Introduction
Epithelial ovarian cancer (EOC) is the leading cause
of mortality from gynecological malignancy. The majority of
patients with EOC are at a late stage upon diagnosis and the 5-year
survival rate of EOC is <30% (1–3). The
standard therapy for EOC for current clinical application is
surgery and platinum-based cytotoxic chemotherapy (4). The clinical management of EOC is
confounded by the difficulty to accurately diagnose EOC at
diagnosis. In addition, approximately 20% of patients will not
respond to platinum based therapy and may relapse (5,6).
Therefore, understanding the molecular mechanism
underlying EOC is important for developing novel therapeutic
targets. Long non-coding RNAs (lncRNAs) refer to RNAs having
>200 nucleotides that cannot be transcribed into proteins.
LncRNAs are evolutionarily conserved and are expressed in a tissue
specific manner (7). They are
involved in various cellular processes and their dysregulation is
emerging as important factors in tumor development and therapy
response (8–10). lncRNAs have been reported to act as
oncogenes or tumor suppressors in cancer development (11,12).
A recent study demonstrated that Lnc SRY-box 4 (SOX4) was able to
promote the self-renewal of liver tumor-initiating cells through
signal transducer and activator of transcription 3-mediated Sox4
expression and may serve as an oncogene in liver cancer (13). Furthermore, it has been reported
that LncSOX4 expression is associated with relapse rate and poor
outcome in EOC (14). The present
study investigated the role of LncSOX4 in EOC. LncSOX4 expression
was significantly upregulated in patients with EOC and in EOC cell
lines. Knocking out LncSOX4 resulted in inhibition of tumor cell
growth, migratory ability and cell proliferation. The cell cycle
was arrested at the G0/G1 phase following LncSOX4 silencing. In
addition, the LncSOX4 expression level was additionally associated
with unfavorable clinical prognostic factors. It was observed that
LncSOX4 may serve as an oncogene in EOC and that downregulating its
expression may be a potential therapeutic target.
Materials and methods
Patients and specimens
Ovarian tissues from 30 female patients (age,
44.9±9.3) with EOC and 18 female control patients (age, 46.2±12.4)
were collected between April 2015 and April 2017 from Hubei Women
and Children's Hospital (Wuhan, China). The study was approved by
the Research Ethics Committee of Hubei Women and Children's
Hospital. All selected patients provided written informed consent.
Patients were sorted into a high LncSOX4 expression group and a low
LncSOX4 expression group, according to the median LncSOX4
expression level (Median relative expression, 3.99). The diagnosis
of EOC was based on the clinical presentation, morphological
criteria and immunohistochemical staining. Patients who had
received radiation, chemotherapy or hormonal therapy were excluded
from the present study. Information about the patients is presented
in Table I.
 | Table I.Association between LncSOX4 expression
levels and clinical data in 30 patients with epithelial ovarian
cancer. |
Table I.
Association between LncSOX4 expression
levels and clinical data in 30 patients with epithelial ovarian
cancer.
|
|
| LncSOX4 expression
level |
|
|---|
|
|
|
|
|
|---|
| Variable | No. patients
(n=30) | Low (n=15) | High (n=15) | P-value |
|---|
| Age |
|
|
|
|
|
>55 | 17 | 7 | 10 | 0.461 |
| ≤55 | 13 | 8 | 5 |
|
| FIGO stage |
|
|
|
|
| I–II | 14 | 12 | 2 | 0.001 |
|
III–IV | 16 | 3 | 13 |
|
| CA125, U/ml |
|
|
|
|
|
≤35 | 17 | 10 | 6 | 0.272 |
|
>35 | 13 | 5 | 9 |
|
| Tumor size, cm |
|
|
|
|
| ≤5 | 14 | 11 | 3 | 0.010 |
|
>5 | 16 | 4 | 12 |
|
| Distant
metastases |
|
|
|
|
|
Yes | 18 | 3 | 10 | 0.027 |
| No | 12 | 12 | 5 |
|
Cell culture and culture
conditions
EOC cell lines including SKOV3, HO8910-PM, OVCAR3
and the human ovarian immortalized non-tumorigenic ovarian surface
epithelial (IOSE-80) cell line were purchased from the American
Type Culture Collection (Manassas, VA, USA). The high grade ovarian
serous adenocarcinoma HEY-A8 cell line was provided by Professor
Zehua Wang from the Wuhan Union Hospital (Wuhan, China). Cells were
cultured in RPM1-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and maintained at 37°C in a
5% CO2 incubator.
