Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a disease
with a poor prognosis. With increasing incidence and mortality,
PDAC is the 6th most common cause of cancer-associated death in
China (1). Despite advances in
surgical and oncological treatment strategies, PDAC exhibits an
extremely poor prognosis, with a 6-month median survival period and
≤1–2% 5-year survival rate (2,3).
The exact molecular mechanism of PDAC remains
undetermined. Pancreatic intraepithelial neoplasia (PanIN) in the
pancreatic duct has been proposed to be the primordial precursor of
PDAC (4). As PanIN progresses to
carcinoma, accumulated mutations may result in the loss of
cyclin-dependent kinase inhibitor 2A/p16 and/or the inactivation of
cellular tumor antigen 53 and mothers against decapentaplegic
homolog 4, in addition to the activation of the KRAS proto-oncogene
(5). Unlike other adenocarcinomas,
the usual composition of pancreatic cancer tumor is a large mass of
fibroinflammatory tissue, which is interspersed with islands of
neoplastic epithelia (6,7). Previous studies have suggested that
pancreatic tumor stroma directly affect the progression and outcome
of disease via the release of factors, including insulin-like
growth factor and platelet-derived growth factor, into the tumor
microenvironment that aid tumor growth and invasiveness (8–10).
However, the association between differentially expressed genes and
tumor cells remains poorly understood, which has limited the
development of effective treatments for patients with PDAC.
The Cancer Genome Atlas (https://cancergenome.nih.gov/) may further the
understanding of the molecular basis of cancer via genome analysis,
including high-throughput genome sequencing. In the present study,
combined analysis using genomic and transcriptomic data from two
PDAC studies was performed and a survival-associated biomarker was
revealed, transient receptor potential cation channel subfamily M
member 2 (TRPM2). Furthermore, the role of TRPM2 in pancreatic
adenocarcinoma cell lines was investigated using an in vitro
gene silencing method. The present study provides a comprehensive
investigation of TRPM2, a promising biomarker for PDAC prognosis,
and investigates how TRPM2 affects tumor progression and
invasion.
Materials and methods
Data analysis
Clinical information, somatic mutation and gene
expression data of patients with pancreatic cancer were obtained
from two previously published cancer genome studies (11,12).
The somatic mutation and clinical information from 483 patients
with pancreatic cancer was obtained from data published by Bailey
et al (11) and Waddell
et al (12). The survival
time of 159 patients of the total 483 patients was available, with
survival status indicated as ‘alive’, ‘deceased’ or ‘loss’ at the
final follow-up. Gene expression data of 96 patients were obtained
from the Gene Expression Omnibus (13) (dataset, GSE36924) (11), while 96 patients have survival
information. The samples with follow-up information were used for
survival analysis.
Survival analysis of genes mutated in ≥10 (out of
159) patients was performed. Patients were divided into two groups
according to the mutation status of each gene. The difference in
overall survival between the two groups was estimated using a
Kaplan-Meier curve and tested for significance using a log-rank
test. A gene was considered to be associated with patient clinical
outcomes when a P<0.05 value was obtained. Following this,
patients were additionally divided into two groups, based on
mutation and gene expression levels, according to the median
expression values of each gene. Survival analysis was performed
using gene-expression data obtained from the Gene Expression
Omnibus (dataset, GSE36924) (11).
It was identified that somatic mutation and gene expression of
TRPM2 were significantly associated with the patient's overall
survival (Tables I and II). Thus, further experimental assays
were performed to investigate the role of TRPM2 in pancreatic
cancer.
 | Table I.Survival analysis of genes associated
with the patient's overall survival. |
Table I.
Survival analysis of genes associated
with the patient's overall survival.
| Gene | Mutation
P-value | Expression
P-value |
|---|
| TRPM2 | 0.0104 | 0.0111 |
| COL18A1 | 0.0384 | 0.0138 |
| THSD7B | 0.144 | 0.166 |
| FRAS1 | 0.0259 | 0.395 |
| PTPRT | 0.00367 | 0.426 |
| SCN5A | 0.102 | 0.886 |
| PXDN | 0.0301 | 0.903 |
 | Table II.Somatic mutations of transient
receptor potential cation channel subfamily M member 2 in
patients. |
Table II.
Somatic mutations of transient
receptor potential cation channel subfamily M member 2 in
patients.
| Hugo symbol | Chromosome | Start position | End position | Variant
classification | Reference
allele | Tumor seq
allele1 | Tumor seq
allele2 | Tumor sample
barcode | Canonical base
change |
|---|
| TRPM2 | 21 | 45820197 | 45820197 | Missense
mutation | G | A | G | ICGC_0021 | 2477G/A |
| TRPM2 | 21 | 45833788 | 45833788 | Missense
mutation | C | A | C | ICGC_0054 | 3190C>A |
| TRPM2 | 21 | 45845642 | 45845642 | Missense
mutation | C | C | T | ICGC_0114 | 3940C>T |
| TRPM2 | 21 | 45789057 | 45789057 | Splice site | C | C | T | ICGC_0121 | NA |
| TRPM2 | 21 | 45810792 | 45810792 | Missense
mutation | C | C | T | ICGC_0242 | 1537C>T |
| TRPM2 | 21 | 45811433 | 45811433 | Silent | G | A | G | ICGC_0315 | 1932G>A |
| TRPM2 | 21 | 45811283 | 45811283 | Silent | C | C | T | ICGC_0326 | 1782C>T |
It was hypothesized that genes involved in a similar
biological process may exhibit similar expression patterns. Gene
expression correlation analysis was performed to reveal genes
functionally associated with TRPM2 in patients with pancreatic
cancer. Pearson's correlation coefficient between TRPM2 and each of
the genes of interest was calculated. Following this, the top genes
with a correlation coefficient of >0.50 were retained (Table III).
 | Table III.Gene expression correlation analysis
with transient receptor potential cation channel subfamily M member
2. |
Table III.
