Introduction
Strabismus, which is an imbalance of both eyes, has
been reported to be a common ocular disorder in childhood
populations (1). Serious and
persistent strabismus may lead to both exterior abnormality and
impaired visual functions, including binocularity, stereo vision
and visual activity. Uncorrected strabismus is also one of the most
common causes of visual impairment and some cases were accompanied
with amblyopia (2). Considering
the infaust consequences of strabismus, early diagnosis and
treatments were required. Nowadays, the treatments of strabismus
included both surgical treatment and visual function training. Even
operation could improve the strabismal status in most patients, the
long-term prognosis of certain sub-type strabismus, including
intermittent strabismus and AV signs, were quite unsatisfactory.
Residual strabismus and impaired visual functions were present in
some cases and would conduct an advanced surgical or rehabilitative
interventions. Better understanding of the pathogenesis of
strabismus would help in the improved diagnosis and treatments in
the future.
According to a generally accepted theory, the
incidence of strabismus was caused by the impairment of central
neural pathways and maladjusted extraocular muscles (EOMs) would
behave strabismal symptoms (3).
EOM, which demonstrated crucial roles in the control of eye
position, certainly played a key role in the development of
strabismus. Initial opinion demonstrated that the EOMs in
strabismus cases were not pathologically altered, however, this
opinion was challenged by most recent studies. In a previous study
based on immunofluorescence multiple-marker method, it was found
that the number of Pax7-positive cells/satellite cells in anterior
portion of EOMs was higher (4).
Differently disputation of subtype cells would provide both
understanding of the function of EOM and guide for the strabismus
surgery. Our team also focused on the ultrastructure and
pathological changes in patients with strabismus. In a study based
on clinical EOM samples of intermittent exotropia, it was found
that the significantly higher levels of myosin and actin was
detected in adolescent group comparing with the adult group. When
the ultrastructure was considered, electron microscopy was
conducted to reveal sarcomere destruction, myofilament
disintegration, collagen proliferation, and fibrosis between
different age groups.
Considering that both molecular biomarkers and
microstructure abnormality were detected in the EOMs in strabismus
cases, it was quite important to detect the detailed pathogenesis
of strabismus (5). Besides, it
also demonstrated significant potential importance in the detection
of molecular and structural biomarkers in the diagnosis,
classification and prognosis of strabismus management. A study by
Altick et al was conducted to detect the gene expression
profile in the EOMs from strabismal cases and normal controls
(6). A total of 604 genes in
strabismal EOM samples were detected based on the microarray
analysis and advanced PCR array identified the significant
muscle-specific genes expression pattern. However, all the previous
studies focused on the coding RNA expression pattern. Noncoding
RNAs, especially long non-coding RNAs (lncRNAs) which was the
noncoding RNA transcripts of above 200 nucleotides that do not
encode proteins, were also reported to play key roles in different
biological progresses. As showed in previous studies, lncRNAs were
reported to be involved in cancer occurrence, organ development and
homeostasis maintenance (7–9). As
showed in previous studies, lncRNAs were also involved in ocular
disorders, such as diabetic retinopathy, choroidal
neovascularization and age-related macular degeneration (10–12).
Considering that lncRNAs could also regulate the function
maintenance of muscles, it was quite important and interesting to
detect the pathogenic roles of lncRNAs in the development of
strabismus. As there was high-throughput data available in public
databases, the re-annotations and data mining would provide us
updated knowledge on the development of strabismus. The aim of this
study to determine the expression pattern of both coding and
lncRNAs in the EOM samples from strabismus cases and thus provide
new understanding in the pathogenesis of strabismus with public
data. Comprehensive analyses and updated knowledge on the roles of
RNAs in the development of strabismus would provide potential clues
for the detection of diagnostic, therapeutic and prognostic
targets.
Materials and methods
Microarray data
Gene expression profiles of four strabismic and four
normal EOM samples were downloaded from the Gene Expression Omnibus
(GEO) database (http://www.ncbi.nlm.nih.gov/geo/). All the strabismic
samples were from independent samples, while one of the controls
were repeated samples of the other three samples. There were 3
females and 1 male in the strabismus group while 2 females and 2
males in the control group. No significant difference was detected
in the age distribution between the case (22.5±27.11) and control
(16.75±15.09) group (P=0.4493). The deviation angles of four
strabismic cases were approximately 12°-14°, 30°, 45° and
approximately 45°-55°. All the microarray analyses were conducted
using Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix;
Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Data preprocessing and lncRNAs
re-annotations
Expression data of both cases and controls in cel
document were downloaded from GEO database with a serial accession
number GSE38780 and used for advanced analyses. Considering that
the dataset were based on the GPL570, the gene symbol as well as
annotation information (refGene) were added into the downloaded
datasets. Differently expressed genes (DEGs) between strabismic and
control samples were identified using the LIMMA package (Linear
Models for Microarray Data) in R software. To detect the DEGs, the
adjusted P-value <0.01 and |logFC|>1 cutoff criterion were
obtained in the screening. All the DEGs were presented in heat map
and volcano graph. Among all the DEGs, the refGene (NM, mRNA. NR,
ncRNA. XM, predicted mRNA model. XR, predicted ncRNA model) was
used in the annotation of differently expressed lncRNAs.
Bioinformatics analyses and functional
enrichment
To conduct the bioinformatics analyses based on the
detected DEGs, the Gene Ontology (GO) functional enrichment based
on Database for Annotation, Visualization and Integrated Discovery
(DAVID) (13) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analyses (14) were both obtained for the functional
analyses. GO analyses could be divided into three independent
parts, including molecular function, biological process and
cellular component. The detected protein or gene can match the
corresponding GO number and then GO term would demonstrate the
functional category or the cell location. KEGG is a database that
integrates functional information about genomes, biochemical and
organismal system. Application of KEGG pathway database would help
in the comprehensive inferences for pathway mapping of DEGs. In
this study, KOBAS 3.0 was used to carry on the KEGG pathway
enrichment analysis and P<0.01 was set as the screening
condition.
Protein-protein interactions
Protein-protein interactions analyses were conducted
using an a online bioinformatics tool, string (15) and a graphical presentation of the
interaction network. Both the analyses were conducted by the
Cytoscape 3.5.1 software (16).
The functional node points of the interacting proteins were
analyzed in advance. A total of eight evidence points that
demonstrate the relationship between different nodes were obtained
in the analyses and the associations with a combined_score >0.9
were listed in the network association list. Besides, the hub
nodes, which demonstrated most significant potential reputational
function, in this study were also detected.
lncRNA-mRNA co-expression network and
functional enrichment of lncRNAs
For the moment, there was no available functional
enrichment tool for high-throughput lncRNAs data. In general,
functional related genes may demonstrate similar expression
profiles and related expression pattern of lncRNAs-co-expressed
mRNA would provide clues for the functional enrichments of lncRNAs.