Cell transfection
The SKOV3 cells (1×106/well) and OVCAR3
cells (1×106/well) were transfected with small
interfering (si)-LncSOX4 and the negative control (NC) using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.).
si-LncSOX4 and NC were purchased from Shanghai GenePharma Co., Ltd.
(Shanghai, China). The expression level of LncSOX4 was detected by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis according to the protocol detailed below in
order to determine transfection efficiency. The sequence of
si-LncSOX4 was 5′-GGATGACAAGAAGTACAAA-3. The sequence of the
negative control was: Sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and
antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. Cells were incubated for 48
h after transfection and then used for further analysis.
RT-qPCR analysis
Total RNA was extracted from human samples and
ovarian cell lines using TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc.). A reverse transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) was used to transform RNA into
cDNA. The RT reaction was performed at 37°C for 15 min and 85°C for
5 sec. The level of LncSOX4 was measured using a SYBR Premix kit
(Takara420a; Takara Bio, Inc., Otsu, Japan) using GAPDH as the
endogenous control. The qPCR reaction was performed in an Applied
Biosystems 7500 Fast Real-Time PCR system (Thermo Fisher
Scientific, Inc.) at 95°C for 3 min, followed by 40 cycles at 95°C
for 15 sec, and finally 60°C for 1 min. The relative expression of
LncSOX4 was calculated using the comparative Cq (2−ΔΔCq)
method (15). The primers for
LncSOX4 included (forward) 5′-AGCGACAAGATCCCTTTCATTC-3′ and
(reverse) 5′-CGTTGCCGGACTTCACCTT-3′. The primers for GAPDH included
(forward) 5′-GGCTGAGAACGGGAAGCTTGTCAT-3′ and (reverse)
5′-CAGCCTTCTCCATGGTGGTGAAGA-3′.
In situ hybridization (ISH)
ISH was performed using a ISH Detection Kit III (AP;
Boster Biological Technology, Pleasanton, CA, USA) according to the
manufacturer's protocol. Samples were digested with proteinase K
for 5 min, fixed in 4% paraformaldehyde for 1 h at room temperature
and hybridized with a 5′-digoxin-labeled probe for LncSOX4 with a
sequence of 5′-CATGGGATCTTACTACAGGA-3′, at 55°C overnight.
Paraffin-embedded sections of 7 µm thickness were deparaffinized
with xylene and washed with ethanol at concentrations of 100, 95,
85 and 75%. Samples were incubated with horseradish peroxidase at
4°C for 30 min. The results were observed using a light microscope
(magnification, ×200) following the addition of hematoxylin at 15°C
for 5 min and eosin at 15°C for 2 min.
Transwell assay
Following 48 h of transfection, SKOV3 cells were
harvested using RPMI-1640 without serum. 150 µl cell suspension
(2.5×104 cells) was added into the upper chamber
(Corning Incorporated, Corning, NY, USA) and 600 µl culture medium
containing 10% FBS was added into the lower chamber. The membrane
was pre-coated with Matrigel (Corning Incorporated) for 6 h prior
to the invasion assay. Following incubation for 12 h, the cells
were fixed with methanol for 10 min at 4°C and stained with 0.1%
crystal violet at room temperature for 5 min. The results were
captured using an inverted light microscope (magnification, ×200;
TS100; Nikon Corporation, Tokyo, Japan).
Cell proliferation assay
The cell proliferation assay was carried out using a
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan), according to the manufacturer's protocol. SKOV3
and OVCAR3 cells were seeded into 96-well plates, with each well
containing 3,000–5,000 cells. Cells were incubated for 24, 48 and
72 h, respectively. Subsequently, 10 µl CCK-8 was added to the
wells and the optical density was read at 450 nm with a microplate
reader once every 30 min for 4 h in total.
Western blotting
Following 48 h of si-LncSOX4 and NC transfection,
cells were lysed on ice in radioimmunoprecipitation assay lysis
buffer (Beyotime Institute of Biotechnology, Haimen, China). A
bicinchoninic acid protein assay kit (Goodbio Biotechnology Co.,
Ltd., Wuhan, China) was used for protein determination. Protein
samples of 20 µg/lane were separated on 8–12% SDS-PAGE gels
(Goodbio Biotechnology Co., Ltd.) and transferred onto
nitrocellulose membranes. Membranes were blocked in Tris-buffered
saline with 0.05% Tween-20 and 5% non-fat milk for 1 h at 37°C.