Gene expression correlation analysis
with transient receptor potential cation channel subfamily M member
2.
| Ensembl | Gene | Correlation
coefficient |
|---|
|
ENSG00000104043 | ATP8B4 | 0.574108 |
|
ENSG00000156414 | TDRD9 | 0.526303 |
|
ENSG00000198879 | SFMBT2 | 0.520375 |
|
ENSG00000138964 | PARVG | 0.511799 |
|
ENSG00000196664 | TLR7 | 0.503315 |
TRPM2 knockdown and
overexpression
PANC-1 cells (American Type Culture Collection,
Manassas, VA, USA) were seeded in 6-well plates until 80%
confluency was reached following 24 h of culture at 37°C.
Hilymax-Trpm2 siRNA complex (20 pmol/l), Hilymax-Trpm2 OverExp
vector (4 µg/well), empty vector and scramble siRNA (Generay
Biotech Co., Ltd., Shanghai, China) were added to the cell culture,
using the transfection agent Hilymax (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) well as per the manufacturer's
instructions, and incubated at 37°C in a 5% CO2 incubator for 4 h.
The medium was subsequently replaced with fresh medium (RPMI 1640;
10% fetal bovine serum, Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and incubated for a further 48 h. The
untransfected group was subjected to normal culture without
transfection reagent, the empty vector represented the control for
TRPM2 overexpression, and scramble siRNA represented the control
for TRPM2 silencing. Cells were prepared for the subsequent reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
Transwell assays. The Trpm2 siRNA and Trpm2 OverExp plasmid were
designed and synthesized by Generay Biotech Co., Ltd. (Shanghai,
China; Table IV).
 | Table IV.Primer and siRNA information. |
Table IV.
Primer and siRNA information.
| Primer | Sequence
(5′-3′) | Amplicon length,
base pairs |
|---|
| Homo-TRPM2-F |
ATTGTGAAGCGGATGATGAAGGA | 158 |
| Homo-TRPM2-R |
ATGGTGAGGTAGGAGTGGTAGAC | 158 |
| Homo-GAPDH-F |
TGGACCTGACCTGCCGTCTA | 149 |
| Homo-GAPDH-R |
GGAGTGGGTGTCGCTGTTGA | 149 |
| TRPM2-siRNA
sense |
GAAAGAAUGCGUGUAUUUUTT | N/A |
| TRPM2-siRNA
antisense |
AAAAUACACGCAUUCUUUCTT | N/A |
| Scramble siRNA
sense |
UUCUUCGAACGUGUCACGUTT | N/A |
| Scramble siRNA
antisense |
ACGUGACACGUUCGGAGAATT | N/A |
RNA extraction and RT-qPCR
Total RNA from cells was extracted using Cell
Culture and Tissue Total RNA Extraction and Preparation Mini kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The quantity and quality of RNA were confirmed using
a NanoDrop 1000. The primers were designed using Primer Premier 5.0
software (Premier Biosoft International, Palo Alto, CA, USA) and
synthesized by Generay Biotech Co., Ltd. (Table IV). RT-qPCR was performed using
the KAPA SYBR Green Supermix PCR kit with the AriaMx Real-Time PCR
System (both Agilent Technologies, Inc., Santa Clara, CA, USA). RT
was performed at 50°C for 30 min. qPCR conditions: Denaturing at
95°C, 10 min, then 40 cycles of denaturing at 95°C for 15 sec,
annealing at 60°C for 1 min, and elongation at 95°C for 15 sec and
60°C for 15 sec. The relative expression levels among the different
genes were determined using the 2−ΔΔCq method (14).
Cell proliferation assay
PANC-1 cell proliferation was investigated using the
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.).
PANC-1 cells were seeded onto a 96-well microplate at a density of
3×104 cells/well, and the cells were transfected with either
Hilymax-Trpm2 OverExp vector (0.2 µg/well), Hilymax-Trpm2 siRNA (20
pmol/l), empty vector (0.2 µg/well) and scramble siRNA (20 pmol/l).
The cells were subsequently cultured for 0, 24 and 48 h. Following
this, 5 µl CCK-8 solution was added to each well and incubated at
37°C for a further 2 h. Optical density was determined at 450 nm
using a microplate spectrophotometer (BioTek Instruments, Inc.,
Winooski, VT, USA).
Scratch wound-healing assay
PANC-1 cells were seeded in 24-well plates at a
density of 1×105 cells per well and incubated overnight at 37°C.
Following this, the cells were transfected with Hilymax-Trpm2
OverExp vector (1 µg/well), Hilymax-Trpm2 siRNA (20 pmol/l), empty
vector and scramble siRNA. The cells were subsequently cultured for
24 h. The cells at the bottom of wells were then scratched using a
10 µl tip and the floating cells were gently washed twice using
RPMI 1640 medium. Images of each well were captured using a
microscope (magnification, ×100) at 0 and 24 h time intervals
post-injury (Olympus X51; Olympus Corporation, Tokyo, Japan).