Thus it was an optional method for the functional enrichment of
lncRNA by analyzing co-expressed mRNAs. In this study, WGCNA was
used in the construction of lncRNA-mRNA co-expression network
(17) and adjacency threshold was
set at 0.85. Cytoscape software was obtained in the network
formation as well. For advanced functional enrichments, both GO and
KEGG pathway analyses of lncRNA-co-expressed mRNA were conducted in
this study.
Results
DEGs and dysregulated lncRNAs
To detect the DEGs in the strabismic cases, the
microarray data of 4 cases and 4 controls were used for advanced
analyses. Using LIMMA with a P-value <0.01 and |logFC|>1.0, a
total of 790 DEGs were screened (648 upregulated and 142
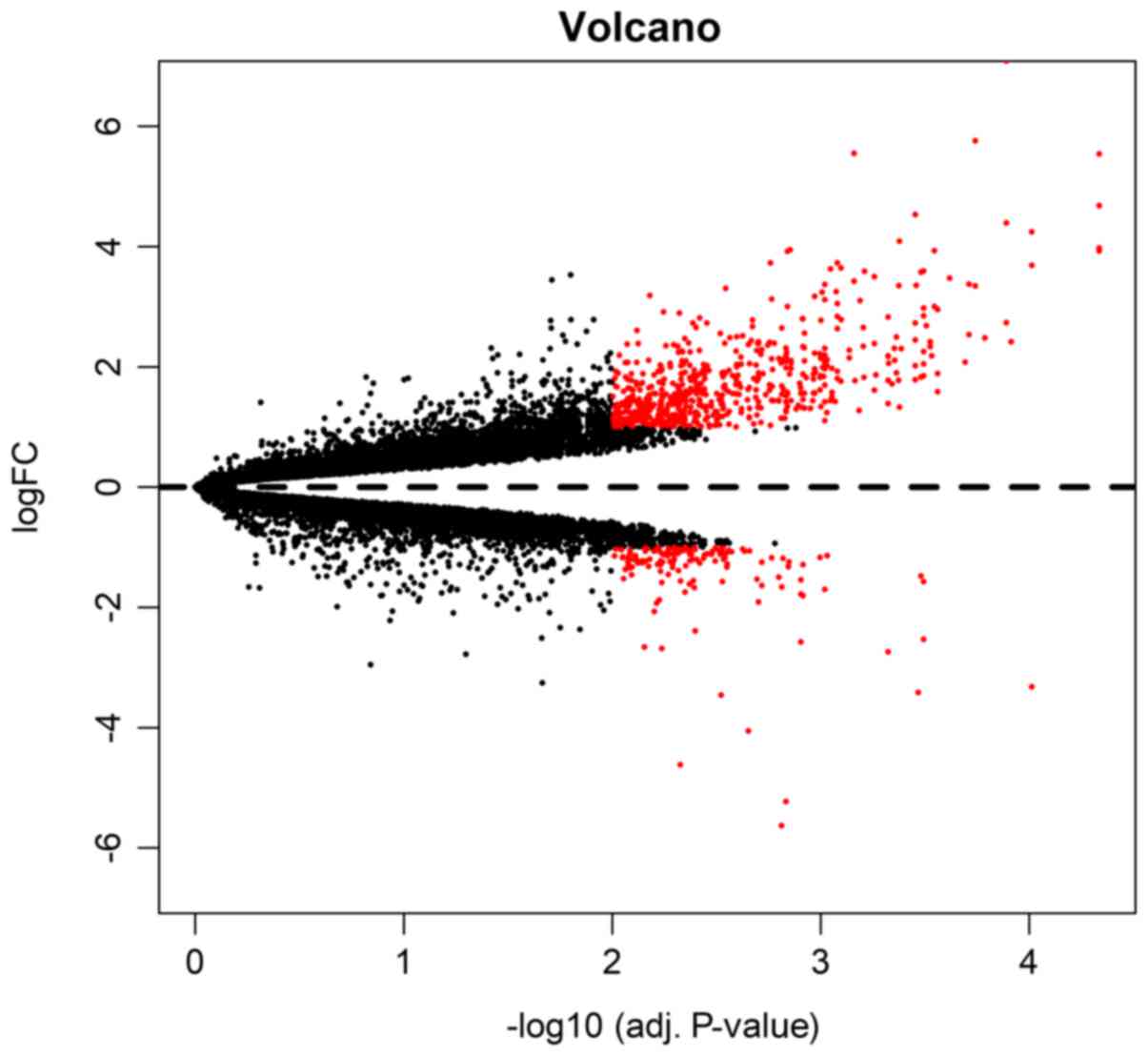
downregulated). Both volcano plot (Fig. 1) and clustering heatmap (Fig. 2) were used in the presentation of
the DEGs. In the volcano plot, the DEGs were marked red among all
the detected genes. In another way, clustering heatmap provided a
graphical review of DEGs with upregulated (marked red) and
downregulated (marked green) genes. To detect the most dysregulated
genes of significance, we presented the top 10 DEGs in Table I. Among the 10 DEGs (TNMD, HBB,
FNDC1, PTHLH, CRISPLD1, NPTX2, COL1A2, CHRDL1, CYS1 and SFRP2),
only NPTX2 demonstrated a downregulation pattern.
 | Table I.Differently expressed genes between
strabismic cases and normal controls. |
Table I.
Differently expressed genes between
strabismic cases and normal controls.