Membranes were subsequently incubated with antibodies against 72
kDa type IV collagenase (MMP2; cat. no. ab37150; 1:1,000; Abcam,
Cambridge, UK), matrix metalloproteinase 9 (MMP9; cat. no. ab38898;
1:1,000; Abcam) and GAPDH (cat. no. ab9485; 1:2,500; Abcam)
overnight at 4°C. Immunodetection of the bound antibodies was
conducted using a goat anti-rabbit immunoglobulin G horseradish
peroxidase conjugated antibody (cat. no. 7074; 1:2,000; Cell
Signaling Technology, Inc., Danvers, MA, USA) for 1 h at room
temperature. Immunostaining was detected using an enhanced
chemiluminescence substrate (Beyotime Institute of Biotechnology)
and signal intensities were determined using Quantity One image
software (version 4.6.8; Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Flow cytometric analysis
Following transfection with si-LncSOX4 or NC for 48
h, SKOV3 and OVCAR3 cells were harvested for flow cytometric
analysis. Cells were stained with the Annexin V-Fluorescein
Isothiocyanate/Propidium Iodide Apoptosis Detection kit (cat. no.
556547; BD Biosciences, Franklin Lakes, NJ, USA), according to the
manufacturer's protocol. Subsequently, the cell cycle was detected
using a FACSCanto II (BD Biosciences) and analyzed using FlowJo
software (version 7.6.1; FlowJo LLC, Ashland, OR, USA).
