Introduction
Systemic lupus erythematosus (SLE) is one of the
common autoimmune diseases, mostly occurs in Asian women. The main
cause of this disease is that the autoimmune system attacks its own
tissues, resulting in tissue damage (1–3). The
typical symptoms of SEL include specific lesions with butterfly
erythema, subacute skin lupus erythematosus and discoid erythema,
and non-specific lesions with light allergy, hair loss, mouth
ulcers, skin vasculitis (purpura), pigmentation or depigmentation,
livedo reticularis, Raynaud's phenomenon, urticaria-like rash and
rare lupus lipid film Inflammation or deep lupus and bullous lupus
erythematosus (4). Numerous
studies have shown that the abnormalities in genetic, endocrine,
infection, immune and some environmental factors are associated
with the incidence of SLE (3,5).
However, the pathogenesis of SLE has not yet been elucidated.
The main pathological manifestation of autoimmune
diseases is that the patient produces a high level of
autoantibodies to identify different autoantigens. It was
demonstrated that autoantibodies associated to several autoantigens
and involved in SLE including anti-double stranded antibody
(anti-dsDNA antibody), anti-nuclear antibody (ANA), anti-soluble
antigen antibodies (anti-ENA antibody) including anti-Jo-1,
anti-U1RNP antibody (anti-nRNP antibody), anti-ribosomal P antibody
(anti-rRNR antibody), anti-Scl-70, anti-Sm antibody, anti-SSA/Ro
antibody and anti-SSB/La antibody, anti-nucleosome antibodies, and
anti-phospholipid antibodies (6–12).
The use of autoantigens to detect autoantibodies is an important
technique for the diagnosis of autoimmune diseases. However, the
intact autoantigen profiling is difficult to obtain and purify, and
the stability of the antigens is poor. In order to avoid the high
production cost and poor stability of autoantigens, some attempts
have been made to detect autoantibodies by alternative. To achieve
the goal of diagnosis, some studies have been attempted to detect
the autoantibodies in SLE using protein chip (13–17).
However, the protein chip is also limited by the difficult in
expression and purification the intact antigens. Researchers try to
find a more desirable alternative such as peptide microarray.
In recent years, peptide microarray has been
developed rapidly (18–20). Peptide microarrays integrate many
peptide active molecules on a very small surface area, so as to
detect the expression and function of different biomolecules
(21). Peptide microarrays has
advantages in simple and fast, high-throughput and accuracy, and
low-cost, compared with traditional protein chips. Using peptide
microarrays, the diagnostic accuracy of lung cancer indicators was
93.1% (22). The diagnostic
accuracy of plasma in lung cancer patients using peptide
microarrays was also reached 92% (23,24).
It was also found that peptide microarrays are useful in the
detection of p53 autoantibodies, and have potential application
value in head and neck cancer patients (25,26).
However, thus far, the use of the peptide microarray technique in
diagnosis of autoimmune diseases, particularly the early diagnosis
of SLE have not been well studied yet.
In the present study, we predicted the SLE-related
epitopes in 14 autoantigens using the antigenic epitope prediction
software DNA star, and designed the peptide microarray for SLE
detection. Then, the autoantibodies in 120 SLE patients and 110
healthy subjects were analyzed and an optimal set of epitopes were
screened out. The sensitivity and specificity of the optimal set of
epitopes in diagnosis of SLE were evaluated.
Materials and methods
Patients
Samples from 120 patients with SLE (including 15
males and 105 females, average age was 34.5 years) who underwent
SLE treatment in the Southern China Hospital were collected.
Samples from 110 healthy volunteers (including 15 males and 95
females, average age was 30.2 years) were also collected. There
were no significant differences in the sex and age between the SLE
patients and healthy subjects. The diagnostic criteria for SLE were
acute or subacute cutaneous lupus manifestations, manifestations of
chronic cutaneous lupus, arthritis, serositis, renal disorder,
blood-hematologic diseases, oral or nasopharyngeal ulcers,
immunological disorder, and alopecia. The diagnosis of SLE should
include four of the above criteria, one clinical criterion and one
immunological criterion. The immunological criteria were as
follows: i) higher titers of ANA than the laboratory reference
standard; ii) higher titers of anti-dsDNA than the laboratory
reference standard; iii) positive anti-Sm antibody; iv)
anti-phospholipid antibodies (positive anti-lupus
anticoagulant/false positive serological test for
syphilis/anticardiolipin antibody at twice the normal level or
increased anti-B2GPI above titer); v) decreased level of complement
proteins (C3, C4 and CH50); and vi) no hemolytic anemia, but Coombs
test is positive. Renal disorder was confirmed as lupus nephritis
(diagnosed by lupus nephritis with ANA or anti-dsDNA-positive). The
exclusion criteria included patients with viral hepatitis,
tuberculosis or SLE combined with other primary organ diseases. In
the present study, we did not exclude the patients with other
infections including influenza, EBV and HIV, because those
infections in China are very low. All participants signed informed
consent. The present study was approved by the Ethics Committee of
Southern China Hospital.
Epitopes prediction by DNA star
software
The DNA star (NIAID, USA) was used to predict the
epitopes on 14 autoantigens. In the DNA STAR online analysis
system, the parameters ‘Epitope’ and ‘assay’ were set as ‘any
epitopes’ and ‘all’. Followed by searching and querying, the
parameters ‘MHC Restriction’, ‘Host’, and ‘Disease’ were set as
‘any MHC Restriction’, ‘Humans’, and ‘Autoimmune Disease’ for each
antigen indicator. After further narrowed the search range by the
peptide information and the linked literatures, we obtained the
peptide sequences associated with SLE.