| Gene | Id | Log value of fold
change | Average expression
value | t | P-value | Adjusted
P-value |
|---|
| TNMD | 64,102 | 5.539642628 | 8.654139143 | 18.94555632 |
2.53×10−09 |
4.61×10−05 |
| HBB | 3,043 | 3.924910079 | 10.86418424 | 17.3483421 |
6.10×10−09 |
4.61×10−05 |
| FNDC1 | 84,624 | 4.682337268 | 7.527809329 | 17.00657939 |
7.44×10−09 |
4.61×10−05 |
| PTHLH | 5,744 | 3.976736723 | 6.135764953 | 16.68278379 |
9.01×10−09 |
4.61×10−05 |
| CRISPLD1 | 83,690 | 4.244618812 | 7.381617504 | 15.04795712 |
2.50×10−08 |
9.72×10−05 |
| NPTX2 | 4,885 | −3.319432337 | 7.137493269 | −14.61908535 |
3.32×10−08 |
9.72×10−05 |
| COL1A2 | 1,278 | 3.687990231 | 10.9966917 | 14.617825 |
3.33×10−08 |
9.72×10−05 |
| CHRDL1 | 91,851 | 2.416155203 | 10.50052948 | 14.09099772 |
4.77×10−08 | 0.000121962 |
| CYS1 | 192,668 | 2.734788648 | 7.593811022 | 13.67185162 |
6.41×10−08 | 0.000128488 |
| SFRP2 | 6,423 | 4.393543094 | 9.280684423 | 13.63393902 |
6.58×10−08 | 0.000128488 |
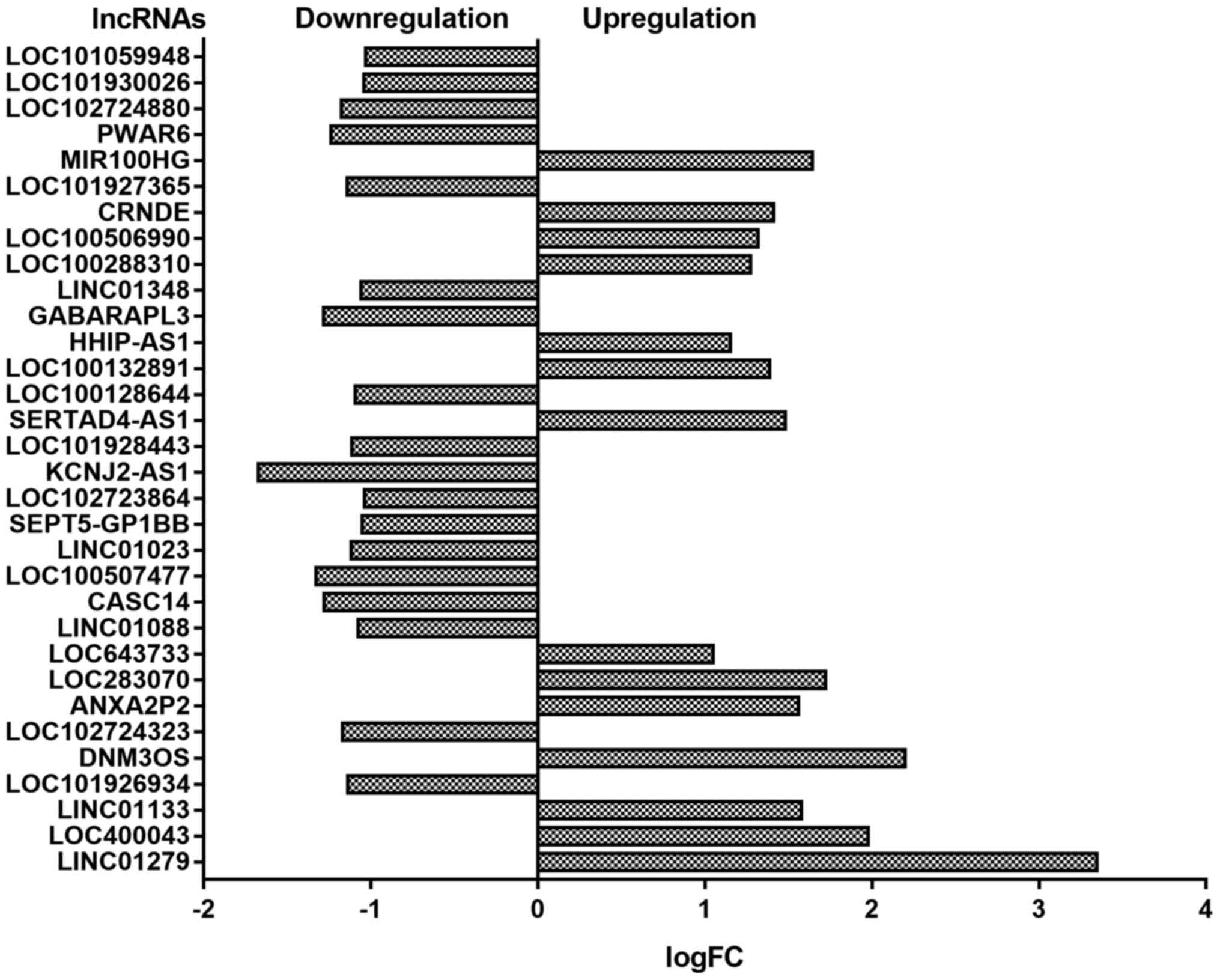
Considering the important reputational role of
lncRNAs in different biological progress, we also conducted
advanced analyses to detect differently expressed lncRNAs. In this
study, a total of 32 differently expressed lncRNAs were detected.
Among all the detected lncRNAs, 14 lncRNAs were upregulated and the
rest 18 were downregulated. The detected lncRNAs were presented in
Fig. 3.