Statistical analysis
The data obtained are expressed as the mean ±
standard deviation. Two-way analysis of variance was performed when
comparing continuous variables between groups, and Scheffe's post
hoc test was performed where significant differences were observed.
A χ2 test was used to compare enumerated data. Each
experiment was repeated in triplicate. All analyses were performed
using R Bioconductor (version 3.3.2; https://www.bioconductor.org/). P<0.05 was
considered to indicate a statistically significant difference.
Results
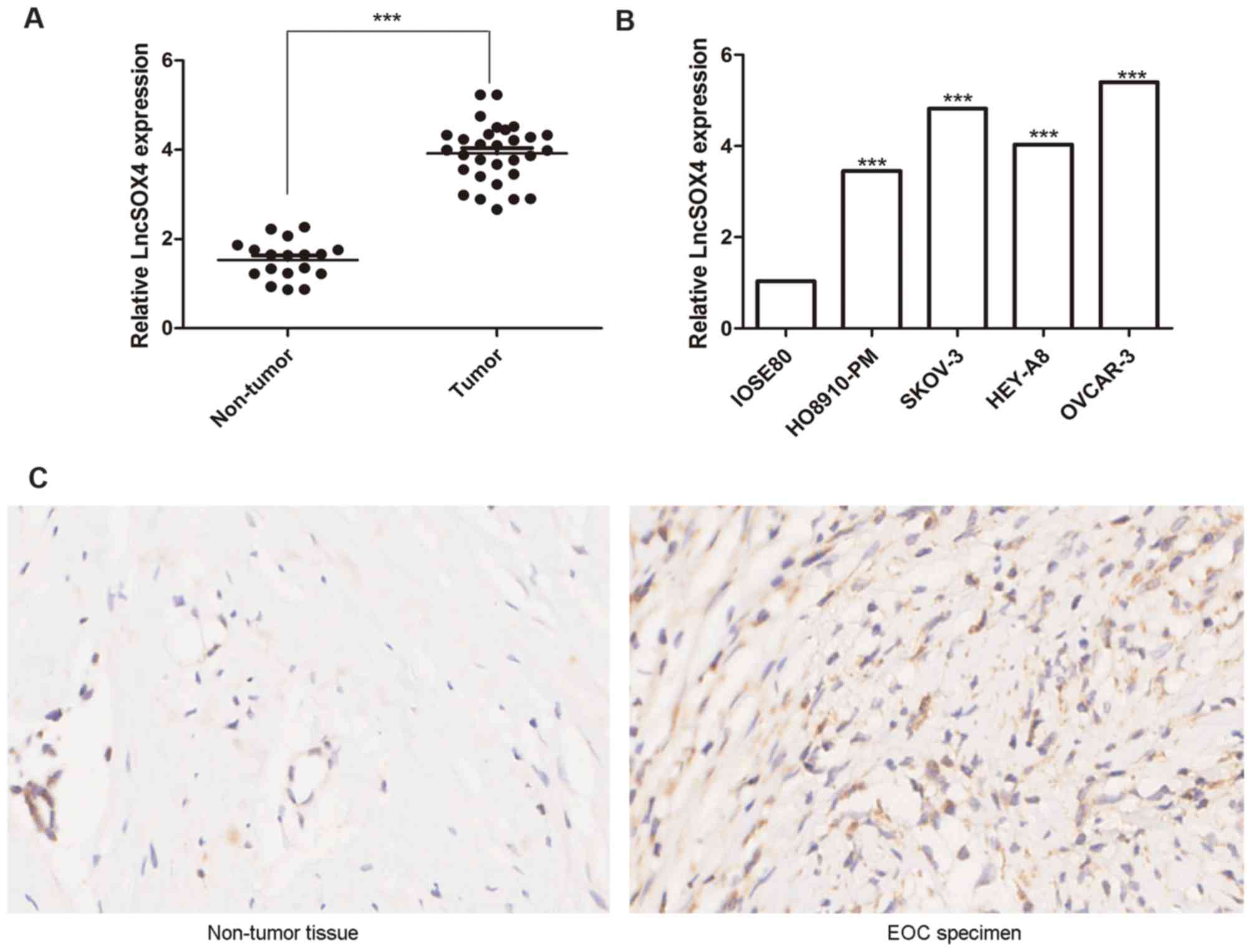
LncSOX4 is upregulated in EOC tissues
and cell lines
The expression levels of LncSOX4 in the ovarian
tissues of 30 patients with EOC and 18 control patients were
detected via RT-qPCR. Relative LncSOX4 expression levels in
patients with EOC were significantly upregulated compared with
non-tumor samples (3.98 vs. 1.71; P<0.001), the results of which
are presented in Fig. 1A. To
further confirm these results, LncSOX4 expression levels were
detected in the EOC cell lines HO8910-PM, SKOV-3, HEY-A8 and OVCAR3
and the non-tumor cell line IOSE-80. Significant differences in
LncSOX4 expression levels were observed between these five groups
(P<0.05). The results demonstrated that the expression levels of
