(12), and reported as the
fold-change in gene expression normalized to the endogenous control
gene hypoxanthine phosphoribosyltransferase 1 (HPRT1), and relative
to cells without treatment. The primers used were: HPRT1 forward,
5′-GAACCTCTCGGCTTTCCCG-3′ and reverse, 3′-CACTAATCACGACGCCAGGG-5′;
homeobox protein nanog (nanog) forward, 5′-ACCTCGCTGATTAGGCTCCAA-3′
and reverse, 3′-AGTCTGGACACTGGCTAATCC-5′; octamer binding protein 4
(oct-4) forward, 5′-CAGGCCCGAAAGAGAAAGC-3′ and reverse,
3′-CCACACTCGGACCACATCCT-5′; and sox-2 forward,
5′-GCTAGTCTCCAAGCGACGAAA-3′ and reverse,
3′-AATTCAGCAAGAAGCCTCTCCTT-5′. The annealing temperature was 60°C
for all reactions.
Protein extraction and western blot
analysis of nanog, sox-2 and oct-4
Unsorted whole population MDA-MB-231 cells
(4×105) were cultured in 25 cm2 flasks and
treated with ivermectin (or its vehicle control) for 72 h. Once the
cells had reached a density of ~70%, the cells were washed once
with PBS and then harvested with 0.05% trypsin/0.025% EDTA.
Detached cells were washed with PBS, proteins were extracted with
radioimmunoprecipitation buffer (150 mM NaCl; 1.0% IGEPAL CA-630;
0.5% sodium deoxycholate; 0.1% SDS; 50 mM Tris, pH 8.0) in the
presence of proteinase inhibitors (catalog no. p8340;
Sigma-Aldrich; Merck KGaA). Protein concentration was determined
using a bicinchoninic acid assay and the integrity was assessed by
coomassie staining. A total of 30 µg protein was separated by 10%
SDS-PAGE and transferred onto a polyvinylidene difluoride membrane
(cat no. 162-0177; Bio-Rad Laboratories, Inc.). The membrane was
blocked with 3% skim milk in PBS for 1 h at room temperature and
subsequently incubated with antibodies against nanog (catalog no.
sc-134218; 1:500); sox-2 (catalog no. sc-17320; 1:200); oct-4
(catalog no. sc-9081; 1:200) (all from Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), and anti-actin peroxidase (A3854; 1:10,000;
Sigma-Aldrich; Merck KGaA) in blocking solution (5% skim milk in
TBS + 0.1% Tween-20), overnight at 4°C. The following secondary
antibodies were used: Oct4, bovine anti-rabbit, sc-2370; sox-2,
donkey anti-goat, sc-2020; nanog, anti-mouse, sc-2371 (all from
Santa Cruz Biotechnology, Inc). Secondary antibodies were diluted
1:1,000 and the incubation was performed for 1 h at room
temperature. Protein bands were visualized using the chromogenic
substrate Clarity Western Enhanced Chemiluminescence Substrate
(catalog no. 1705060; Bio-Rad Laboratories, Inc.). Bands were
quantified densitometrically using Image J version 1.50f (National
Institutes of Health, Bethesda, MD, USA).
Statistical analyses
Analyses were performed with GraphPad Prism software
(version 6.0; GraphPad Software, Inc., La Jolla, CA, USA). The
experiments were conducted with at least three independent
triplicates. P-values were calculated using the one-way analysis of
variance test followed by the Bonferroni post-hoc test. P<0.01
was considered to indicate a statistically significant
difference.
Results
Ivermectin exhibits high structural
similarity to salinomycin
Compounds identified in DrugBank possessed a low
molecular similarity with salinomycin (median and mean similarity
values of 0.34±0.14). Subsequent results from the similarity
searching using salinomycin as a query molecule identified 25
top-ranked compounds with a high similarity value. To define a
compound with ‘high’ similarity value, molecules were selected as
those with similarity values two standard deviations above the mean
similarity of all DrugBank compounds to salinomycin. Following
visual inspection of the chemical structures of the identified
compounds, the drugs with the highest similarity scores were
ivermectin followed by natamycin (a food preservative) and narasin
(an antibiotic for veterinary use). Ivermectin had the highest
similarity value of 0.78, and following the principle of molecular
similarity, which states that similar compounds demonstrate similar
properties (13) (with the
exception of the so-called activity cliffs) (14), it was hypothesized that the
antiparasitic drug ivermectin may also possess similar biological
properties as salinomycin. In addition, ivermectin is a Food and
Drug Administration (FDA)-approved drug for clinical use, whereas
natamycin and narasin are not. Notably, using this method of
screening that is based on structural similarity rather than
biological activity upon CSC cells, as reported by Gupta et
al (7), did not identify
avermectin as a candidate compound. Two- and three-dimensional
structures of salinomycin and ivermectin are provided in Fig. 1.