Transwell assay
PANC-1 cells were transfected with TRPM2 Vector,
TRPM2 siRNA, empty vector and scramble siRNA. A total of 24 h
post-transfection, the cells were seeded into Matrigel-plated upper
wells with RPMI 1640 medium without serum (5×104 cells/well), and
the lower wells contained 500 µl complete medium (RPMI 1640 and 10%
fetal bovine serum), following a routine procedure. Following
incubation for 48 h at 37°C, cells that did not migrate through the
pores were gently removed with a cotton swab. Cells on the lower
side of the insert filter were fixed by 5% glutaraldehyde for 10
min and stained with 1% crystal violet in 2% ethanol at room
temperature for 20 min. Numbers of cells on the underside of the
filter from five randomly selected microscopic views
(magnification, ×100) were counted.
Statistical analysis
The difference of overall survival between the two
groups was estimated using Kaplan-Meier curve and tested for
significance using log-rank test by SPSS 17.0 software (SPSS, Inc.,
Chicago, IL, USA). GraphPad Prism 5 (GraphPad Software, USA) was
used to perform statistical analysis for the results of RT-qPCR,
CCK-8, wound healing and Transwell assays. Significance between
groups was evaluated by one way analysis of variance followed by a
Bonferroni post hoc test. P<0.05 was considered statistically
significant.
Results
TRPM2 is significantly correlated with
patient survival
Following the data analysis of genes using the
Kaplan-Meier curve and the log-rank test, TRPM2 was selected as a
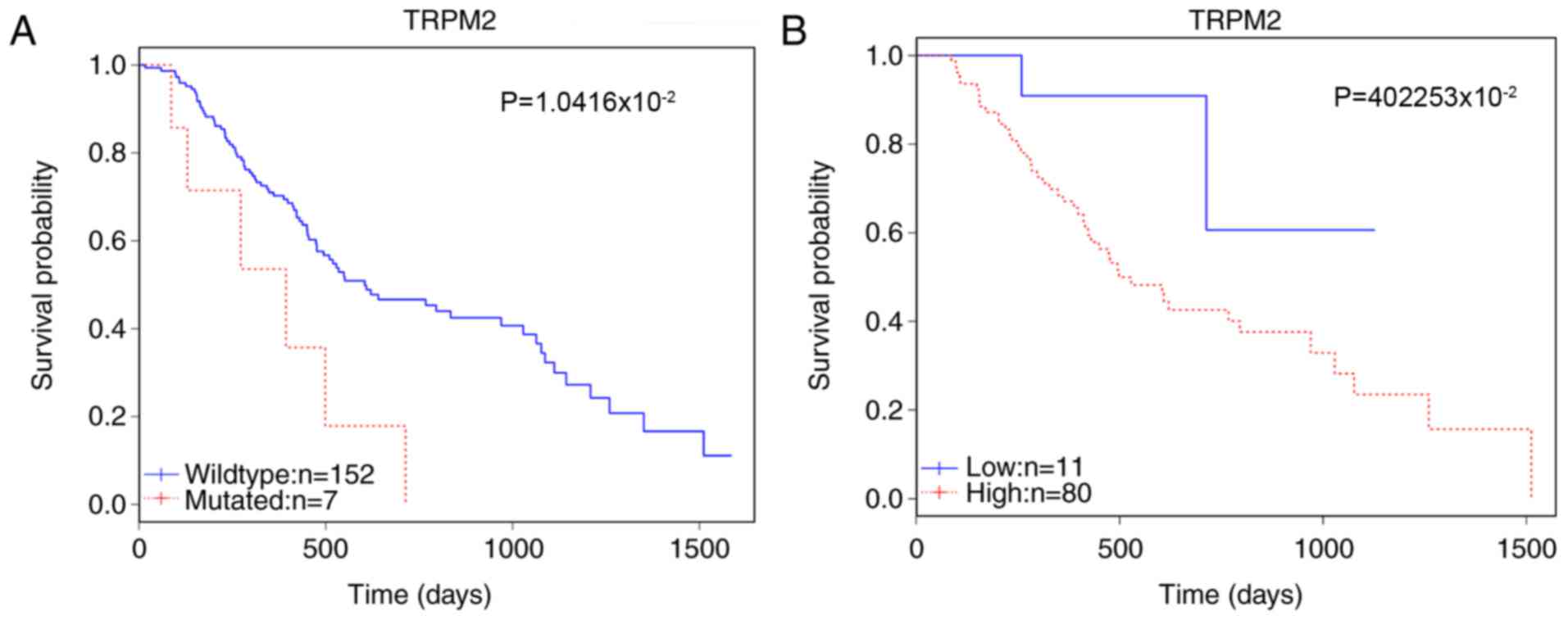
potential candidate marker of prognosis. The survival analysis
revealed that in 159 patients, the mutation status of TRPM2 was
significantly correlated with patient survival (P=1.0416×10-2;
Table I). The somatic mutations of
TRPM2 in this cohort are detailed in Table II. Furthermore, the enhanced
expression level affected survival among 91 patients
(P=4.2253×10-2; Fig. 1). Gene
expression correlation analysis revealed that TRPM2 was strongly
correlated with the expression of probable
phospholipid-transporting ATPase IM (ATP8B4), γ-parvin (PARVG),
tudor domain containing 9 (TDRD9), Toll-like receptor (TLR7) and
Scm-like with four MBT domains protein 2 (SFMBT2) (Table III).