Peptide microarray preparation
All peptides (purity >98%) were synthesized by
Sangon Biotech, Shanghai, China. The peptide indicators were
prepared using a biochip spotting instrument (AD3200; BioDot,
Irvine, CA, USA). The peptide microarray was prepared in a clean
slide. After the peptide microarray was soaked in a 5% ammonia
silane anhydrous ethanol solution for 30 min. After washed 3 times
with anhydrous ethanol and deionized water for 5 min, the peptide
microarray was air-dried and soaked in a phosphate-buffer solution
(PBS) containing 2.5% glutaraldehyde for 30 min. After washes with
anhydrous ethanol and deionized water for 5 min, three times, the
peptide microarray (384-well plate) was prepared for peptide
loading. The peptide was well-diluted to 0.5 mg/ml in PBS and
loaded into the peptide microarray at 20 µl/well. After centrifuged
2 min at 2,000 rpm, the peptide microarrays were placed overnight,
stored in a slide box, and sealed within hermetic bags at 4°C (or
−20°C with humidity <50% for long-term storage). The bags were
exposed to room temperature for 3–4 h before pick out the peptide
microarray.
Diagnosis of SLE by peptide
microarray
For screening, the peptide microarray was blocked
with 0.1% bovine serum albumin (BSA) in PBS for 30 min, incubated
with serum specimens for 4 h, followed by washes with PBS with
Tween-20 5 times, and PBS 5 times. Then, the peptide microarray was
incubated with a 555-Streptavidin fluorescein for 1 h at room
temperature, and then washes with PBST and PBS 5 times,
respectively. A biotinylated anti-human IgG was used for
autoantibody detection. After air-drying, the peptide microarray
was measured by Jingxin LuxScan™ 10K-B Microarray Scanners
(CapitalBio Corporation, Beijing, China) with 532 nm excitation
wavelength. Finally, data analysis was performed by GenePro
software version 6 (GenePro, Fitchburg, WI, USA) and GraphPad Prism
software version 6 (GraphPad Software, Inc., La Jolla, CA,
USA).
ELISA detection
In order to verify the validity of epitopes, we
selected the 2 most commonly used SLE autoantibodies in clinic
including Sm, and RNP. A complete antigen of each indicator was
used to detect the autoantibodies in sera of the SLE patients using
the Human peripheral blood anti-Sm IgG (cat. no. EA1593-9601G) and
anti-nRNP IgG (cat. no. EA1591-9601G) ELISA Kit (EUROIMMUN
Medizinische Labordiagnostika AG, Lubeck, Germany) following the
kit instruction. The cut-off value was 20 RU/ml. The results of
ELISA were compared with those of peptide microarray. For Sm, the
peptides including SMD1-2, SMD2-1, SMD2-2 and SMD3-1 were compared;
for RNP, the peptides including U1-SnrnpA-2 and U1-SnRNP 68/70 kDa
were compared.
Statistical analysis
Peptide microarray data were measured using GenePro
software version 6.0 (GenePro), and analyzed by GraphPad Prism
software v 6.0 (GraphPad) using Student's t-test. The ROC curves
and area calculated were performed by SPSS software version 16.0
(SPSS, Inc., Chicago, IL, USA). A value of P<0.05 was considered
statistically significant.
Results
Prediction of antigens
Using the DNA star software, a total of 73 potential
epitopes were obtained from 14 autoantigens were predicted. The 14
autoantigens included acidic ribosomal phosphoprotein (P0), acidic
ribosomal phosphoprotein (P1), acidic ribosomal phosphoprotein
(P2), DNA topoisomerase 1 (full length 0, DNA topoisomerase 1
(truncated), nucleolin, proliferating cell nuclear antigen (PCNA),
SMD1, SMD2, SMD3, snRNP-B/B', U1-snRNP 68/70 kDa, U1-snRNP-A, and
U1-snRNP-C. The detailed information of the epitopes are shown in
Table I.
 | Table I.Predicted epitopes on 14 antigens. |
Table I.
Predicted epitopes on 14 antigens.