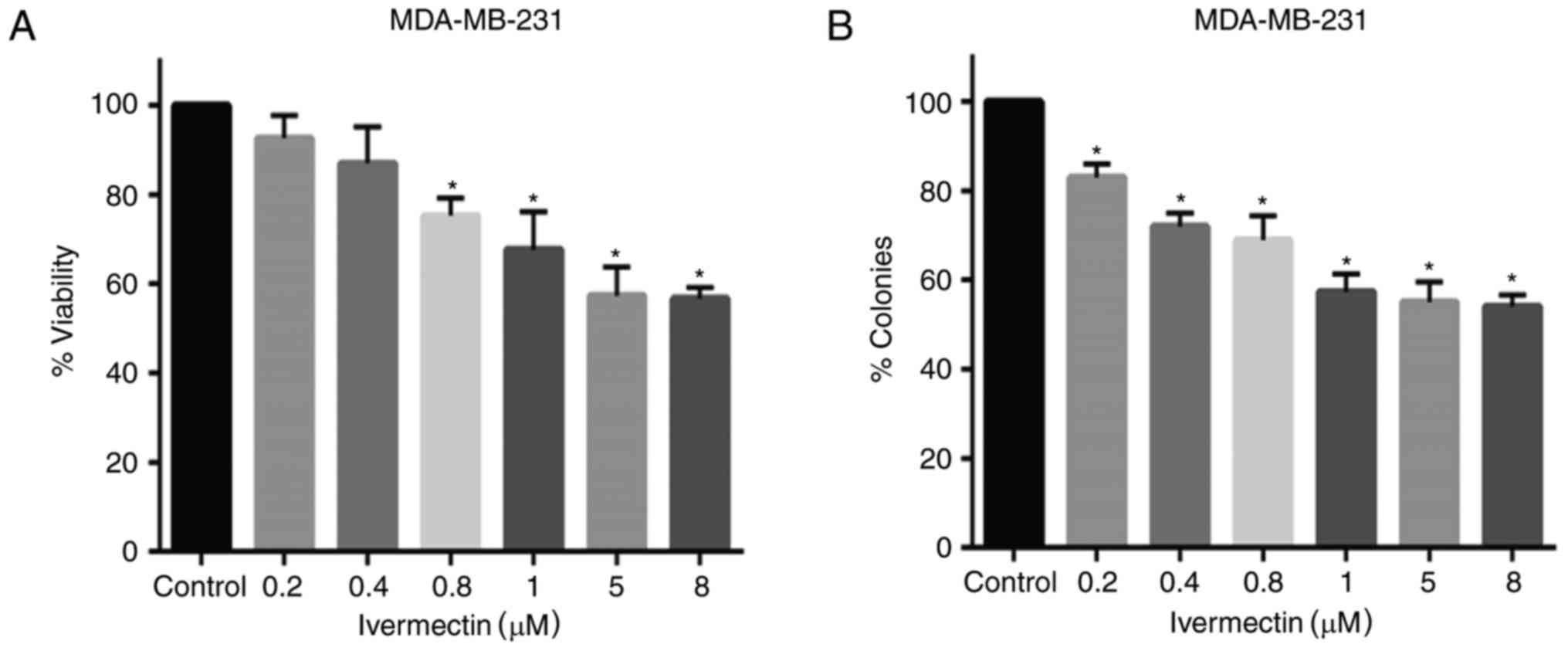
Ivermectin partially inhibits cell
viability and clonogenic capacity
To evaluate the antitumoral effects of ivermectin on
MDA-MB-231 cells, cell viability and clonogenicity was evaluated. A
mild-to-moderate dose-dependent effect was observed in cells
treated with various concentrations of ivermectin (Fig. 2A); however, cell viability did not
decrease beyond 45%, even at 8 µM of ivermectin. Statistically
significant differences were observed at concentrations between 0.8
and 8 µM, compared with untreated controls (P<0.0001). Colony
forming ability was also significantly reduced with ivermectin
treatment with doses as low as 0.2 µM (Fig. 2B); however, these effects appear to
be similar at doses between 1 and 8 µM (P<0.0001).
Inhibitory effects of ivermectin and
paclitaxel in stem cell enriched MDA-MB-231 cells
The CD44+/CD24− subpopulation
of MDA-MB-231 cells has been previously reported to possess
stem/progenitor cell properties, and this subpopulation in patients
with breast cancer exclusively retains the ability to form novel
tumors in a non-obese diabetic-severe compromised immunodeficiency
mouse xenograft model (15,16).