TRPM2 is successfully overexpressed in
the overexpression group and suppressed in the siRNA group
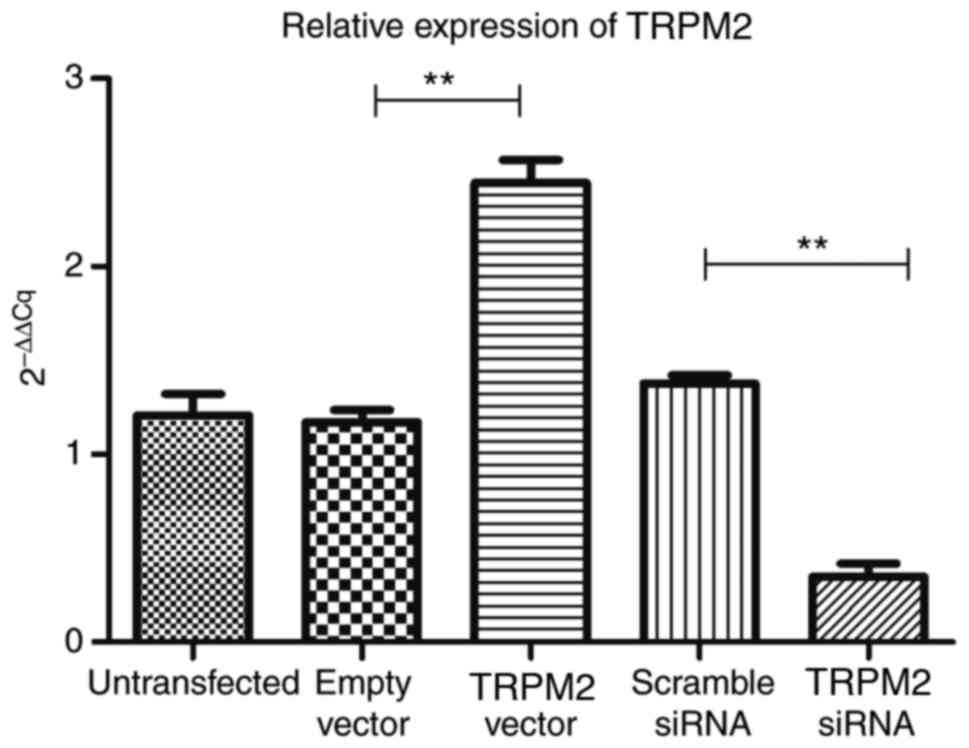
RT-qPCR analysis demonstrated that compared with the
empty vector group, TRPM2 was successfully overexpressed in the
Hilymax-TRPM2 OverExp group (P<0.05; Fig. 2). In addition, the expression level
of TRPM2 was successfully knocked down in the Hilymax-TRPM2 siRNA
group compared with the scramble siRNA group (P<0.05; Fig. 2).
Overexpression of TRPM2 enhances the
proliferation of PANC-1 cells
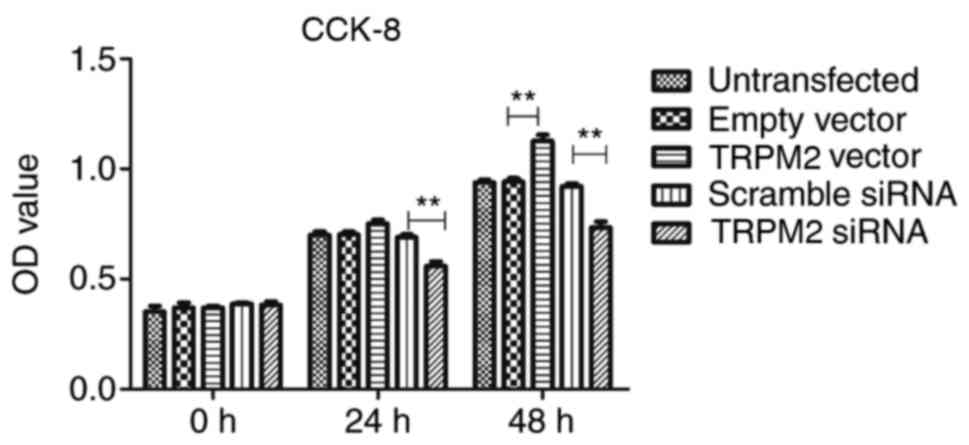
The results of the CCK-8 assay demonstrated that
TRPM2 successfully enhanced PANC-1 cell proliferation in the
Hilymax-Trpm2 OverExp group compared with the empty vector group
(P<0.05; Fig. 3) at the 48-h
time interval, whereas the Hilymax-Trpm2 siRNA group exhibited an
inhibitory effect compared with the scramble siRNA group
(P<0.05; Fig. 3). There were no
significant differences between the vector group and the empty
vector group at the 24 h time interval, however there was a
significant difference between the TRPM2 siRNA group compared with
the scramble siRNA group (Fig.
3).