| Number | Start | End | Peptide |
|---|
| SMD1 (Accession:
CAE11897.1) |
|
|
|
| 1 | 83 | 119 |
VEPKVKSKKREAVAGRGRGRGRGRGRGRGRGRGGPRR |
| 2 | 41 | 57 |
KAVKMTLKNREPVQLET |
| 3 | 12 | 26 | HETVTIELK |
| SMD2 (Accession:
AAC13776.1) |
|
|
|
| 1 |
1 | 19 |
MSLLNKPKSEMTPEELQKR |
| 2 | 112 | 118 | NPLIAGK |
| 3 | 76 | 90 | EVPKSGKGKKKSKPV |
| 4 | 93 | 98 | DRYISK |
| 5 | 22 | 27 | EEFNTG |
| SMD3 (Accession:
AAA57034.1) |
|
|
|
| 1 | 120 | 126 | NIFQKRR |
| 2 | 110 | 117 | RGRGRGMG |
| 3 | 43 | 61 |
MSNITVTYRDGRVAQLEQV |
| 4 | 96 | 108 | GRGKAAILKAQVA |
| 5 | 32 | 39 | LIEAEDNM |
| Proliferating cell
nuclear antigen (PCNA) (Accession: NP_872590.1) |
|
|
|
| 1 | 253 | 261 | PKIEDEEGS |
| 2 | 57 | 67 | FDTYRCDRNLA |
| 3 | 149 | 157 | RDLSHIGDA |
| 4 | 80 | 87 | KCAGNEDI |
| 5 |
1 |
8 | MFEARLVQ |
| Acidic ribosomal
phosphoprotein (P1) (Accession: AAA36471.1) |
|
|
|
| 1 | 18 | 30 | DDEVTVTEDKINA |
| Acidic ribosomal
phosphoprotein (P2) (Accession: AAA36472.1) |
|
|
|
| 1 | 44 | 61 |
SELNGKNIEDVIAQGIGK |
| 2 | 12 | 18 | LGGNSSP |
| snRNP-B/B'
(Accession: P14678.2) |
|
|
|
| 1 |
1 | 11 | MTVGKSSKMLQ |
| 2 | 48 | 66 |
FRKIKPKNSKQAEREEKRV |
| 3 | 90 | 99 | TGIARVPLAG |
| 4 | 34 | 40 | FDKHMNL |
| 5 | 70 | 78 | VLLRGENLV |
| 6 | 223 | 231 | PPPGMRGPP |
| 7 | 22 | 45 |
LQDGRIFIGTFKAFDKHMNLILCD |
| U1-snRNP-C
(Accession: NP_003084.1) |
|
|
|
| 1 | 82 | 91 | SLPGPPRPGM |
| 2 | 69 | 80 | APPPAGAMIPPP |
| 3 | 35 | 47 | KDYYQKWMEEQAQ |
| U1-snRNP-A
(Accession: NP_004587.1) |
|
|
|
| 1 |
1 | 10 | MAVPETRPNH |
| 2 | 60 | 77 |