The proportion of CD44+/CD24− cells in the
cell line MDA-MB-231 is 85±5% (17). In addition, spheroids of cell lines
growing in nonadherent conditions are also reported to be enriched
in stem cells (18). Therefore,
these two conditions were used to determine whether ivermectin
treatment preferentially inhibited the CSC population compared with
treatments with the cytotoxic drug paclitaxel. At concentrations ≤2
µM ivermectin, there were no significant differences in viability
in the total MDA-MB-231 population compared with cultures of cells
grown in spheroids and those sorted by
CD44+/CD24− (Fig. 3A). However, cells treated with 4 or
8 µM ivermectin exhibited a significant decrease in viability in
the CSC-enriched populations compared with the total cell
population (P<0.001); whereas viability in the total cell
population was inhibited by ≤30%. As expected, the two
subpopulations (spheroids and CD44+/CD24−)
exhibited less inhibition with paclitaxel treatments compared with
ivermectin treatments, and no significant differences were
identified between either of these groups compared with the
inhibition exhibited in the total MDA-MB-231 cell population
(Fig. 3B). Ivermectin treatment
exhibited an increased inhibitory effect upon CSC-enriched
subpopulations compared with cells treated with paclitaxel.
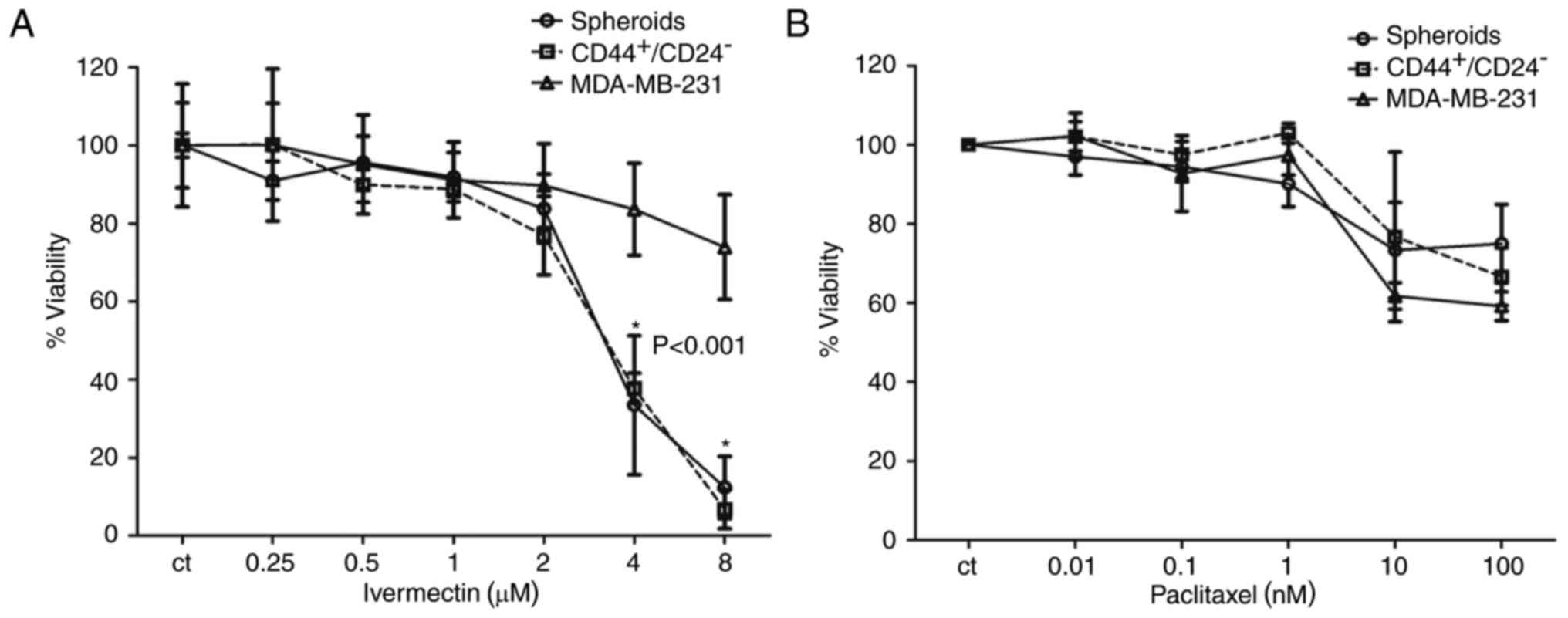 | Figure 3.Effects of treatment with ivermectin
or paclitaxel on the CSC-enriched populations of MDA-MB-231 cells.