Overexpression of TRPM2 enhances the
migratory ability of PANC-1 cells
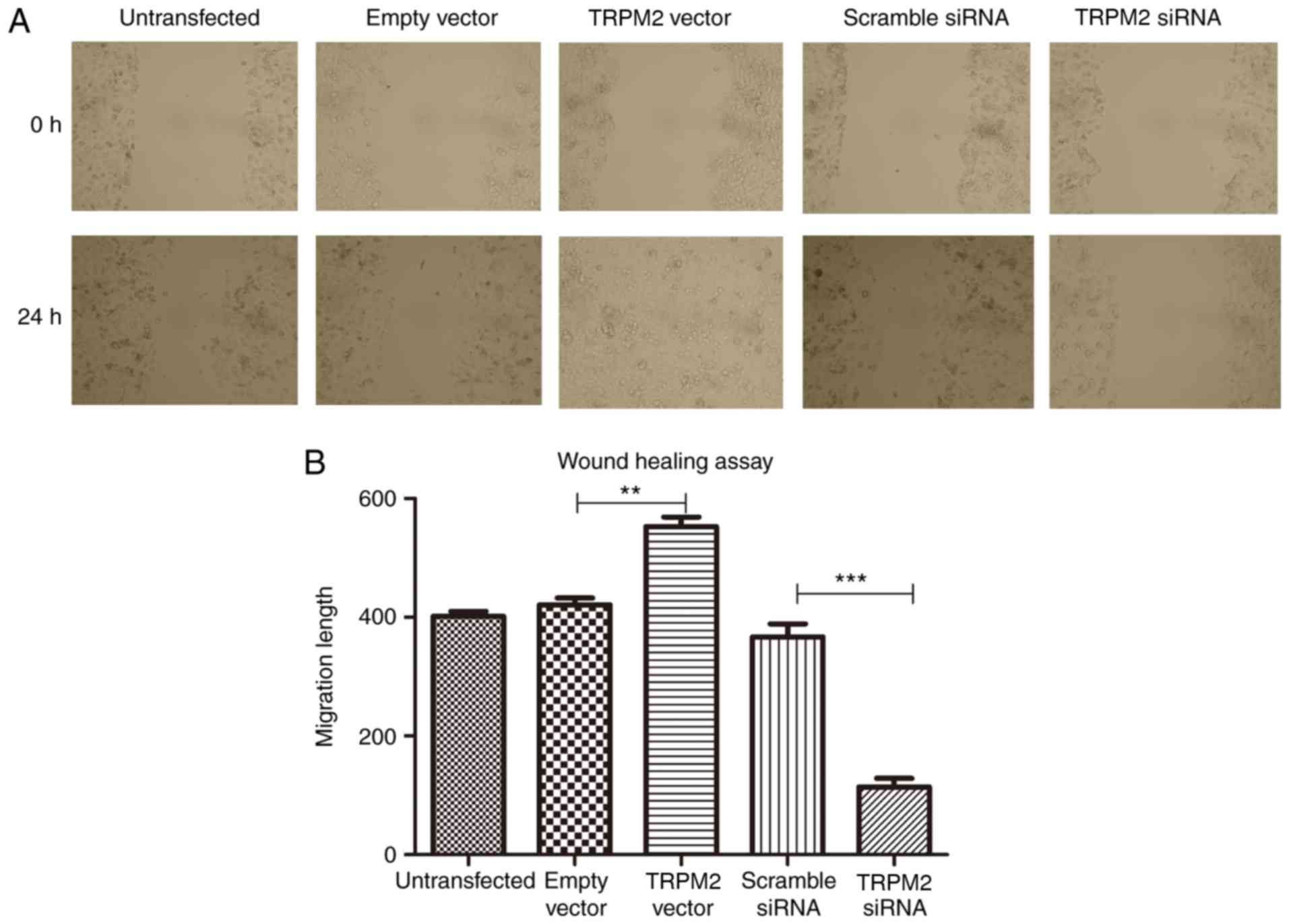
The results of the scratch wound-healing assay
revealed that the migratory ability of PANC-1 cells was
significantly enhanced following overexpression of TRPM2
(P<0.01; Fig. 4), however, this
was significantly suppressed in the RNA interfering group
(P<0.001; Fig. 4).
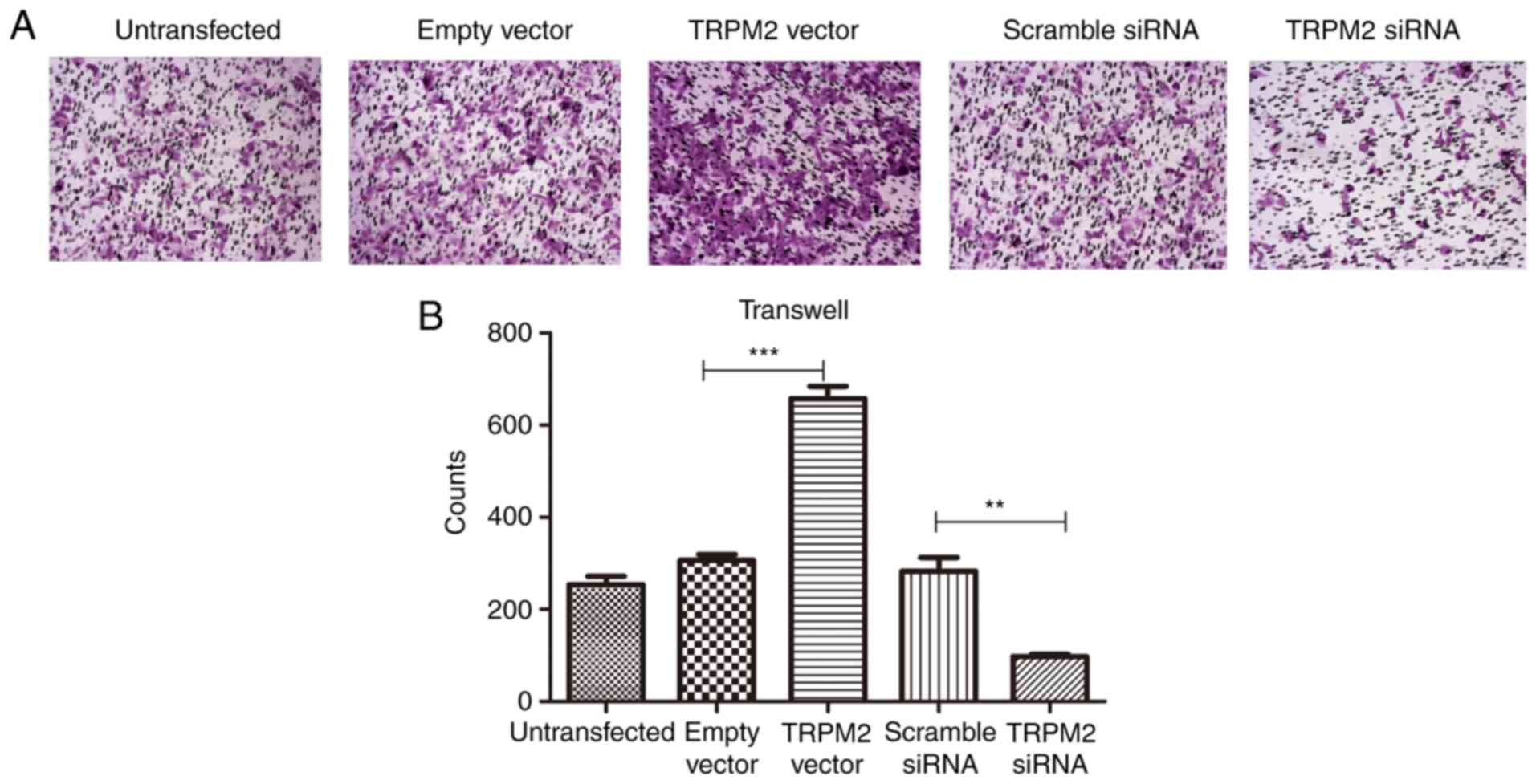
Overexpression of TRPM2 enhances the
invasive ability of PANC-1 cells
Invasive ability was determined via Transwell assays
using Matrigel, and the results demonstrated a significantly
enhanced invasive ability in the OverExp group compared with the
empty vector group (P<0.01; Fig.
5). The siRNA group exhibited fewer invaded cells in the lower
chamber of the microwell (P<0.01; Fig. 5).
Discussion
The TRPM2 gene encodes a tetrameric cation channel
that is permeable to sodium, calcium and potassium, and is
activated by free intracellular adenosine 5′-diphosphate
(ADP)-ribose (15,16). The encoded protein, also activated
by oxidative stress, may induce susceptibility to cell death
(17,18). According to previous studies, TRPM2
may induce apoptosis in different types of cancer (19,20).
However, Huang et al (21)
reported opposite effects of TRPM2 in pancreatic cancers.
According to the data analysis performed in the
present study, it was revealed that the mutated TRPM2 gene exhibits
a marked negative correlation with patient survival rate compared
with the normal control group. The higher TRPM2 is expressed in
cancerous tissue, the shorter the survival time exhibited by PDAC
patients. The results of the in vitro analyses in the
present study revealed that the overexpression of TRPM2 enhanced
cell proliferation and invasive ability, and these findings are
consistent with the results of the data analysis. Therefore, it may
be suggested that TRPM2 expression levels are markedly associated
with proliferation, invasive ability and poor prognosis in patients