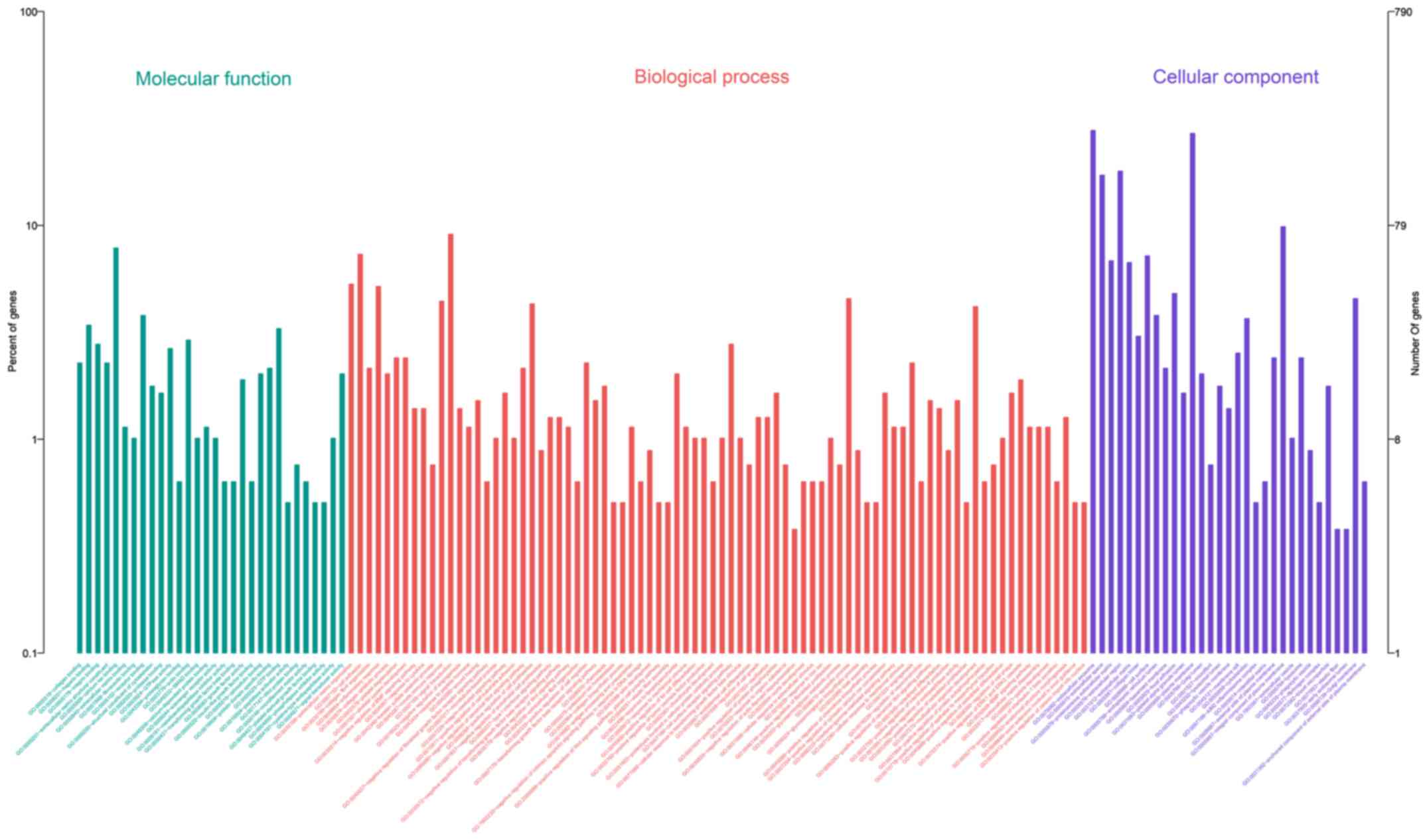
GO enrichment analysis
To further detect the roles of DEGs in the
pathogenesis of strabismus, GO enrichment analysis was conducted in
this study. By dividing all the related GO terms in to different
progresses, it was found that a total of 143 GO terms were
identified in this study. Among all the detected terms, there were
82 in biological process, 31 in cellular component and 30 in
molecular function. The detailed constituent of all the GO terms of
significance were displayed in Fig.
4 and the top 10 most important terms were listed in Table II.
 | Table II.The top 10 identified Gene ontology
terms of all the DEGs. |
Table II.
The top 10 identified Gene ontology
terms of all the DEGs.
| Terms | Count | P-value |
|---|
|
GO:0070062~extracellular exosome | 220 |
2.99×10−26 |
|
GO:0005615~extracellular space | 136 |
3.14×10−25 |
|
GO:0005578~proteinaceous extracellular
matrix | 54 |
5.07×10−23 |
|
GO:0005576~extracellular region | 142 |
8.77×10−21 |
|
GO:0031012~extracellular matrix | 53 |
3.43×10−20 |
|
GO:0030198~extracellular matrix
organization | 42 |
4.77×10−19 |
| GO:0007155~cell
adhesion | 58 |
7.67×10−15 |
| GO:0030199~collagen
fibril organization | 17 |
3.92×10−13 |
| GO:0005581~collagen
trimer | 24 |
6.19×10−13 |
| GO:0009986~cell
surface | 57 |
2.95×10−11 |
KEGG pathway analysis
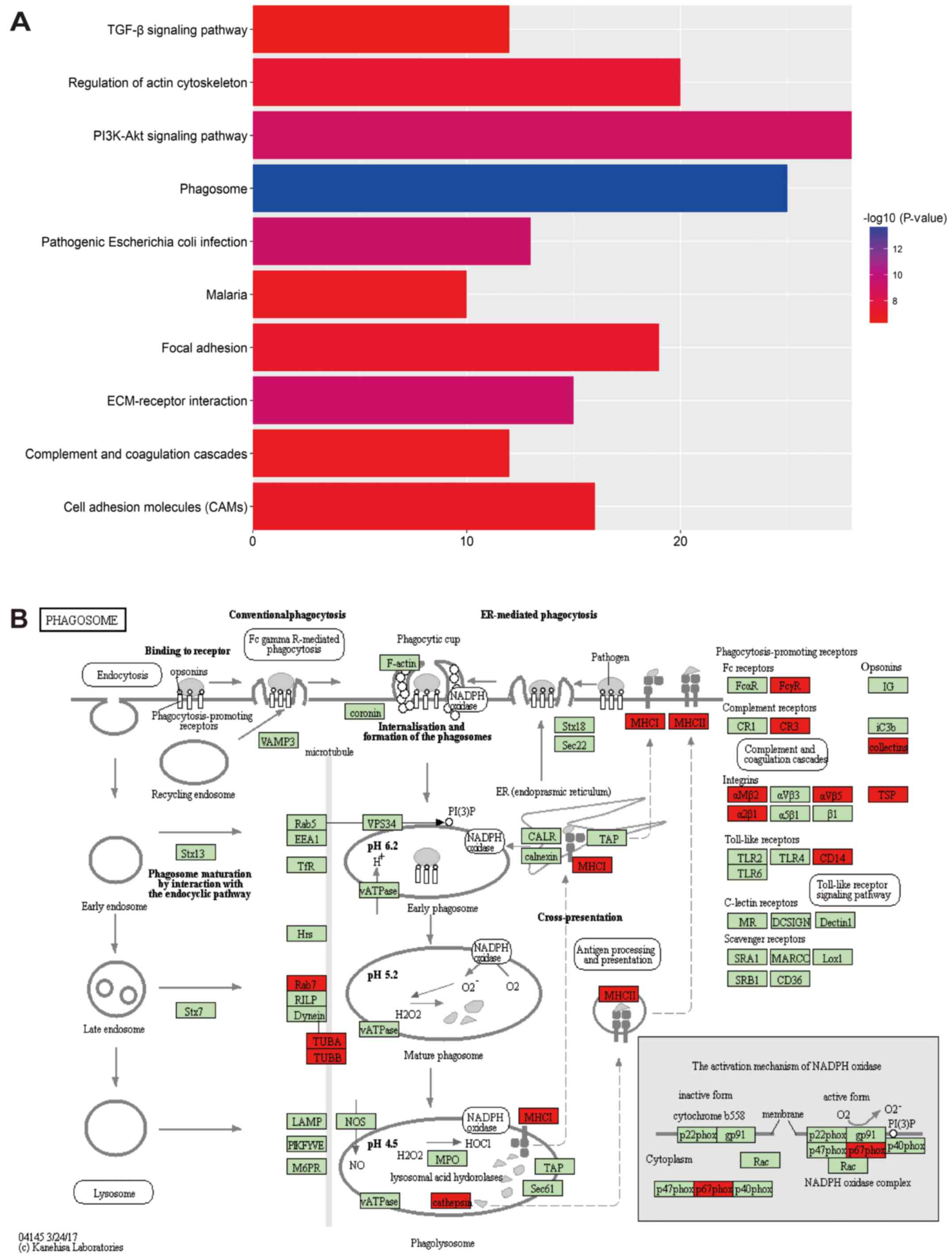
KEGG enrichment analysis was also conducted to map
the DEGs into regulation pathways. With the analyses by KOBAS 3.0
and P<0.01 as screening criteria, a total of 57 evaluated
pathways demonstrated statistical significance. Among all the
evaluated pathways, phagosome, pathogenic Escherichia coli
infection, ECM-receptor interaction, PI3K-Akt signaling pathway,
regulation of actin cytoskeleton, focal adhesion, cell adhesion
molecules, malaria, complement and coagulation cascades and TGF-β
signaling pathway were the top 10 significantly important pathways
(Fig. 5A). While the phagosome
pathway, which was labeled as hsa004145, demonstrated the most
bioinformatics importance and may be related with the development
of strabismus. The detailed pathway information of hsa004145 was
presented in Fig. 5B.