KEVSSATNALRSMQGFPF |
| 3 | 94 | 104 | IAKMKGTFVER |
| 4 | 80 | 91 | KPMRIQYAKTDS |
| 5 | 94 | 104 | IAKMKGTFVER |
| Nucleolin
(Accession: AAA59954.1) |
|
|
|
| 1 | 118 | 127 | VATPGKKGA |
| 2 | 214 | 233 | TPAKGKKAAK |
| 3 | 315 | 327 | NFNKSAPELKTGI |
| 4 | 331 | 337 | FAKNDLA |
| 5 | 347 | 353 | RKFGYVD |
| 6 | 421 | 430 | LVSKDGKSKG |
| 7 | 514 | 526 | VPQNQNGKSKGYA |
| Acidic ribosomal
phosphoprotein (P0) (Accession: AAA36470.1) |
|
|
|
| 1 |
1 | 13 | MPREDRATWKSNY |
| 2 | 21 | 27 | LDDYPKC |
| 3 | 32 | 41 | ADNVGSKQMQ |
| 4 | 45 | 51 | MSLRGKA |
| 5 | 91 | 100 | TKEDLTEIRD |
| 6 | 125 | 136 | AQNTGLGPEKTS |
| 7 | 146 | 152 | KISRGTI |
| 8 | 162 | 171 | KTGDKVGASE |
| 9 | 202 | 213 | EVLDITEETLH |
| 10 | 215 | 220 | FLEGVR |
| 11 | 243 | 249 | NGYKRVL |
| 12 | 296 | 312 |
AKVEAKEESEESDEDMG |
| DNA topoisomerase1
(truncated) (Accession: NP_003277.1) |
|
|
|
| 1 | 52 | 66 |
YDGKVMKLSPKAEEV |
| 2 | 89 | 103 |
FKDWRKEMTNEEKNI |
| 3 |
1 | 12 | KKKKPKKEEEQK |
| 4 | 128 | 136 | QMSKEEKLK |
| 5 | 415 | 422 | LTAPDENI |
| DNA topoisomerase 1
(full length) (Accession: NP_003277.1) |
|
|
|
| 1 | 459 | 476 |
NQYREDWKSKEMKVRQRA |
| 2 | 674 | 682 | VMKDAKTKK |
| 3 | 491 | 498 | NEKEEGET |
| 4 | 504 | 510 | CCSLRVE |
| 5 | 365 | 376 | GNHPKMGMLKRR |
| U1-SnRNP 68/70 KDa
(Accession: P08621.2) |
|
|
|
| 1 | 424 | 437 | LAPENGYLMEAAPE |
| 2 | 375 | 383 | DREHKRGER |
| 3 | 120 | 134 |
RREFEVYGPIKRIHM |
| 4 | 282 | 294 | KDKDRDRKRRSSR |
| 5 | 303 | 315 | RERKEELRGGGGD |
| 6 | 1 | 8 | MTQFLPPN |
| 7 | 211 | 217 | SGRDDTS |