with PDAC. Thus, TRPM2 represents a potential therapeutic target
and prognostic marker for the treatment of patients with PDAC.
The results of the gene expression correlation
analysis demonstrated that TRPM2 is strongly correlated with the
expression levels of ATP8B4, PARVG, TDRD9, TLR7 and SFMBT2. Among
these genes, TLR7 has previously been suggested to be associated
with the progression of pancreatic cancer (22–25).
TLRs are expressed in pancreatic cancer cell lines and numerous
human cancer cell lines; however, TLRs are not expressed in the
healthy pancreas (26). The
expression levels of phosphorylated-extracellular signal-regulated
kinase (12) 1/2 are upregulated
following TLR7 activation (20),
suggesting that the activation of TLR7 is at least partially
associated with the mitogen-activated protein kinase-ERK1/2 pathway
in BxPC-3 pancreatic cancer cells (22). Furthermore, Grimmig et al
(23) revealed that chronic
inflammation-mediated TLR7/TLR8 signaling may result in pancreatic
cancer cell growth and chemoresistance.
The ATP8B4 gene can encode a P4-ATPase flippase
complex, a member of the cation transport ATPase (P-type) family
and type IV subfamily (27). Ni
et al (27) suggested that
ATP8B4 may be a potential prognostic marker and a therapeutic
target in the treatment of patients with multiple myeloma who are
B-Cell chronic lymphocytic lymphoma/small lymphocytic Lymphoma
(BCL)-1/JHt(11;14)(q13;q32) translocation positive.
PARVG, a member of the parvin protein family, is an
actin-binding protein associated with focal adhesion (28). Chen et al (29) revealed that four DNA methylation
signatures (phospholipase C β2, Rac family small GTPase 2, vav
guanine nucleotide exchange factor 1 and PARVG) are strongly
associated with the prognosis of renal clear cell carcinoma.
TDRD9, a protein coding gene, is associated with
pathways including mitotic prophase and PIWI-interacting RNA
biogenesis (30–32). At present, there is no evidence to
suggest that TDRD9 has an association with cancer.