(A) The effect of ivermectin (0.25, 0.5, 1, 2, 4 and 8 µM) on cell
viability of spheroids, CD44+/CD24− selected
and total MDA-MB-231 cells. A statistically significant effect of
ivermectin was observed in the CSC-enriched populations at 4 and 8
µM compared with the total MDA-MB-231 cell population. (B) The
effects of paclitaxel (0.01, 0.1, 1, 10 and 100 nM) on cell
viability of spheroids, CD44+/CD24− selected
and total MDA-MB-231 cells. No statistically significant
differences were observed among the paclitaxel treatments. CD,
cluster of differentiation. |
To further evaluate the relative selectivity of
ivermectin upon the stem cell population,
CD44+/CD24− and
CD44+/CD24+ subpopulations of MDA-MB-231
cells were sorted and cultured in the presence of either ivermectin
or paclitaxel. Cells treated with 4 and 8 µM ivermectin exhibited a
significant reduction in viability of the
CD44+/CD24− stem cell population; 8 µM
ivermectin treatment reduced the CD44+/CD24−
viability to 0%, compared with CD44+/CD24+
cells (Fig. 4A), whereas the
reduction in viability in CD44+/CD24+ cells
treated with 8 µM ivermectin approached 25%. The non-CSC
CD44+/CD24+ cell populations were sensitive
to paclitaxel from 0.01 nM; whereas inhibition of the
CD44+/CD24− stem cell subpopulation was only
observed at ≥10 nM. Statistical differences were observed at all