Protein-protein interactions and
function module analysis
Protein interactions analysis between DEGs were
would provide the function enrichment as well as detect the hot
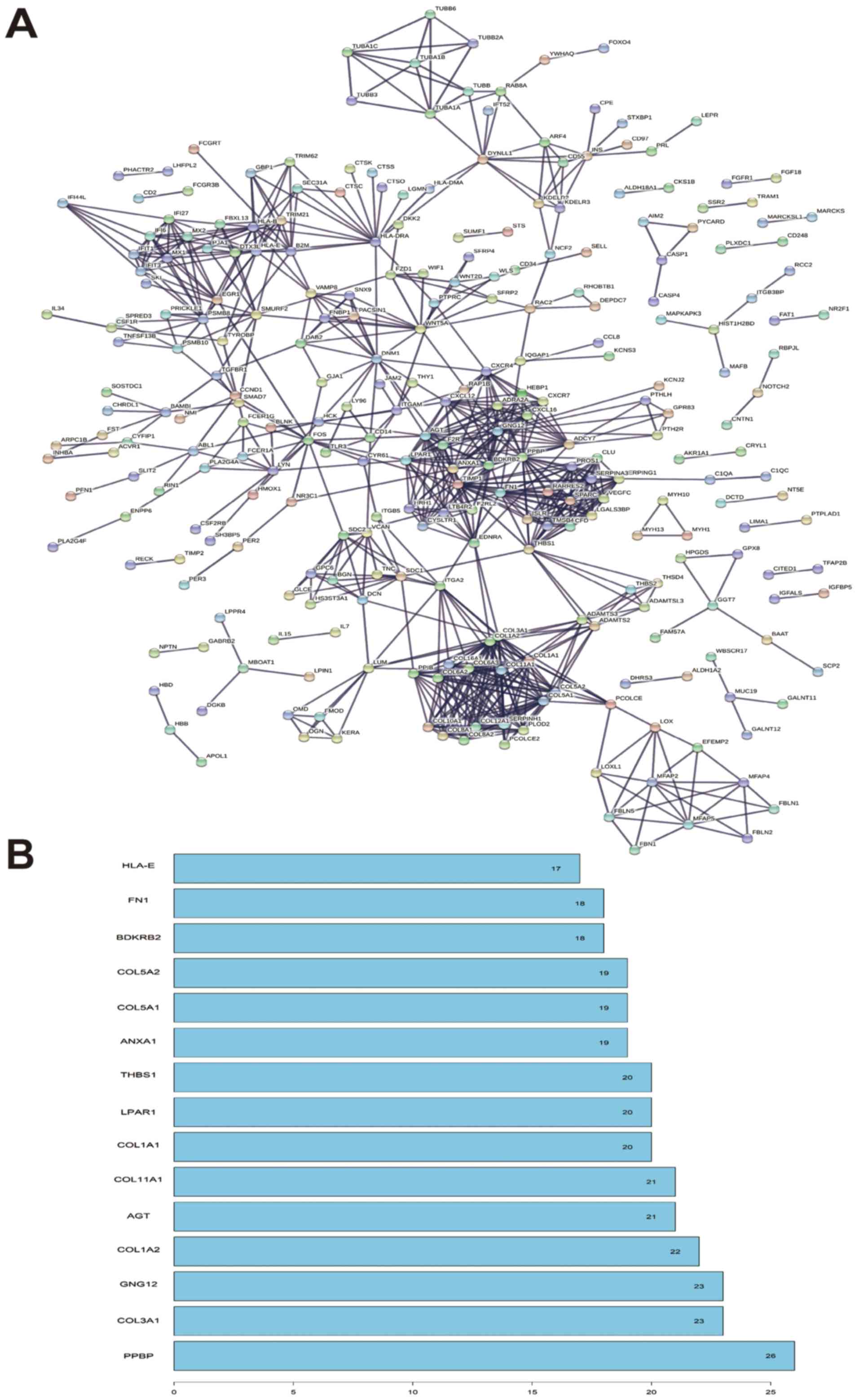
point of significance. In this study, the detailed PPI network was
showed in Fig. 6A. In the function
module analysis, it was found that PPBP, COL3A1, GNG12, COL1A2 and
AGT were reported to be the hub mode of the PPI network and the
detailed interaction modes were presented in Fig. 6B.