Mammalian SFMBTs have been suggested to be polycomb
group repressors (33). Lee et
al (33) demonstrated that
transcriptional repression was associated with human SFMBT2, which
binds preferentially to methylated histone H3 and H4. In addition,
it has been revealed that SFMBT2 regulates cell proliferation via
epigenetic regulation of homeobox B13 gene expression in the DU145
prostate cancer cell line (34).
Further research is required to investigate the
association between TRPM2 and these five genes in the mechanism of
promoting pancreatic cancer.
In addition, Li et al (35) demonstrated that H2O2-mediated
activation of the TRPM2 pathway enhanced the migratory ability of
HeLa and prostate cancer cells by inducing filopodia formation,
inducing the degradation of actin fibers and inducing the
decomposition of focal adhesions. Bauer et al (36) reported that the expression of
ADP-ribose, a calcium channel activator, was increased by
NAD-dependent protein deacetylase sirtuin-6 (SIRT6), which in turn
enhanced the expression of interleukin 8 and tumor necrosis factor
pro-inflammatory cytokines and promoted pancreatic cancer cell
migration. Thus, TRPM2 may promote PDAC metastasis via the
aforementioned mechanisms, however, further investigation is
required to determine the underlying molecular mechanisms.
The results of the survival analyses regarding TRPM2
mutations suggest a poor prognosis for the TRPM2 mutation group
compared with in the wild type group. The results of the present
study have demonstrated that overexpression of TRPM2 promotes
PANC-1 cell growth, migration and invasion ability. Furthermore,
the results of the present study hypothesize that TRPM2 may mediate
cell proliferation via regulation of TLR7 and SFMBT2. In addition,
TRPM2 may be associated with the promotion of invasion and
migration via the regulation of PARVG and SIRT6, or by inducing
filopodia formation, however, the mechanism underlying this process
remains to be determined.
Acknowledgements
We would like to thank Professor Wei Zuo for
generously providing us with lab space and facilities, Dr. Yu Ma,
Timing Zhen and Sibo Zhu for technical assistance, and for
assisting with manuscript preparation.
Funding
The present study was supported by Core Department
Supporting Funding from Tongji Hospital, Tongji University (Tonji,
China).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BMS designed the research. RL and YFW conducted the
research. QNC and ZYL helped collect and interpret the data. SX and
BYW helped culturing cells, collecting reference articles and
writing the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lin QJ, Yang F, Jin C and Fu DL: Current
status and progress of pancreatic cancer in China. World J
Gastroenterol. 21:7988–8003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Warshaw AL and Fernández-del Castillo C:
Pancreatic carcinoma. N Engl J Med. 326:455–465. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li D, Xie K, Wolff R and Abbruzzese JL:
Pancreatic cancer. Lancet. 363:1049–1057. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hruban RH, Maitra A and Goggins M: Update
on pancreatic intraepithelial neoplasia. Int J Clin Exp Pathol.
1:306–316. 2008.PubMed/NCBI
|
|
5
|
Feldmann G, Beaty R, Hruban RH and Maitra
A: Molecular genetics of pancreatic intraepithelial neoplasia. J
Hepatobiliary Pancreat Surg. 14:224–232. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Korc M: Pancreatic cancer-associated
stroma production. Am J Surg. 194 4 Suppl:S84–S86. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Miyamoto H, Murakami T, Tsuchida K, Sugino
H, Miyake H and Tashiro S: Tumor-stroma interaction of human
pancreatic cancer: Acquired resistance to anticancer drugs and
proliferation regulation is dependent on extracellular matrix
proteins. Pancreas. 28:38–44. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
De Wever O and Mareel M: Role of tissue
stroma in cancer cell invasion. J Pathol. 200:429–447. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding K, Lopez-Burks M, Sanchez-Duran JA,
Korc M and Lander AD: Growth factor-induced shedding of syndecan-1
confers glypican-1 dependence on mitogenic responses of cancer
cells. J Cell Biol. 171:729–738. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kleeff J, Ishiwata T, Kumbasar A, Friess
H, Büchler MW, Lander AD and Korc M: The cell-surface heparan
sulfate proteoglycan glypican-1 regulates growth factor action in
pancreatic carcinoma cells and is overexpressed in human pancreatic
cancer. J Clin Invest. 102:1662–1673. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bailey P, Chang DK, Nones K, Johns AL,
Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC,
et al: Genomic analyses identify molecular subtypes of pancreatic
cancer. Nature. 531:47–52. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Waddell N, Pajic M, Patch AM, Chang DK,
Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al:
Whole genomes redefine the mutational landscape of pancreatic
cancer. Nature. 518:495–501. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ishii M, Shimizu S, Hagiwara T, Wajima T,
Miyazaki A, Mori Y and Kiuchi Y: Extracellular-added ADP-ribose
increases intracellular free Ca2+ concentration through Ca2+
release from stores, but not through TRPM2-mediated Ca2+ entry, in
rat beta-cell line RIN-5F. J Pharmacol Sci. 101:174–178. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Inamura K, Sano Y, Mochizuki S, Yokoi H,
Miyake A, Nozawa K, Kitada C, Matsushime H and Furuichi K: Response
to ADP-ribose by activation of TRPM2 in the CRI-G1 insulinoma cell
line. J Membr Biol. 191:201–207. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang W, Chu X, Tong Q, Cheung JY, Conrad
K, Masker K and Miller BA: A novel TRPM2 isoform inhibits calcium
influx and susceptibility to cell death. J Biol Chem.