concentrations between CD44+/CD24+ and
CD44+/CD24− (Fig. 4B).
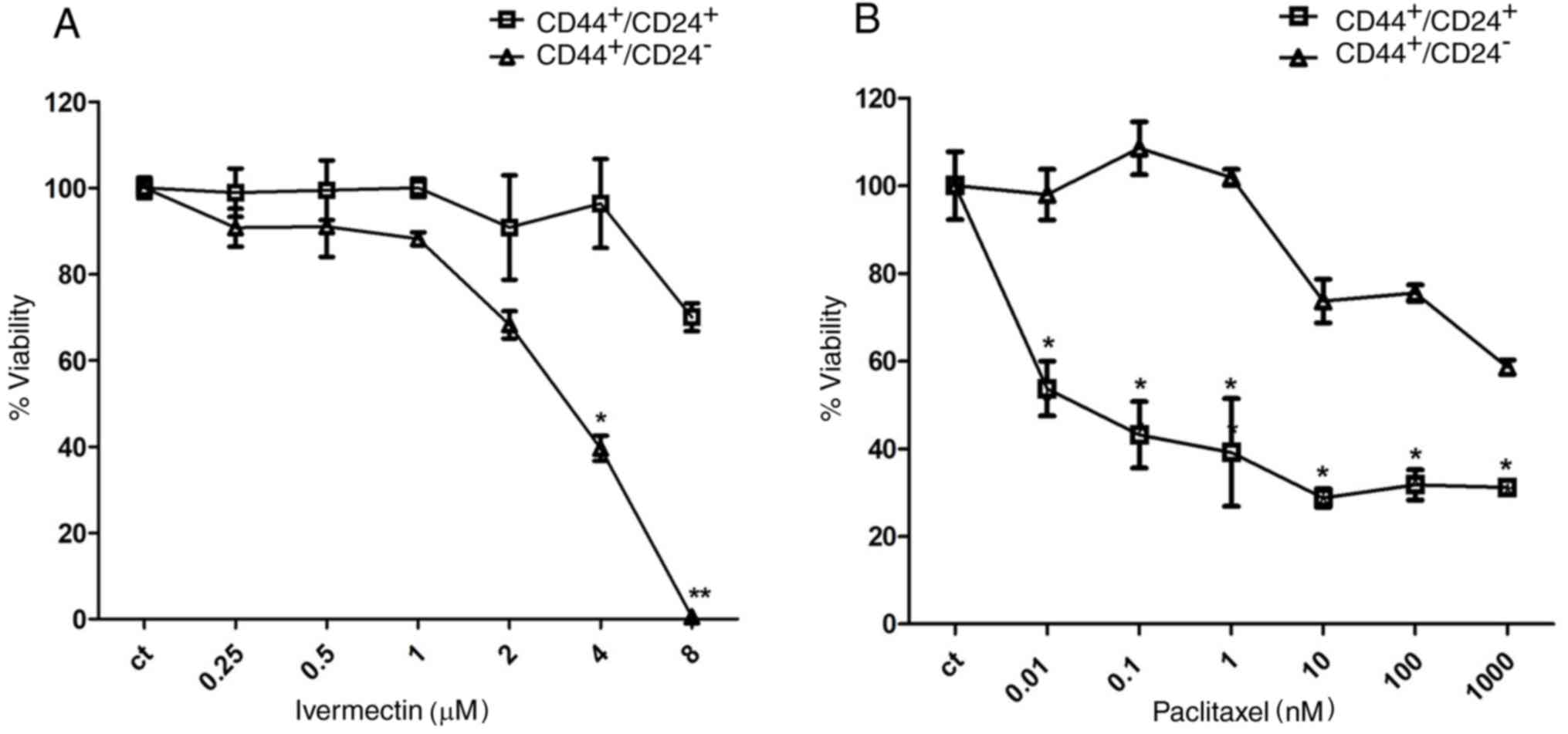 | Figure 4.Effects of ivermectin treatment on
CD44+/CD24− and
CD44+/CD24+ MDA-MB-231 cells. (A) Effects of
ivermectin treatment (0.25, 0.5, 1, 2, 4 and 8 µM) on cell
viability of CD44+/CD24− and
CD44+/CD24+ MDA-MB-231 cells. A statistically
significant effect of ivermectin was observed in
CD44+/CD24− cells at 4 and 8 µM, compared
with CD44+/CD24− cells. (B) Effects of
paclitaxel (0.01, 0.1, 1, 10, 100 and 1,000 nM) on cell viability
of CD44+/CD24− and
CD44+/CD24+ MDA-MB-231 cells. The opposite
pattern was observed with paclitaxel at all concentrations tested
*P<0.01 and **P<0.001 vs. CD44+/CD24+
cells. CD, cluster of differentiation. |
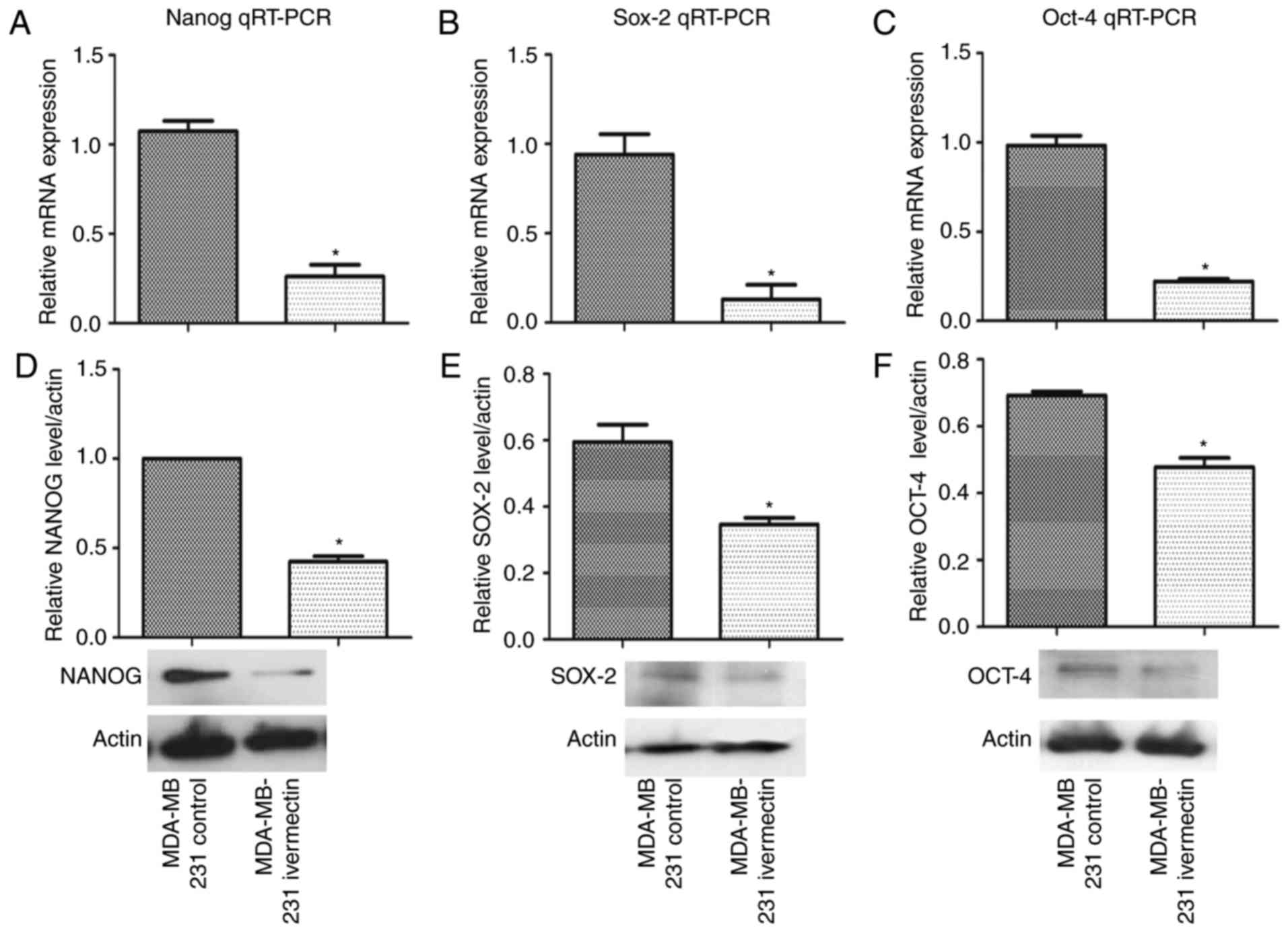
Ivermectin reduces the expression of
stemness genes
As salinomycin has been demonstrated to decrease the
expression of stemness genes including nanog (19), MDA-MB-231 cells cultured in normal
(adherent) conditions were treated with ivermectin at 4 µM for 72 h
and the mRNA expression levels of nanog, sox-2 and oct-4 were
evaluated by RT-qPCR and western blotting. As demonstrated in
Fig. 5, ivermectin significantly
reduced the expression of these three genes at both the mRNA and
the protein level (P<0.01).