lncRNA-mRNA co-expression network and
functional analyses
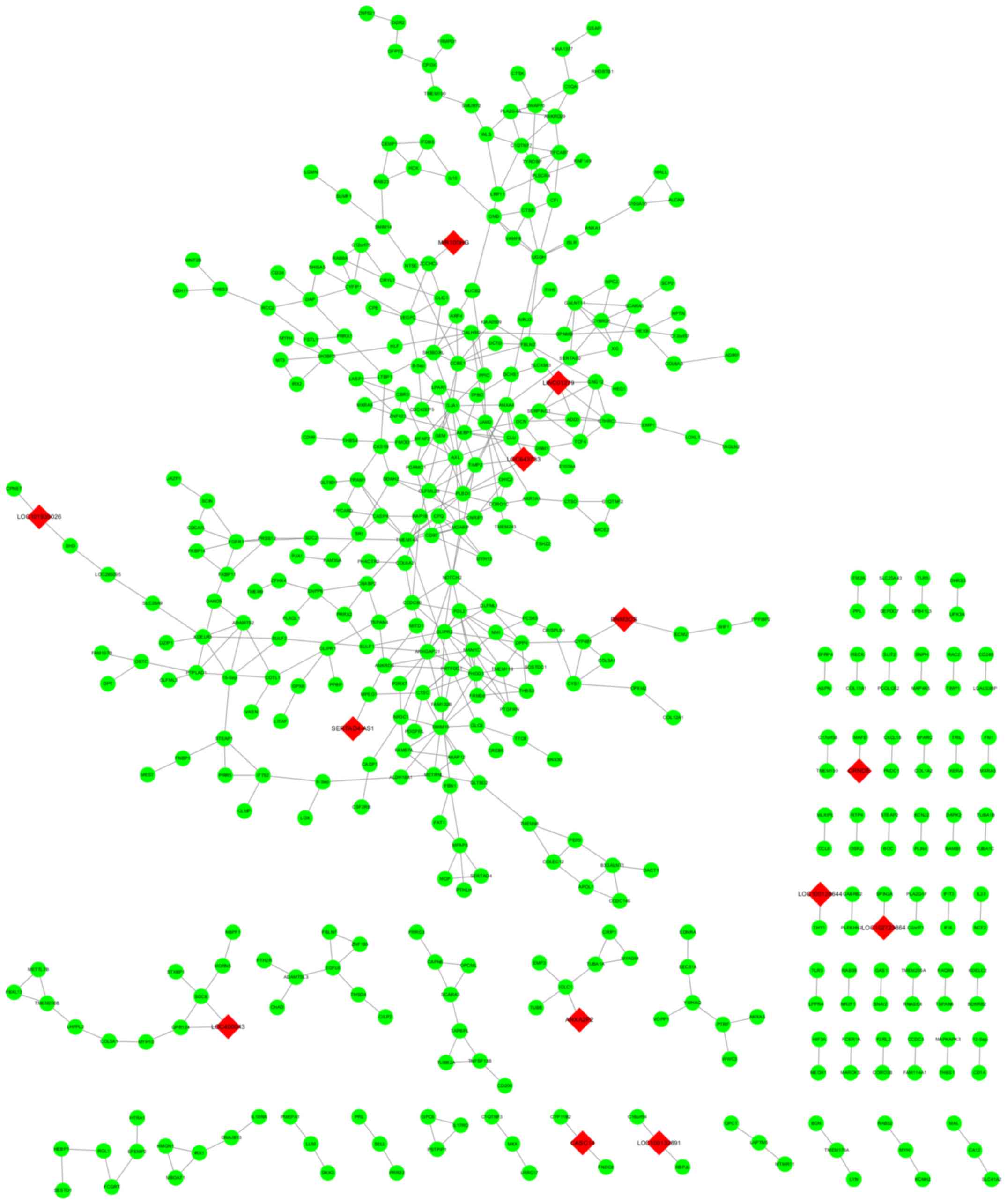
Through WGCNA software, the top 10 most significant
lncRNA-mRNA co-expression relationships detected and presented in
Fig. 7. Among all the detected
co-expression relationships, the related mRNAs were selected for
advanced functional analyses. Most lncRNAs, except LINC01279 and
LOC643733, indicated less than 3 target mRNAs and were not suitable
for advanced bioinformatics analyses. Through advanced GO
enrichment and KEGG pathway analysis, it was found that no
potential KEGG pathways could be detected in the related DEGs.
However, there was a GO term for each lncRNA (proteinaceous
extracellular for LINC01279 and cell surface for LOC643733) and
they were displayed in Table
III.
 | Table III.Gene Ontology terms of the
lncRNAs. |
Table III.
Gene Ontology terms of the
lncRNAs.
| lncRNAs | Category | Term | Count | P-value | Genes |
|---|
| LINC01279 | Cellular
component |
GO:0005578~proteinaceous extracellular
matrix | 2 | 0.057543258 | CTHRC1, FBLN2 |
| LOC643733 | Cellular
component | GO:0009986~cell
surface | 2 | 0.058599058 | TIMP2, ANXA4 |
Discussion
The detailed mechanism for the development of
strabismus was quite poorly understood. Even certain work has been
conducted, the molecular pathogenesis of strabismus was litter
detected in previous studies (18). In the present study, we re-analyzed
the public microarray data and detected the effects of both coding
and non-coding RNAs in the development of strabismus. By analyzing
the RNA expression pattern of four strabismal and their paired
controls, it was found that both coding and long non-coding RNAs
were differently expressed in the EOM of strabismal patients.
Advanced bioinformatics analyses provided updated evidence and clue
in the understanding of strabismus.
Even different causes, including genetic, nerve
regulated and structural modification, were reported to be
associated with the development of strabismus (19–21),
the EOM dysregulation was regarded as the most direct effector
organ among various causes. Through scientific and systematic mRNA
detection, the results would provide abundant knowledge on the
incidence of strabismus. In 2012, Altick et al and
colleagues conducted a microarray analyses based four pairs of
strabismus cases and controls (6).
It was found that a total of 604 genes were differently expressed
in strabismic EOMs and advanced functional analyses demonstrated
that DEGs involved in extracellular matrix structure (upregulated)
and contractility (downregulated) demonstrated the most important
effect. In our study, we modified the screening condition to
adjusted P<0.01 and |logFC|>1. Through adjustments of both
P-value and |logFC|, more DEGs with less variance and statistically
significance would be detected in this study. In our study, more
DEGs were detected comparing with the primary analyses (790 DEGs
vs. 604 DEGs). Based on the updated DEG data and GO analyses tool,
it was reported that extracellular exosome, extracellular space and
proteinaceous extracellular matrix were the top three GO terms of
significance. The previous detected pathways (extracellular matrix
structure and contractility) were also significant in the updated
analyses. It was also found that both conclusions indicated that
the most important pathogenesis of strabismus was the modification
of extracellular structure. It was quite easy to understand this
conclusion demonstrating that the pathological study of strabismus
demonstrated noticeable extracellular structure modification,
including increased content of collagen fiber, and the gap between
fibrous tissue, adipose tissue and muscle fiber widened (22). This phenomenon was also proved by a
recent study in different study design. As reported by Agarwal
et al, the difference in protein and RNA level of EOM
samples from strabismic cases were detected proteomics, standard
and customized PCR arrays, and microarrays (23). It was also reported that expression
of collagens and regulators of collagen synthesis and degradation
was upregulated in both RNA and protein level. These findings
suggest that more work should be focused on the extracellular
matrix modification in the development of strabismus.