LncSOX4 in EOC cell lines were significantly higher compared with
IOSE-80 (HO8910-PM vs. IOSE80, P<0.001; SKOV-3 vs. IOSE80,
P<0.001; HEY-A8 vs. IOSE80, P<0.001; OVCAR-3 vs. IOSE80,
P<0.001; Fig. 1B). A
representative EOC specimen and surrounding non-tumor tissue was
used for ISH analysis, and elevated expression levels of LncSOX4
were observed only in EOC tumor cells (Fig. 1C).
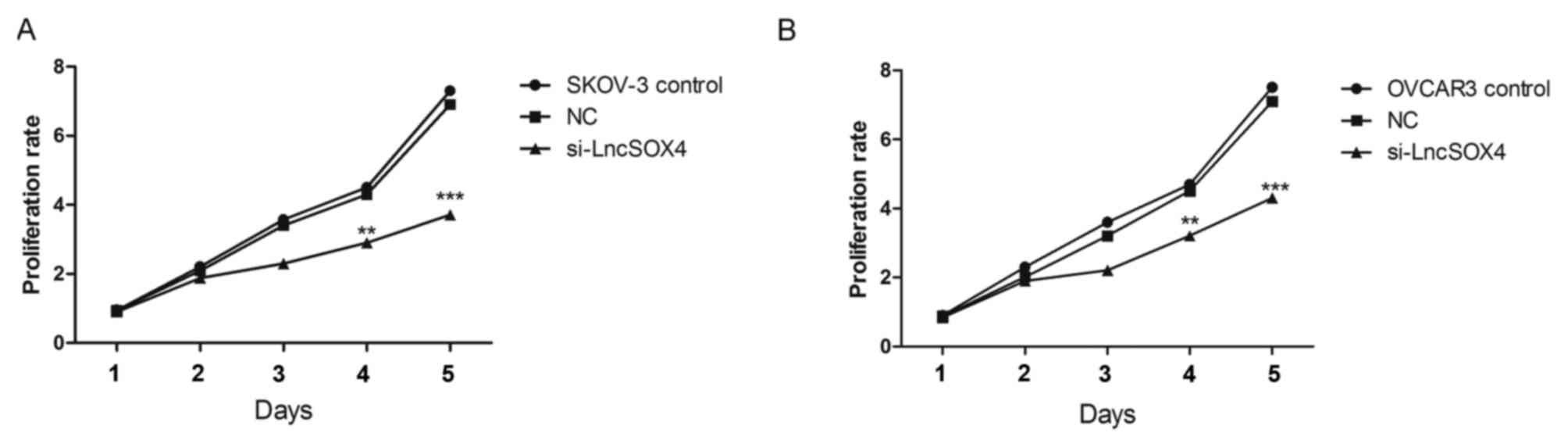
Knocking out LncSOX4 inhibits cell
proliferation in EOC cell lines
To further study the function of LncSOX4 in EOC,
siRNA targeting LncSOX4 was transfected to knock out LncSOX4 from
EOC cell lines. SKOV-3 and OVCAR3 were used for transfection and
further analysis. The transcription levels of LncSOX4 were
decreased by 58 and 69% following transfection with si-LncSOX4 in
SKOV-3 and OVCAR3 cells, respectively (si-LncSOX4 vs. Control,
P<0.001; si-LncSOX4 vs. NC, P<0.001; si-LncSOX4 vs. Control,
P<0.001; si-LncSOX4 vs. NC, P<0.001; Fig. 2), which confirmed the high
efficiency of si-LncSOX4. CCK-8 assays were performed following
transfection. In the first 72 h, the cell proliferation rate did
not exhibit significant differences between the SKOV-3, NC and
si-LncSOX4 groups, according to the results of analysis of variance