Discussion
In the field of drug repurposing for cancer therapy,
major efforts have been undertaken to identify drugs that may
selectively or preferentially target the CSC population of tumors.
The results of the present study demonstrated that ivermectin, an
antiparasitic drug for approved for human use, preferentially
targeted the CSC-enriched subpopulation of the breast cancer cell
line MDA-MB-231. Higher reductions in cell viability were observed
for the CD44+/CD24− subpopulation and
spheroids treated with ivermectin compared with paclitaxel. These
results were accompanied by the decreased expression of stemness
genes nanog, sox-2 and oct-4, previously reported to be highly
expressed in CSCs (20).
Failure to successfully eradicate tumors no longer
amenable to local treatments is at least partly due to the
existence of CSCs, which are characterized by tumorigenic
properties, such as self-renewal, formation of differentiated
progeny and development of resistance to therapy (21). It has been established that Notch,
hedgehog (Hh) and Wnt signaling pathways are involved in the
regulation of proliferation and differentiation of CSCs; therefore,
these pathways may be key targets for the development of more
efficient cancer therapeutics (22). However, the development of drugs
that target crucial steps in the Wnt, Notch and Hh signaling
pathways may not be effective or can be toxic, owing to the
signaling crosstalk among these pathways. An alternative strategy
may be to look in an unbiased and systematic manner at
pharmacological entities that demonstrate preferential toxicity in
CSCs over the total tumoral cell population (7,23). A
previous study performed a screen for agents with epithelial
CSC-specific toxicity and identified four compounds that
consistently reduced the proportion of CSCs, including salinomycin,
etoposide, abamectin and nigericin (7). Among these, salinomycin was the most
effective, exhibiting a >100-fold reduction in viability
compared with paclitaxel. Further investigation demonstrated that
salinomycin inhibited mammary tumor growth in vivo and
induced an increase in epithelial differentiation of tumor cells,
which is associated with the loss of stemness genes (7). That salinomycin is currently only
approved for veterinary use, prompted the authors of the present
study to search for drugs approved for human use by screening
DrugBank to identify structurally similar, clinically approved
drugs. The results of the present study adhered to the principle of
molecular similarity that states that similar compounds demonstrate
similar properties (13,14); therefore, the antiparasitic drug
ivermectin should exhibit similar biological effects to those
reported for salinomycin (7,24,25).
This is advantageous in terms of drug repositioning, as the
clinical use of ivermectin as antiparasitic is extensive (25), therefore, clinical trials in cancer
may proceed in the near future.
Avermectins are 16-membered pentacyclic lactone
compounds that are derived from polyketides and are linked to a
disaccharide of the methylated deoxysugar L-oleandrose (26). Ivermectin was FDA-approved for
human use in 1987 (http://www.centerwatch.com/drug-information/fda-approved-drugs/drug/250/stromectol-ivermectin),
and is an avermectin derivative of major interest as a parasitic
drug, and is a mixture of 80% 22,23-dihydro-avermectin-B1a and 20%
of the -B1b homolog (27). As
ivermectin is an ionophore, it induces chloride-dependent membrane
hyperpolarization and apoptosis in leukemia (28) and induces mitochondrial dysfunction
and oxidative stress (29). The
antitumor effects of ivermectin have also been associated with its
ability to inactivate the p21-activated kinase 1/protein kinase B
axis, inducing cytostatic autophagy and modulation of P2X
purinoceptor (P2X) 4/P2X7/Pannexin-1 sensitivity to extracellular
adenosine triphosphate (30,31).