KEGG pathway and PPI analyses were effective tools
in the detection of key pathways and core regulators. By analyzing
the information of updated DEGs, there was several interesting
findings. TGF-β signaling pathway, which was one of the top 10
related pathways, may demonstrate certain effects in the formation
of strabismus. TGF-β, which is one of the most important growth
factors in the pathogenesis of fibrotic diseases, demonstrated
important role in the fibration formation and extracellular matrix
modification (24). In the ocular
disorders, TGF-β was also reported to produce important regulative
effect in corneal scarring, conjunctiva fibrosis, fibrosis of the
lens capsule, strabismus development and proliferative
vitreoretinopathy (25).
Remarkable TGF-β1 expression was observed in areas with excessive
collagen deposition in the post-operative adhesion after strabismus
surgery in experimental rabbit model and it indicated the effect of
TGF-β on the effects of postoperative recovery in strabismus
surgery (26). Considering that
local application of agents, including insulin-like growth factor-1
(IGF-1) and botulinum toxin (27,28),
for the treatment of strabismus demonstrated remarkable
improvements, it was an interesting and potential important aspect
in the application of anti-TGF-β in the treatment of strabismus
itself. It was phagosome pathway that demonstrate the most
significant importance in this study, however, no previous study
focused on this point. Phagosome, which was defined as the
regulated uptake of large particles into cytosolic, membranebound
vacuoles, demonstrated immunoregulatory for long (29). Nowadays, it has been reported that
phagosome would provide important role in the organ development,
homeostasis maintenance of inner environment and infection
responses (30,31). As autophagy was regarded as an
important regulative progress in age related disorders, phagosomes
pathway was reported to be involved in the development of
age-related macular degeneration (32). Strabismus, which was a
neurodevelopmental disease, may be regulated the regulation of
phagosomes pathway and more work was required to be conducted in
the future. However, it should be noticed that the bioinformatics
analyses were based on the analyses of co-expressed mRNAs of the
related lncRNAs. As we know, the lncRNAs usually played a role
through DNA, RNA, protein and miRNAs, however no available
bioinformatics tool could be used to demonstrate the annotations of
the differently expressed lncRNAs. The co-expressed mRNAs with
lncRNAs could only explain a part of the function of related
lncRNAs and thus the bioinformatics analyses were just part of the
global function of lncRNAs. The conclusion of this part should be
considered with cautions.
Apart from coding RNAs, we also pay attention on the
effect of lncRNAs on the incidence of strabismus in this study. As
lncRNAs were reported to be involved in kinds of diseases,
including cancer, cardiovascular disorders, diabetes and immune
disorders (33–35). It was also reported that lncRNAs
would demonstrate certain effects in the ocular disorders (36–38)
and may produce potential therapeutic effects. However, no previous
study focused the effect of lncRNAs in the development of
strabismus by now. Considering EOM demonstrated direct pathogenic
modification and provided primary therapy, the study on the effect
of lncRNAs in the EOMs may provide important knowledge in this
field. Previous studies demonstrated the effect of lncRNAs in the
muscle function maintenance. A previous study based on in
vivo and in vitro studies showed that a lncRNA, LncMyoD,
demonstared regulative effects in skeletal muscle differentiation
through blocking the translation of mRNA (39). Another study showed that s a novel
lncRNA lnc133b, could regulate bovine skeletal muscle satellite
cell, which was significantly actived in strabismus (40), proliferation and differentiation by
mediating miR-133b (41). This
study provide a potential thread in the research of lncRNAs on the
pathopoiesis of strabismus. As mentioned in the above, TGF-β was
one of the most important regulator of strabismus development and
treatment, a recent study by Tang et al indicated the
detailed mechanism through which the lncRNA GAS5 regulated
TGF-β-induced smooth muscle cell differentiation (42). The cross-talk between important
lncRNAs and mRNA provided us abundant in the study of lncRNAs in
the development of strabismus. Thus we conducted a relevant
research on the expression of lncRNAs in the EOM samples of
strabismus cases. Even many differently expressed lncRNAs were
detected, however, no previous available literature demonstrated
potential relation between lncRNAs and strabismus. Besides, the
function enrichment analyses of lncRNAs was quite hard to conduct
by now. In this study, we chose to conduct the functional analyses
by analyzing the function of lncRNA-co-expressed mRNA. No
satisfactory outcome was gained in this study. Besides, there was
potential bias in this strategy because lncRNAs may demonstrate the
effect through interaction with DNA or protein. Further in
vitro and in vivo study as well as advanced
bioinformatics analyses would provide better understanding of the
effect of lncRNAs in the strabismus.
These results in this study demonstrated both coding
and lncRNA produced certain effects in the development of
strabismus. Functional enrichment analyses provide updated
knowledge on the understanding of this disorder and thus provide
potential therapeutic methods. However, the evidence of lncRNAs
affecting the development should be proved in advanced studies.
Further studies will be needed to conclusively demonstrate and
elucidate the precise role of lncRNAs in strabismus.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WXM and JYY conceived and designed the experiments;
WXM, TKY and JYY performed the experiments; WXM and XGH analyzed
the data; WXM, XGH and JYY contributed reagents/materials/analysis
tools and WXM and JYY wrote the manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lambert SR: Population-based incidence of
strabismus: Why is it important? JAMA Ophthalmol. 135:1053–1054.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shapira Y, Machluf Y, Mimouni M, Chaiter Y
and Mezer E: Amblyopia and strabismus: Trends in prevalence and
risk factors among young adults in Israel. Br J Ophthalmol.
2017.doi: 10.1136/bjophthalmol-2017-310364. PubMed/NCBI
|
|
3
|
Nelson LB: Macular changes following
strabismus surgery confirmed by the use of optical coherence
tomography. J Pediatr Ophthalmol Strabismus. 53:102016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lindstrom M, Tjust AE and Domellof Pedrosa
F: Pax7-positive cells/satellite cells in human extraocular
muscles. Invest Ophthalmol Vis Sci. 56:6132–6143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gong HM, Wang J, Xu J, Zhou ZY, Li JW and
Chen SF: Identification of rare paired box 3 variant in strabismus
by whole exome sequencing. Int J Ophthalmol. 10:1223–1228.