| 8 | 138 | 145 | KRSGKPRG |
Samples detection by peptide
microarray
The peptide microarray based on the 73 epitopes were
used to test the serum autoantibodies in 120 SLE patients and 110
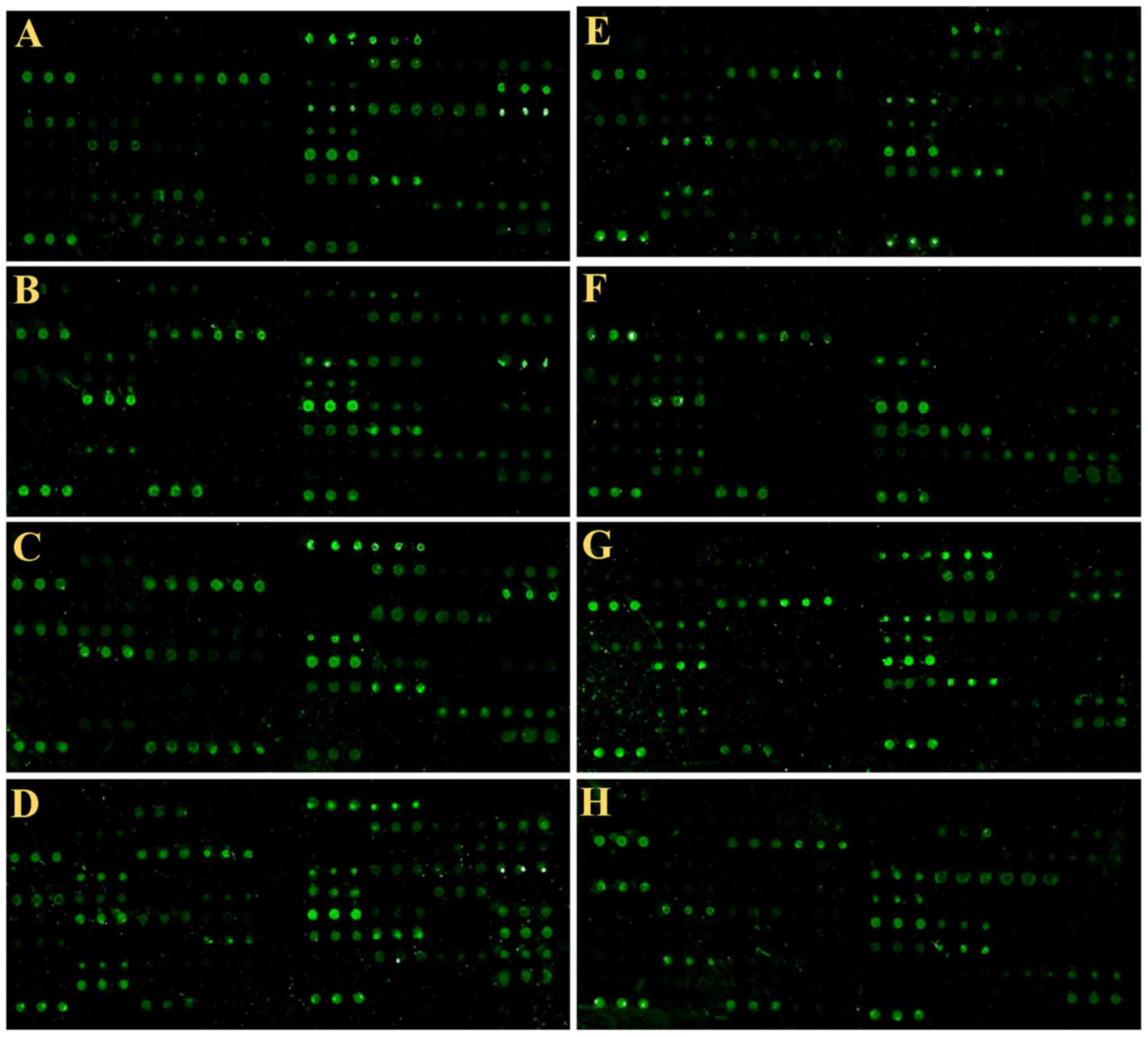
matched healthy subjects (Fig.
1).
The results showed that the autoantibodies that
produced by different individuals recognized and bound different
epitopes. If the signal information larger than the negative
control, the average data plus 10 mines of SD was sued to assess
the positive rate. The positive controls used in the present study
was biotinylated random linear 12-peptide. The negative control was
a random linear 12-peptide. We analyzed the positive rate and false
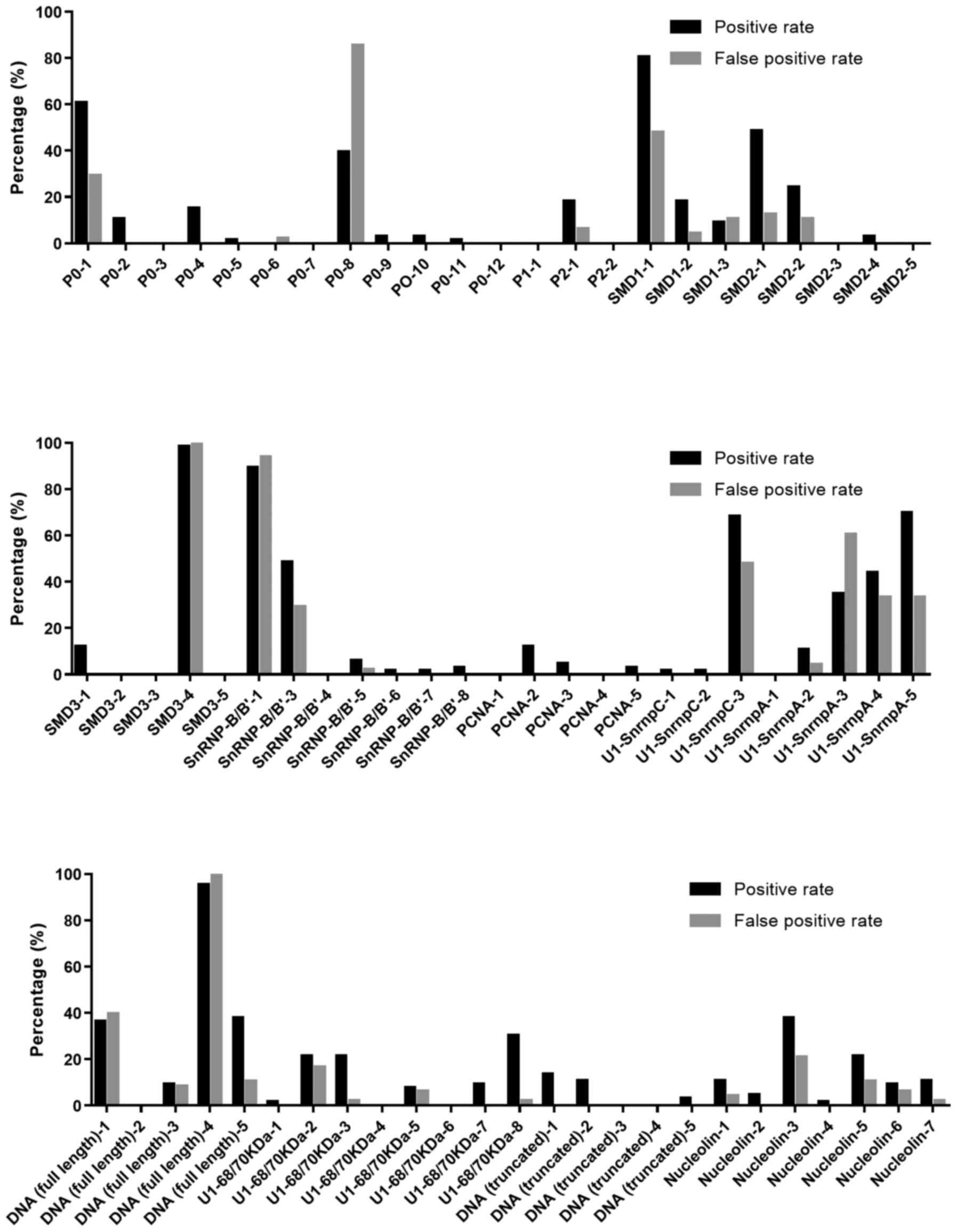
positive rate of each peptide (Fig.
2). It can be seen there were 18 peptides had high positive
rate with low false positive rate, including P0-2, P0-4, P2-1,
SMD1-2, SMD2-1, SMD2-2, SMD3-1, PCNA-2, U1-SnrnpA-2, DNA (full
length)-5, U1-68/70kDa-3, U1-68/70kDa-7, U1-68/70kDa-8, DNA
(truncated)-1, DNA (truncated)-2, Nucleolin-1, Nucleolin-5, and
Nucleolin-7. All the positive rate and false positive rate of the
peptides were no less and no >10%, respectively. The ratio of
positive rate to false positive rate was >5, indicating all the
18 peptides are valuable epitopes.
ROC curve plotting and area
analysis
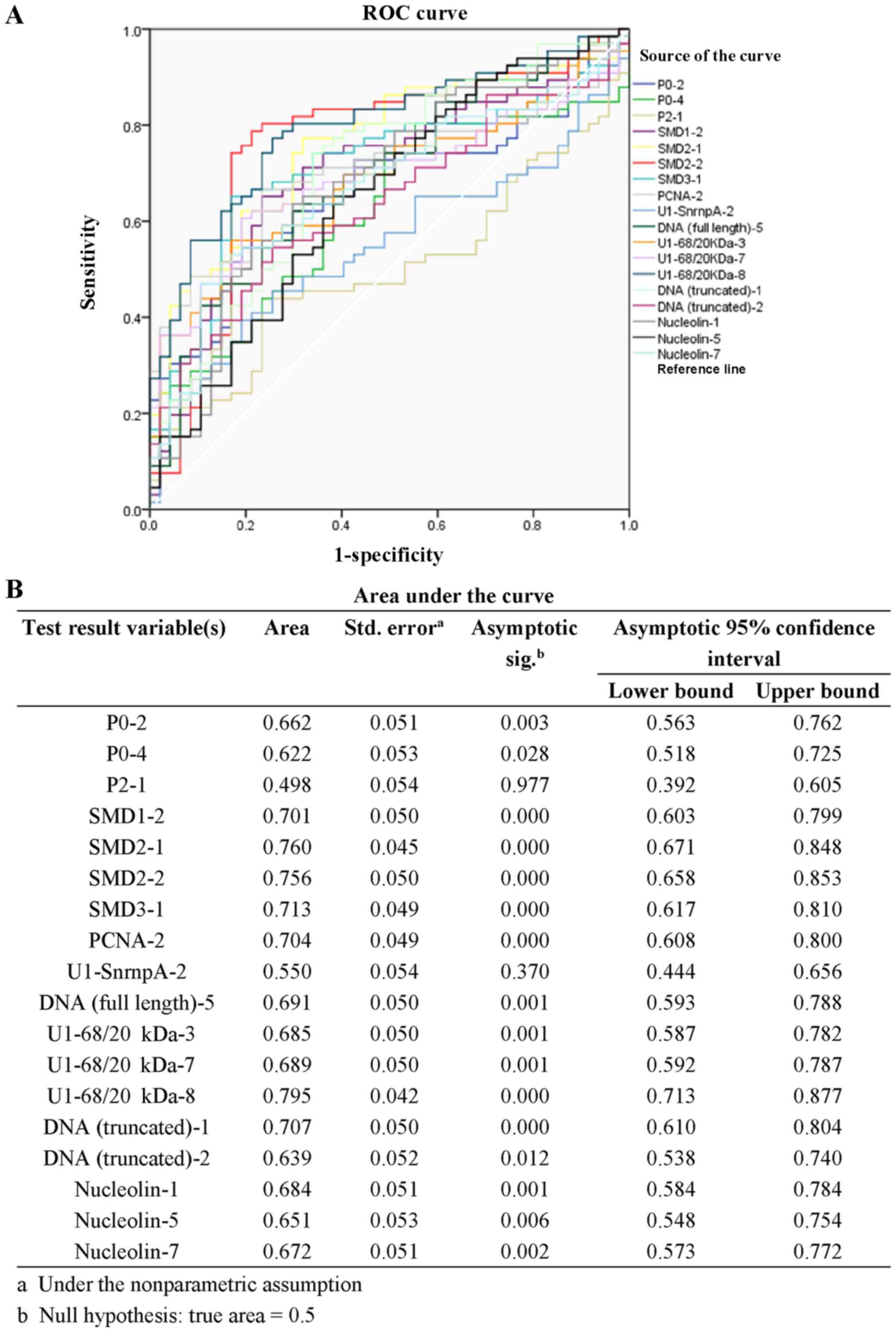
The specificity and 1-specificity of all cut-off
points in data from the 18 screened peptides were calculated and
the ROC curve was plotted (Fig.