(P>0.05). Significant inhibition of cell proliferation was
observed following 72 h between the three groups (P<0.05): Day
4, SKOV-3 vs. NC, 2.7 vs. 4.1, P=0.013; OVCAR3 vs. NC, 3.2 vs. 4.3,
P=0.007; Day 5, SKOV-3 vs. NC, 3.8 vs. 7.1, P<0.001; OVCAR3 vs.
NC, 7.2 vs. 4.3, P<0.001; Fig.
3), which indicated that LncSOX4 promoted cell proliferation
and acted as an oncogene in EOC.
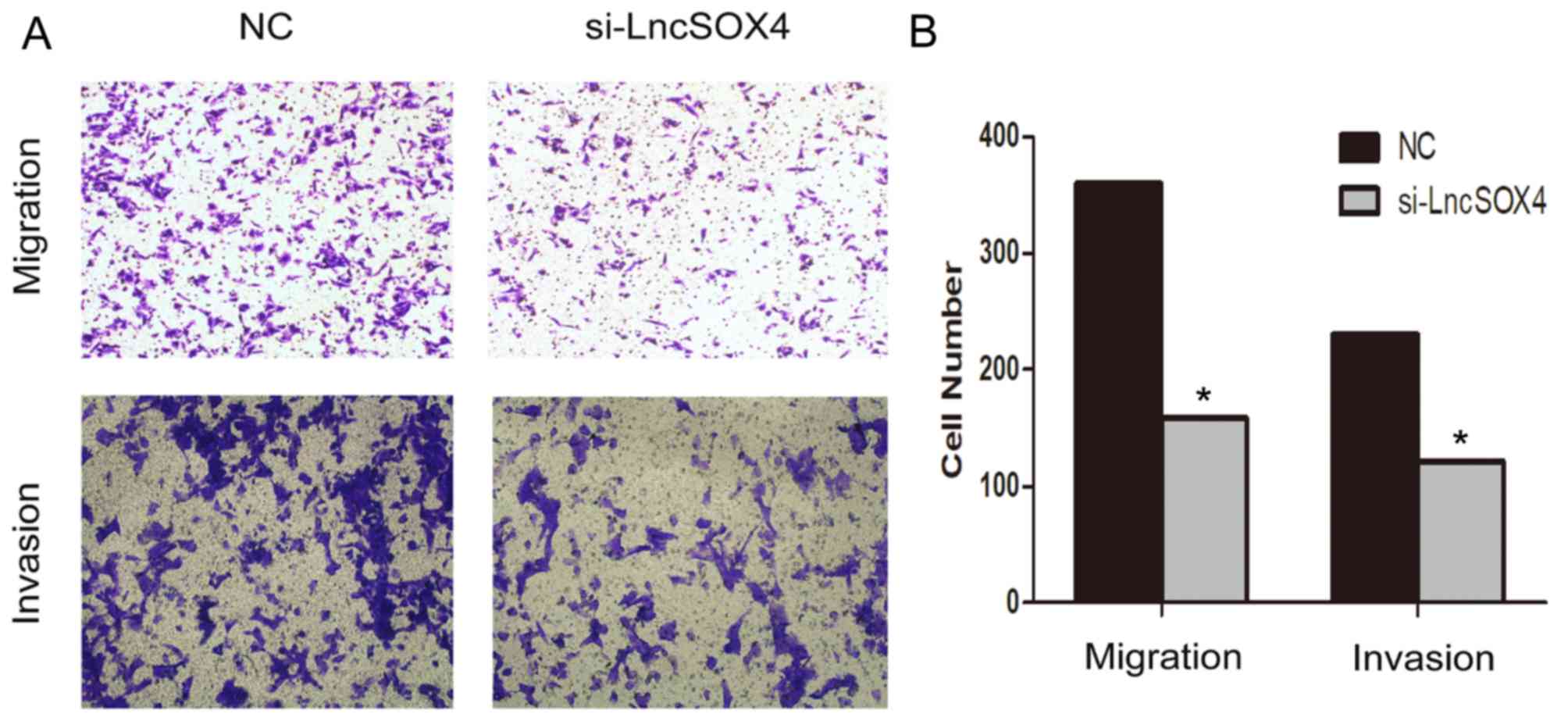
Knocking out LncSOX4 inhibits
migration and invasion in the SKOV3 cell line
A Transwell assay was performed to observe the role
of LncSOX4 in the SKOV3 cell line. Migratory and invasive abilities
were visually decreased following transfection with si-LncSOX4
(Fig. 4A). Migratory ability
decreased by up to 56% in the SKOV3 cell line and invasive ability
was inhibited by up to 47% (Fig.
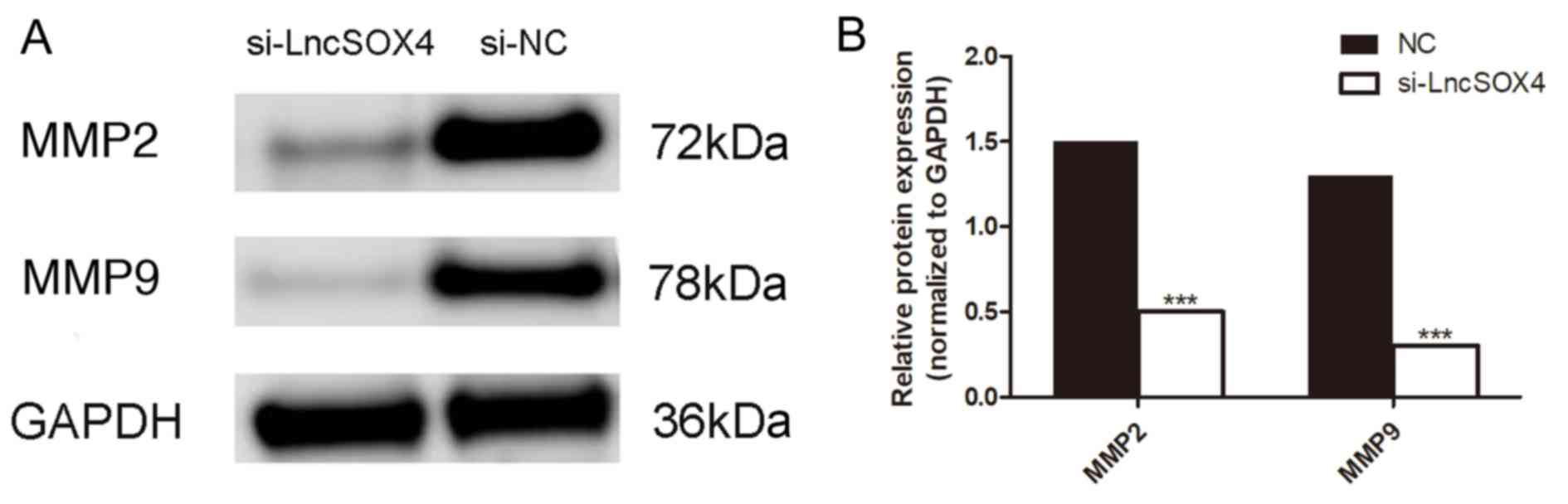
4B). In addition, the protein expression levels of MMP2 and
MMP9 were assessed in the si-LncSOX4 and NC groups in the SKOV3
cell line (Fig. 5A). Consistent
with the results of the Transwell assay, decreased expression of
MMP2 and MMP9 was identified in the si-LncSOX4 group (Fig. 5B), indicating that LncSOX4 may
upregulate MMP2 and MMP9 protein expression. These data suggested
that LncSOX4 promoted cell migration and invasion in an ovarian
cancer cell line.