278:16222–16229. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ishii M, Hagiwara T, Mori Y and Shimizu S:
Involvement of TRPM2 and L-type Ca2* channels in
Ca2* entry and cell death induced by hydrogen peroxide
in rat β-cell line RIN-5F. J Toxicol Sci. 39:199–209. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tao KY, Li XX, Xu WZ, Wang Y, Zhu SM, Xie
HX, Luo WH, Xu YJ and Xu XL: Prognostic role of apoptosis-related
gene functional variants in advanced non-small-cell lung cancer
patients treated with first-line platinum-based chemotherapy. Onco
Targets Ther. 8:147–155. 2015.PubMed/NCBI
|
|
20
|
Orfanelli U, Jachetti E, Chiacchiera F,
Grioni M, Brambilla P, Briganti A, Freschi M, Martinelli-Boneschi
F, Doglioni C, Montorsi F, et al: Antisense transcription at the
TRPM2 locus as a novel prognostic marker and therapeutic target in
prostate cancer. Oncogene. 34:2094–2102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang C, Qin Y, Liu H, Liang N, Chen Y, Ma
D, Han Z, Xu X, Zhou X, He J and Li S: Downregulation of a novel
long noncoding RNA TRPM2-AS promotes apoptosis in non-small cell
lung cancer. Tumour Biol. 39:10104283176911912017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang F, Jin R, Zou BB, Li L, Cheng FW, Luo
X, Geng X and Zhang SQ: Activation of Toll-like receptor 7
regulates the expression of IFN-λ1, p53, PTEN, VEGF, TIMP-1 and
MMP-9 in pancreatic cancer cells. Mol Med Rep. 13:1807–1812. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Grimmig T, Matthes N, Hoeland K, Tripathi
S, Chandraker A, Grimm M, Moench R, Moll EM, Friess H, Tsaur I, et
al: TLR7 and TLR8 expression increases tumor cell proliferation and
promotes chemoresistance in human pancreatic cancer. Int J Oncol.
47:857–866. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Eigenbrod T and Dalpke AH: TLR7
inhibition: A novel strategy for pancreatic cancer treatment?
JAKSTAT. 2:e230112013.PubMed/NCBI
|
|
25
|
Ochi A, Graffeo CS, Zambirinis CP, Rehman
A, Hackman M, Fallon N, Barilla RM, Henning JR, Jamal M, Rao R, et
al: Toll-like receptor 7 regulates pancreatic carcinogenesis in
mice and humans. J Clin Invest. 122:4118–4129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vaz J and Andersson R: Intervention on
toll-like receptors in pancreatic cancer. World J Gastroenterol.
20:5808–5817. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ni IB, Ching NC, Meng CK and Zakaria Z:
Translocation t(11;14)(q13;q32) and genomic imbalances in
multi-ethnic multiple myeloma patients: A Malaysian study. Hematol
Rep. 4:e192012.PubMed/NCBI
|
|
28
|
Danger R, Thervet E, Grisoni ML, Puig PL,
Pallier A, Tregouet D, Lecorre D, Giral M, Legendre C, Soulillou JP
and Brouard S: PARVG gene polymorphism and operational renal
allograft tolerance. Transplant Proc. 44:2845–2848. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen G, Wang Y, Wang L and Xu W:
Identifying prognostic biomarkers based on aberrant DNA methylation
in kidney renal clear cell carcinoma. Oncotarget. 8:5268–5280.
2017.PubMed/NCBI
|
|
30
|
Zhou J, Leu NA, Eckardt S, McLaughlin KJ
and Wang PJ: STK31/TDRD8, a germ cell-specific factor, is
dispensable for reproduction in mice. PLoS One. 9:e894712014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kotov AA, Akulenko NV, Kibanov MV and
Olenina LV: Dead-box RNA helicases in animal gametogenesis. Mol
Biol (Mosk). 48:22–35. 2014.(In Russian). View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lim AK, Lorthongpanich C, Chew TG, Tan CW,
Shue YT, Balu S, Gounko N, Kuramochi-Miyagawa S, Matzuk MM, Chuma
S, et al: The nuage mediates retrotransposon silencing in mouse
primordial ovarian follicles. Development. 140:3819–3825. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee K, Na W, Maeng JH, Wu H and Ju BG:
Regulation of DU145 prostate cancer cell growth by Scm-like with
four mbt domains 2. J Biosci. 38:105–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gwak J, Shin JY, Lee K, Hong SK, Oh S, Goh
SH, Kim WS and Ju BG: SFMBT2 (Scm-like with four mbt domains 2)
negatively regulates cell migration and invasion in prostate cancer
cells. Oncotarget. 7:48250–48264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li F, Abuarab N and Sivaprasadarao A:
Reciprocal regulation of actin cytoskeleton remodelling and cell
migration by Ca2+ and Zn2+: Role of TRPM2 channels. J Cell Sci.
129:2016–2029. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bauer I, Grozio A, Lasigliè D, Basile G,
Sturla L, Magnone M, Sociali G, Soncini D, Caffa I, Poggi A, et al:
The NAD+-dependent histone deacetylase SIRT6 promotes cytokine
production and migration in pancreatic cancer cells by regulating
Ca2+ responses. J Biol Chem. 287:40924–40937. 2012. View Article : Google Scholar : PubMed/NCBI
|