2017.PubMed/NCBI
|
|
6
|
Altick AL, Feng CY, Schlauch K, Johnson LA
and von Bartheld CS: Differences in gene expression between
strabismic and normal human extraocular muscles. Invest Ophthalmol
Vis Sci. 53:5168–5177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Akhade VS, Pal D and Kanduri C: Long
noncoding RNA: Genome organization and mechanism of action. Adv Exp
Med Biol. 1008:47–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aubert G, Strauss KA, Lansdorp PM and
Rider NL: Defects in lymphocyte telomere homeostasis contribute to
cellular immune phenotype in patients with cartilage-hair
hypoplasia. J Allergy Clin Immunol. 140:1120–1129.e1. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Xu F, Xiao H and Han F: Long
noncoding RNA BDNF-AS inversely regulated BDNF and modulated
high-glucose induced apoptosis in human retinal pigment epithelial
cells. J Cell Biochem. 119:817–823. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu W, Meng YF, Xing Q, Tao JJ, Lu J and
Wu Y: Identification of lncRNAs involved in biological regulation
in early age-related macular degeneration. Int J Nanomedicine.
12:7589–7602. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ye Z, Li Z and He S: Long noncoding RNA
associated competing endogenous RNAs are induced by clusterin in
retinal pigment epithelial cells. Mol Med Rep. 16:8399–8405. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Franz M, Lopes CT, Huck G, Dong Y, Sumer O
and Bader GD: Cytoscape.js: A graph theory library for
visualisation and analysis. Bioinformatics. 32:309–311.
2016.PubMed/NCBI
|
|
17
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ye XC, Pegado V, Patel MS and Wasserman
WW: Strabismus genetics across a spectrum of eye misalignment
disorders. Clin Genet. 86:103–111. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lueder GT: Orbital causes of incomitant
strabismus. Middle East Afr J Ophthalmol. 22:286–291. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Min X, Fan H, Zhao G and Liu G:
Identification of 2 potentially relevant gene mutations involved in
strabismus using whole-exome sequencing. Med Sci Monit.
23:1719–1724. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rajab GZ, Suh SY and Demer JL: Magnetic
resonance imaging in dissociated strabismus complex demonstrates
generalized hypertrophy of rectus extraocular muscles. J AAPOS.
21:205–209. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Haider AS: Unilateral internuclear
ophthalmoplegia, strabismus and transient torsional nystagmus in
focal pontine infarction. BMJ Case Rep. 2016:bcr20162165032016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Agarwal AB, Feng CY, Altick AL, Quilici
DR, Wen D, Johnson LA and von Bartheld CS: Altered protein
composition and gene expression in strabismic human extraocular
muscles and tendons. Invest Ophthalmol Vis Sci. 57:5576–5585. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schwalm S, Beyer S, Frey H, Haceni R,
Grammatikos G, Thomas D, Geisslinger G, Schaefer L, Huwiler A and
Pfeilschifter J: Sphingosine kinase-2 deficiency ameliorates kidney
fibrosis by up-regulating Smad7 in a mouse model of unilateral
ureteral obstruction. Am J Pathol. 187:2413–2429. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saika S, Yamanaka O, Okada Y, Tanaka S,
Miyamoto T, Sumioka T, Kitano A, Shirai K and Ikeda K: TGF beta in
fibroproliferative diseases in the eye. Front Biosci (Schol Ed).
1:376–390. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Choi SU, Kim KW and Moon NJ: Effective
treatment for prevention of post-operative adhesion after
strabismus surgery in experimental rabbit model: 0.5% tranilast
ophthalmic solution. BMC Ophthalmol. 16:1662016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mahan M and Engel JM: The resurgence of
botulinum toxin injection for strabismus in children. Curr Opin
Ophthalmol. 28:460–464. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
McLoon LK, Christiansen SP, Ghose GM, Das
VE and Mustari MJ: Improvement of eye alignment in adult strabismic
monkeys by sustained IGF-1 treatment. Invest Ophthalmol Vis Sci.
57:6070–6078. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Levin R, Grinstein S and Canton J: The
life cycle of phagosomes: Formation, maturation and resolution.
Immunol Rev. 273:156–179. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Steinhauser C, Dallenga T, Tchikov V,
Schaible UE, Schutze S and Reiling N: Immunomagnetic isolation of
pathogen-containing phagosomes and apoptotic blebs from primary
phagocytes. Curr Protoc Immunol. 105:14.36.1–26. 2014. View Article : Google Scholar
|
|
31
|
Russell DG: Phagosomes, fatty acids and
tuberculosis. Nat Cell Biol. 5:776–778. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jiang M, Esteve-Rudd J, Lopes VS, Diemer
T, Lillo C, Rump A and Williams DS: Microtubule motors transport
phagosomes in the RPE and lack of KLC1 leads to AMD-like
pathogenesis. J Cell Biol. 210:595–611. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen YG, Satpathy AT and Chang HY: Gene
regulation in the immune system by long noncoding RNAs. Nat
Immunol. 18:962–972. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zheng Y, Liu L and Shukla GC: A
comprehensive review of web-based non-coding RNA resources for
cancer research. Cancer Lett. 407:1–5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery and classification of lncRNAs. Adv Exp Med Biol.
1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wan P, Su W and Zhuo Y: Precise long
non-coding RNA modulation in visual maintenance and impairment. J
Med Genet. 54:450–459. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu J, Ding X, Yuan L and Zhang X:
Identification of pterygium-related long non-coding RNAs and
expression profiling by microarray analysis. Int J Mol Med.
38:529–536. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li F, Wen X, Zhang H and Fan X: Novel
insights into the role of long noncoding RNA in ocular diseases.
Int J Mol Sci. 17:4782016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gong C, Li Z, Ramanujan K, Clay I, Zhang
Y, Lemire-Brachat S and Glass DJ: A long non-coding RNA, LncMyoD,
regulates skeletal muscle differentiation by blocking IMP2-mediated
mRNA translation. Dev Cell. 34:181–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Antunes-Foschini RS, Miyashita D, Bicas HE
and McLoon LK: Activated satellite cells in medial rectus muscles
of patients with strabismus. Invest Ophthalmol Vis Sci. 49:215–220.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jin CF, Li Y, Ding XB, Li X, Zhang LL, Liu
XF and Guo H: lnc133b, a novel, long non-coding RNA, regulates
bovine skeletal muscle satellite cell proliferation and
differentiation by mediating miR-133b. Gene. 630:35–43. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tang R, Zhang G, Wang YC, Mei X and Chen
SY: The long non-coding RNA GAS5 regulates transforming growth
factor beta (TGF-beta)-induced smooth muscle cell differentiation
via RNA Smad-binding elements. J Biol Chem. 292:14270–14278. 2017.
View Article : Google Scholar : PubMed/NCBI
|