3A). The areas under the ROC curves of peptides P0-2, P0-4,
P2-1, SMD1-2, SMD2-1, SMD2-2, SMD3-1, PCNA-2, U1-SnrnpA-2, DNA
(full length)-5, U1-68/20Da-3, U1-68/20kDa-7, U1-68/20kDa-8, DNA
(truncated)-1, DNA (truncated)-2, Nucleolin-1, Nucleolin-5 and
Nucleolin-7 was 0.662±0.051, 0.622±0.053, 0.498±0.054, 0.701±0.050,
0.760±0.045, 0.756±0.050, 0.713±0.049, 0.704±0.049, 0.550±0.054,
0.691±0.050, 0.685±0.050, 0.689±0.050, 0.795±0.042, 0.707±0.050,
0.639±0.052, 0.684±0.051, 0.651±0.053, and 0.672±0.051 (Fig. 3B). And the area under the curves of
14 peptides was >0.65, including P0-2, SMD1-2, SMD2-1, SMD2-2,
SMD3-1, PCNA-2, DNA (full length)-5, U1-68/20kDa-3, U1-68/20kDa-7,
U1-68/20kDa-8, DNA (truncated)-1, Nucleolin-1, Nucleolin-5 and
Nucleolin-7, suggesting they are of significant diagnostic value in
SLE.
Verification of the validity of
epitopes
In order to verify the validity of the 14 epitopes,
we selected the intact antigens of 2 most commonly used SLE
autoantibodies in clinic, including Sm and RNP to detect the sera
of the SLE patients using direct ELSIA kit. The results of ELISA
were compared with those of peptide microarray (for Sm, the
peptides including SMD1-2, SMD2-1, SMD2-2 and SMD3-1 were compared;
for RNP, the peptides including U1-SnrnpA-2 and U1-SnRNP 68/70 kDa
were compared) (Table II). When
one peptide was positive, the results of peptide microarray was set
as positive. The peptide microarray was higher in sensitivity and
lower in specificity than the direct ELISA. In addition, when
detected an expanded sample set including 120 SLE patients and 110
healthy subjects (Table III).
When two peptides in one sample were positive, the sample was
positive. The sensitivity of peptide microarray in the diagnosis of
SLE was 71.6%, and the specificity was 85.5%.
 | Table II.Comparison between ELISA and peptide
microarray. |
Table II.
Comparison between ELISA and peptide
microarray.
|
| Intact
antigens | Peptide
microarray |
|
|---|
|
|
|
|
|
|---|
| Antigens | Sensitivity | Specificity | Sensitivity | Specificity | Related |
|---|
| Sm | 20.1 | 100 | 57.6 | 79.1 | 0.85 |
| RNP | 41.8 | 100 | 51.5 | 91.6 | 0.88 |
 | Table III.Diagnostic results of SLE using 18
peptides. |
Table III.
Diagnostic results of SLE using 18
peptides.
| Detection | SLE | Healthy | Total |
|---|
| Positive
detection | 86 | 16 | 102 |
| Negative
detection | 34 | 94 | 128 |
| Total | 120 | 110 |
|
Discussion
Autoantibodies are an important feature of
autoimmune diseases and are important indicators of disease
diagnosis and progression monitoring. Autoantibodies can be
detected by intact antigens, but the target epitopes by each
autoantibody cannot be accurately identified. In clinic, the
precise identification of autoantibodies and the detection of
combining epitopes is of great significance in accurate diagnosis
and treatment. The present study uses peptide microarray technology
to detect the autoantibodies in peripheral blood of SLE patients.
The findings suggested epitopes recognized by autoantibodies of
individual SLE patients was different. As individual patient has a
specific autoantibody spectrum, detection of autoantibodies by
peptide microarray is useful for diagnosis of SLE.
The use of autoantigens to detect autoantibodies is
an important diagnostic technique for autoimmune diseases. However,
all the current detection methods have limited by poor positive
rates, sensitivity. Although the sensitivity of ANA in SLE
diagnosis is as high as 97–100%, its specificity is only 10–40%.
Moreover, when ANA is negative, it cannot rule out the SLE
completely, suggesting the diagnosis by ANA should take account the
clinical conditions. The specificity of anti-rRNP antibody in SLE
is ~20–30% (4). The specificity of
anti-SSA antibody in neonatal patients with lupus erythematosus is
100% (27,28). The anti-dsDNA antibody are closely
associated with SLE, which shown a high specificity in the
diagnosis of SLE (29). However,
excessive free DNA antigens in the serum will influent the
diagnosis of SLE in some patients since they could combine with
anti-dsDNA antibodies (29,30).