Ivermectin was first investigated as an antitumor drug in 1999
(32) and, to the best of the of
our knowledge, no studies have been performed on its ability to
preferentially target CSCs. A previous study demonstrated that
ivermectin treatment inhibits the Wnt-transcription factor 4
signaling pathway (33), which is
frequently considered to be altered in CSCs (21). Recently, it was demonstrated that
ivermectin downregulated the expression of stemness genes nanog and
sox2, in addition to reducing the activity of aldehyde
dehydrogenase, all of which are CSC markers (34). The results of the present study
demonstrated preferential inhibition of CSC-enriched MDA-MB-231
cells treated with ivermectin, which agreed with the literature on
the effects of ivermectin upon pathways and markers associated with
CSCs. The present study, however, was not mechanistic; therefore,
further investigations into the molecular basis responsible for the
preferential cytotoxicity of ivermectin on CSCs must be
pursued.
In conclusion, results from the present study
demonstrated that ivermectin preferentially targeted the stem cell
population in MDA-MB-231 human breast cancer cells. Ivermectin has
been demonstrated to be safe, following treatment of millions of
patients with onchocerciasis and other parasitic diseases, which
makes it a strong candidate for further studies investigating its
potential use as a repurposed drug for cancer therapy.
Acknowledgements
The present study was supported by FONSEC, National
Council of Science and Technology, Mexico (grant no. 161915) and
National Autonomous University of Mexico, Support Program for
Research Projects on Science and Innovation (grant no.
IT206611).
References
|
1
|
Stromectol cleared by the U.S. Food and
Drug Administration to treat onchocerciasis. http://www.centerwatch.com/drug-information/fda-approved-drugs/drug/250/stromectol-ivermectin
|
|
2
|
Diawara L, Traoré MO, Badji A, Bissan Y,
Doumbia K, Goita SF, Konaté L, Mounkoro K, Sarr MD, Seck AF, et al:
Feasibility of onchocerciasis elimination with ivermectin treatment
in endemic foci in Africa: First evidence from studies in Mali and
Senegal. PLoS Negl Trop Dis. 3:e4972009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ottesen EA and Campbell WC: Ivermectin in
human medicine. J Antimicrob Chemother. 34:195–203. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dueñas-González A, García-López P, Herrera
LA, Medina-Franco JL, González-Fierro A and Candelaria M: The
prince and the pauper. A tale of anticancer targeted agents. Mol
Cancer. 7:822008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Didier A and Loor F: The abamectin
derivative ivermectin is a potent P-glycoprotein inhibitor.
Anticancer Drugs. 7:745–751. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Driniaev VA, Mosin VA, Krugliak EB,
Sterlina TC, Novik TC, Ermakova NV, Kublik LN, Levitman MKh,
Shaposhnikova VV and Korystov IuN: Modification of antitumor effect
of vincristine by natural avermectins. Antibiot Khimioter. 49:3–5.
2004.(In Russian). PubMed/NCBI
|
|
7
|
Gupta PB, Onder TT, Jiang G, Tao K,
Kuperwasser C, Weinberg RA and Lander ES: Identification of
selective inhibitors of cancer stem cells by high-throughput
screening. Cell. 138:645–659. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Law V, Knox C, Djoumbou Y, Jewison T, Guo
AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al:
Drugbank 4.0: Shedding new light on drug metabolism. Nucl Acids
Res. 42(Database Issue): D1091–D1097. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Durant JL, Leland BA, Henry DR and Nourse
JG: Reoptimization of Mdl Keys for use in drug discovery. J Chem
Inf Comput Sci. 42:1273–1280. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Willett P, Barnard JM and Downs GM:
Chemical similarity searching. J Chem Inf Comput Sci. 38:983–996.
1998. View Article : Google Scholar
|
|
11
|
Molecular Operating Environment (MOE),
version 2010.10. Chemical Computing Group Inc.; Montreal, PQ,
Canada: http://www.chemcomp.com
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Medina-Franco JL and Maggiora GM:
Molecular similarity analysisChemoinformatics for Drug Discovery.
Bajorath J: John Wiley & Sons, Inc.; Hoboken, New Jersey: pp.
343–399. 2014
|
|
14
|
Maggiora GM: On outliers and activity
cliffs - why QSAR often disappoints. J Chem Inf Model. 46:15352006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tanaka H, Nakamura M, Kameda C, Kubo M,
Sato N, Kuroki S, Tanaka M and Katano M: The Hedgehog signaling
pathway plays an essential role in maintaining the CD44+CD24-/low
subpopulation and the side population of breast cancer cells.
Anticancer Res. 29:2147–2157. 2009.PubMed/NCBI
|
|
16
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 100:pp.