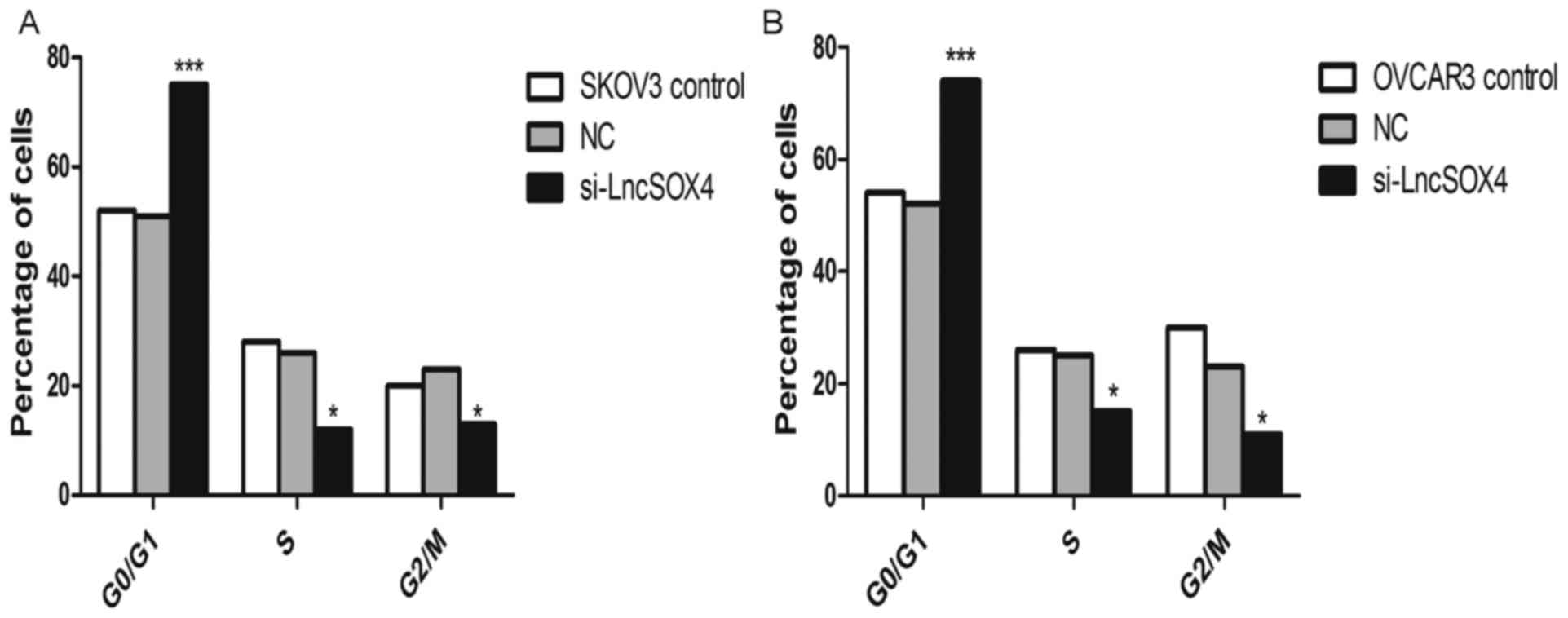
Knocking out LncSOX4 arrests the cell
cycle at the G0/G1 phase in EOC cell lines
The cell cycle was additionally analyzed in SKOV-3
and OVCAR3 cell lines following knockdown of LncSOX4. As presented
in Fig. 6, the percentage of cells
in the G0/G1 phase was significantly increased in the two cell
lines compared with the NC groups (SKOV-3, 77% vs. 50%, P<0.001;
OVCAR3, 73% vs. 52%, P<0.001). In addition, the proportions of
cells in S and G2/M phases decreased significantly in SKOV-3 and
OVCAR3 cells (P<0.05).