It was also demonstrated that the anti-Sm antibodies,
anti-nucleosome antibody could use in the diagnosis of SLE.
Although the positive rate of anti-Sm antibody for SLE was 98%, its
sensitivity was only 20–30% (3,5,31,32).
In addition, the detection of antibodies by ELISA is not only
inefficient but also costly, which is becoming a major obstacle to
the early diagnosis of SLE.
Compared with the intact antigen, we found that
peptide microarray results were negative when the intact antigen
detection was negative. If the intact antigen test result is
positive, at least one peptide detection was positive. Zhu et
al designed arrays containing synthetic peptides and molecular
modified protein which being utilized for identification of
autoantibodies targeting to special antigenic epitopes (33). Our results showed the sensitivity
and specificity of the combination of 73 peptides in the diagnosis
of SLEs can reach 96.9 and 93.8%, but any single index cannot meet
the clinical needs. Therefore, the combined detection of multiple
indictors by peptide microarray has important value in the
detection of SLE.
In the present study, the linear epitopes of 14
antigens were predicted by antigen epitope prediction software and
an optimal set of epitopes for SLE diagnosis was obtained. Although
the current epitope prediction software could predict the spatial
epitopes, short peptide could not be simulated due to the far away
between amino acid in space epitope. Nowadays, the peptide
microarray could not be used to detect the autoantibody by
identified spatial epitope. This is a flaw in this project. In
addition, we found that the spectrum of autoantibodies varied
considerably among different SLE patients. Therefore, a large
number of samples are needed to accurately calculate the positive
rate of each locus, which is another defect of the present study.
Constructing a map of each Person's epitope and analyzing his
association with disease progression are our future research
direction.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 320.6750.16039) and
President Foundation of Nanfang Hospital, Southern Medical
University (grant no. 2017C033).
References
|
1
|
Groot N, de Graeff N, Avcin T,
Bader-Meunier B, Brogan P, Dolezalova P, Feldman B, Kone-Paut I,
Lahdenne P, Marks SD, et al: European evidence-based
recommendations for diagnosis and treatment of childhood-onset
systemic lupus erythematosus: The SHARE initiative. Ann Rheum Dis.
76:1788–1796. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leal GN, Diniz MF, Brunelli J, Lianza AC,
Sallum AM and Silva CA: What are the benefits of two-dimensional
speckle tracking echocardiography for diagnosis and treatment
follow-up of childhood-onset systemic lupus erythematosus
myocarditis? Rev Assoc Med Bras (1992). 62:490–493. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tunnicliffe DJ, Singh-Grewal D, Kim S,
Craig JC and Tong A: Diagnosis, monitoring, and treatment of
systemic lupus erythematosus: A systematic review of clinical
practice guidelines. Arthritis Care Res. 67:1440–1452. 2015.
View Article : Google Scholar
|
|
4
|
Fu SM, Deshmukh US and Gaskin F:
Pathogenesis of systemic lupus erythematosus revisited 2011: End
organ resistance to damage, autoantibody initiation and
diversification and HLA-DR. J Autoimmun. 37:104–112. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kuhn A, Bonsmann G, Anders HJ, Herzer P,
Tenbrock K and Schneider M: The diagnosis and treatment of systemic
lupus erythematosus. Dtsch Arztebl Int. 112:423–432.
2015.PubMed/NCBI
|
|
6
|
Zhao J, Wang K, Wang X, Li T, Guo L, Gu L,
Chen Z, Sun F, Wang H, Li J, et al: The performance of different
anti-dsDNA autoantibodies assays in Chinese systemic lupus
erythematosus patients. Clin Rheumatol. 37:139–144. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ruzieh M, Batizy L, Dasa O, Oostra C and
Grubb B: The role of autoantibodies in the syndromes of orthostatic
intolerance: A systematic review. Scand Cardiovasc J. 51:243–247.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma WT, Chang C, Gershwin ME and Lian ZX:
Development of autoantibodies precedes clinical manifestations of
autoimmune diseases: A comprehensive review. J Autoimmun.
83:95–112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu C, Li M, Liu J, Qian J, Xu D, Zhang S,
Li P, Zhao J, Tian X and Zeng X: Anti-SmD1 antibodies are
associated with renal disorder, seizures, and pulmonary arterial
hypertension in Chinese patients with active SLE. Sci Rep.
7:76172017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bai Y, Tong Y, Liu Y and Hu H: Self-dsDNA
in the pathogenesis of systemic lupus erythematosus. Clin Exp
Immunol. 191:1–10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Arif Z, Neelofar K, Tarannum I, Arfat MY,
Ahmad S, Zaman A, Khan MA, Badar A, Islam SN2 and Iqubal MA: SLE
autoantibodies are well recognized by peroxynitrite-modified-HSA:
Its implications in the pathogenesis of SLE. Int J Biol Macromol.