3983–3988. 2003; View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sheridan C, Kishimoto H, Fuchs RK,
Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R Jr, Badve S and
Nakshatri H: CD44+/CD24- breast cancer cells exhibit enhanced
invasive properties: An early step necessary for metastasis. Breast
Cancer Res. 8:R592006. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Piscitelli E, Cocola C, Thaden FR,
Pelucchi P, Gray B, Bertalot G, Albertini A, Reinbold R and Zucchi
I: Culture and characterization of mammary cancer stem cells in
mammospheres. Methods Mol Biol. 1235:243–262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y: Effects of salinomycin on cancer
stem cell in human lung adenocarcinoma A549 cells. Med Chem.
7:106–111. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang W, Sui Y, Ni J and Yang T: Insights
into the Nanog gene: A propeller for stemness in primitive stem
cells. Int J Biol Sci. 12:1372–1381. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dragu DL, Necula LG, Bleotu C, Diaconu CC
and Chivu-Economescu M: Therapies targeting cancer stem cells:
Current trends and future challenges. World J Stem Cells.
7:1185–1201. 2015.PubMed/NCBI
|
|
22
|
Takebe N, Harris PJ, Warren RQ and Ivy PS:
Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog
pathways. Nat Rev Clin Oncol. 8:97–106. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subedi A, Futamura Y, Nishi M, Ryo A,
Watanabe N and Osada H: High-throughput screening identifies
artesunate as selective inhibitor of cancer stemness: Involvement
of mitochondrial metabolism. Biochem Biophys Res Commun.
477:737–742. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
An H, Kim JY, Oh E, Lee N, Cho Y and Seo
JH: Salinomycin promotes anoikis and decreases the CD44+/CD24-
stem-like population via inhibition of STAT3 activation in
MDA-MB-231 cells. PLoS One. 10:e01419192015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Krotneva SP, Coffeng LE, Noma M, Zouré HG,
Bakoné L, Amazigo UV, de Vlas SJ and Stolk WA: African Program for
Onchocerciasis Control 1995–2010: Impact of annual ivermectin mass
treatment on off-target infectious diseases. PLoS Negl Trop Dis.
9:e00040512015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoon YJ, Kim ES, Hwang YS and Choi CY:
Avermectin: biochemical and molecular basis of its biosynthesis and
regulation. Appl Microbiol Biotechnol. 63:626–634. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barragry TB: A review of the pharmacology
and clinical uses of ivermectin. Can Vet J. 28:512–517.
1987.PubMed/NCBI
|
|
28
|
Sharmeen S, Skrtic M, Sukhai MA, Hurren R,
Gronda M, Wang X, Fonseca SB, Sun H, Wood TE, Ward R, et al: The
antiparasitic agent ivermectin induces chloride-dependent membrane
hyperpolarization and cell death in leukemia cells. Blood.
116:3593–3603. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Y, Fang S, Sun Q and Liu B:
Anthelmintic drug ivermectin inhibits angiogenesis, growth and
survival of glioblastoma through inducing mitochondrial dysfunction
and oxidative stress. Biochem Biophys Res Comun. 480:415–421. 2016.
View Article : Google Scholar
|
|
30
|
Dou Q, Chen HN, Wang K, Yuan K, Lei Y, Li
K, Lan J, Chen Y, Huang Z, Xie N, et al: Ivermectin induces
cytostatic autophagy by blocking the PAK1/Akt axis in breast
cancer. Cancer Res. 76:4457–4469. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Draganov D, Gopalakrishna-Pillai S, Chen
YR, Zuckerman N, Moeller S, Wang C, Ann D and Lee PP: Modulation of
P2X4/P2×7/Pannexin-1 sensitivity to extracellular ATP via
ivermectin induces a non-apoptotic and inflammatory form of cancer
cell death. Sci Rep. 5:162222015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mosin VA, Krugliak EB, Sterlina TS,
Korystov IuN, Shaposhnikova VV, Narimanov AA, Kublik LN, Levitman
MKh, Viktorov AV and Driniaev VA: Cytotoxic and cytostatic effect
of avermectines on tumor cells in vitro. Antibiot Khimioter.
45:10–14. 2000.PubMed/NCBI
|
|
33
|
Melotti A, Mas C, Kuciak M, Lorente-Trigos
A, Borges I and Ruiz Altaba A: The river blindness drug ivermectin
and related macrocyclic lactones inhibit WNT-TCF pathway responses
in human cancer. EMBO Mol Med. 6:1263–1278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kwon YJ, Petrie K, Leibovitch BA, Zeng L,
Mezei M, Howell L, Gil V, Christova R, Bansal N, Yang S, et al:
Selective inhibition of SIN3 corepressor with avermectins as a
novel therapeutic strategy in triple-negative breast cancer. Mol
Cancer Ther. 14:1824–1836. 2015. View Article : Google Scholar : PubMed/NCBI
|