106:1240–1249. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Agmon-Levin N, Dagan A, Peri Y, Anaya JM,
Selmi C, Tincani A, Bizzaro N, Stojanovich L, Damoiseaux J, Cohen
Tervaert JW, et al: The interaction between anti-Ro/SSA and
anti-La/SSB autoantibodies and anti-infectious antibodies in a wide
spectrum of auto-immune diseases: Another angle of the autoimmune
mosaic. Clin Exp Rheumatol. 35:929–935. 2017.PubMed/NCBI
|
|
13
|
Zhang B, Jarrell JA, Price JV, Tabakman
SM, Li Y, Gong M, Hong G, Feng J, Utz PJ and Dai H: An integrated
peptide-antigen microarray on plasmonic gold films for sensitive
human antibody profiling. PLoS One. 8:e710432013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Haddon DJ, Diep VK, Price JV, Limb C, Utz
PJ and Balboni I: Autoantigen microarrays reveal autoantibodies
associated with proliferative nephritis and active disease in
pediatric systemic lupus erythematosus. Arthritis Res Ther.
17:1622015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Price JV, Tangsombatvisit S, Xu G, Yu J,
Levy D, Baechler EC, Gozani O, Varma M, Utz PJ and Liu CL: On
silico peptide microarrays for high-resolution mapping of antibody
epitopes and diverse protein-protein interactions. Nat Med.
18:1434–1440. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Price JV, Haddon DJ, Kemmer D, Delepine G,
Mandelbaum G, Jarrell JA, Gupta R, Balboni I, Chakravarty EF,
Sokolove J, et al: Protein microarray analysis reveals BAFF-binding
autoantibodies in systemic lupus erythematosus. J Clin Invest.
123:5135–5145. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Balboni I, Niewold TB, Morgan G, Limb C,
Eloranta ML, Rönnblom L, Utz PJ and Pachman LM: Interferon-α
induction and detection of anti-ro, anti-la, anti-sm, and anti-rnp
autoantibodies by autoantigen microarray analysis in juvenile
dermatomyositis. Arthritis Rheum. 65:2424–2429. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gori A, Cretich M, Vanna R, Sola L, Gagni
P, Bruni G, Liprino M, Gramatica F, Burastero S and Chiari M:
Multiple epitope presentation and surface density control enabled
by chemoselective immobilization lead to enhanced performance in
IgE-binding fingerprinting on peptide microarrays. Anal Chim Acta.
983:189–197. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
De Plano LM, Carnazza S, Messina GML,
Rizzo MG, Marletta G and Guglielmino SPP: Specific and selective
probes for Staphylococcus aureus from phage-displayed random
peptide libraries. Colloids Surf B Biointerfaces. 157:473–480.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kafi MA, Cho HY and Choi JW: Engineered
peptide-based nanobiomaterials for electrochemical cell chip. Nano
Converg. 3:172016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maksimov P, Zerweck J, Maksimov A, Hotop
A, Gross U, Spekker K, Däubener W, Werdermann S, Niederstrasser O,
Petri E, et al: Analysis of clonal type-specific antibody reactions
in Toxoplasma gondii seropositive humans from Germany by
peptide-microarray. PLoS One. 7:e342122012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhong L, Coe SP, Stromberg AJ, Khattar NH,
Jett JR and Hirschowitz EA: Profiling tumor-associated antibodies
for early detection of non-small cell lung cancer. J Thorac Oncol.
1:513–519. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Woodard KM and Chapman CJ: Lung cancer-can
autoantibodies provide an aid to diagnosis? Expert Opin Med Diagn.
2:911–923. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chapman CJ, Murray A, McElveen JE, Sahin
U, Luxemburger U, Türeci O, Wiewrodt R, Barnes AC and Robertson JF:
Autoantibodies in lung cancer: Possibilities for early detection
and subsequent cure. Thorax. 63:228–233. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saleh J, Brunner C, Gölzer R, Nastainczyk
W and Montenarh M: p53 autoantibodies from patients with head and
neck cancer recognise common epitopes on the polypeptide chain of
p53. Cancer Lett. 233:48–56. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saleh J, Kreissler-Haag D and Montenarh M:
p53 autoantibodies from patients with colorectal cancer recognize
common epitopes in the N- or C-terminus of p53. Int J Oncol.
25:1149–1155. 2004.PubMed/NCBI
|
|
27
|
de Jesus GR, Mendoza-Pinto C, de Jesus NR,
Dos Santos FC, Klumb EM, Carrasco MG and Levy RA: Understanding and
managing pregnancy in patients with lupus. Autoimmune Dis.
2015:9434902015.PubMed/NCBI
|
|
28
|
Marder W, Ganser MA, Romero V, Hyzy MA,
Gordon C, McCune WJ and Somers EC: In utero azathioprine exposure
and increased utilization of special educational services in
children born to mothers with systemic lupus erythematosus.
Arthritis Care Res. 65:759–766. 2013. View Article : Google Scholar
|
|
29
|
Crampton SP, Morawski PA and Bolland S:
Linking susceptibility genes and pathogenesis mechanisms using
mouse models of systemic lupus erythematosus. Dis Model Mech.
7:1033–1046. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gottschalk TA, Tsantikos E and Hibbs ML:
Pathogenic inflammation and its therapeutic targeting in systemic
lupus erythematosus. Front Immunol. 6:5502015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ramos-Casals M, Sanz I, Bosch X, Stone JH
and Khamashta MA: B-cell-depleting therapy in systemic lupus
erythematosus. Am J Med. 125:327–336. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ching KH, Burbelo PD, Tipton C, Wei C,
Petri M, Sanz I and Iadarola MJ: Two major autoantibody clusters in
systemic lupus erythematosus. PLoS One. 7:e320012012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu H, Luo H, Yan M, Zuo X and Li QZ:
Autoantigen microarray for high-throughput autoantibody profiling
in systemic lupus erythematosus. Genomics Proteomics
Bioinformatics. 13:210–218. 2015. View Article : Google Scholar : PubMed/NCBI
|