LncSOX4 expression is associated with
unfavorable prognostic factors in patients with EOC
The cell experiments indicated that LncSOX4 may
serve an oncogenic role in EOC cell lines. To assess whether the
expression of LncSOX4 was associated with clinicopathological
parameters, a comparative study was performed by sorting patients
into a high and low LncSOX4 expression group, according to the
median LncSOX4 expression level of 3.99. Table I summarizes the association between
LncSOX4 expression and clinical parameters. It was demonstrated
that the expression level of LncSOX4 was positively associated with
International Federation of Gynecology and Obstetrics stage
(16), tumor size and distant
metastases, with P-values of 0.000, 0.010 and 0.027, respectively,
and not associated with CA125 levels.
Discussion
Epithelial ovarian cancer, as the commonest type of
ovarian cancer, usually occurs in women >50 years old. Among all
cancer types in female patients, ovarian cancer has the highest
mortality rate, and the majority of patients with EOC are diagnosed
at a late stage due to its nonspecific signs and symptoms (17,18).
As a result, the 5-year survival rate of EOC is <30% (19). An increasing number of studies have
been conducted to elucidate the pathogenesis of this disease and to
identify circulating biomarkers for the early detection of EOC
(20–24).
lncRNAs consist of >200 nucleotides and have a
limited protein-coding capacity. lncRNAs have been reported to have
important effects on cell proliferation, apoptosis and
differentiation of various kinds of human cancer (25–27),
including EOC (28,29). LncSOX4 was first reported to
promote liver cancer progression (13). To determine whether this lncRNA
additionally served an oncogenic role in the tumorigenesis of EOC,
relevant experiments were performed in the present study, and it
was identified that the transcription levels of LncSOX4 were
significantly upregulated in EOC tissues and cell lines compared
with non-tumor controls. In addition, lncRNAs may promote cell
proliferation, migration and invasion. Cell cycle analysis
additionally demonstrated that inhibition of LncSOX4 was able to
arrest the cell cycle at the G0/G1 phase.
Deficiencies of diagnostic and prognostic factors
are responsible for the poor outcome of ovarian cancer. A number of
studies have been conducted to identify efficient serological
biomarkers of ovarian cancer (30–33).
However, there is no reliable biomarker at present. Recently,
lncRNAs have been analyzed in various studies on EOC, and certain
among them may have prognostic effects (14). It was reported that lncSOX4 was
associated with relapse and poor outcomes in EOC (14). The results of the present study are
consistent with previous findings that the high expression level of
LncSOX4 was associated with unfavorable clinical prognostic factors
(14). This indicated that LncSOX4
may serve as an unfavorable biomarker for EOC patients.
The present study investigated the role of LncSOX4
in EOC cell lines in vitro, although the molecular mechanism
remains unknown. It was previously reported that Pvt1 oncogene
(PVT1), an lncRNA which exhibits the potential to predict poor
prognosis for patients with stage I EOC, was upregulated by
transforming growth factor (TGF)β1 in hepatocellular carcinoma
(34). Martini et al
(14) isolated the most highly
correlated genes to perform computed pathway enrichment for
LncSOX4, and used the microGraphite pipeline to identify circuits
associated with LncRNAs. They reported that in LncSOX4 circuits the
upregulation of TGFβ1 was associated with poor prognosis for
patients with EOC, suggesting a potential molecular association
between PVT1 and LncSOX4.
In conclusion, LncSOX4 was upregulated in tissues
from patients with EOC and EOC cell lines, and knocking out LncSOX4
was able to inhibit cell proliferation and arrest the cell cycle at
the G0/G1 phase. Cellular migratory and invasive abilities
decreased following transfection with si-LncSOX4. Following
analysis of the clinical data, it was identified that LncSOX4 may
serve as a novel biomarker and therapeutic target for EOC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YL and DC designed the study. YL prepared the
manuscript and performed the statistical analysis. YW researched
the literature and contributed to manuscript revisions. YL and DY
performed the experiments. DC critically revised and edited the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Hubei Women and Children's Hospital.
Consent for publication
The patients provided written informed consent for
the publication of associated data and accompanying images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Clarke-Pearson DL: Clinical practice.
Screening for ovarian cancer. N Engl J Med. 361:170–177. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Diaz-Gil D, Fintelmann FJ, Molaei S, Elmi
A, Hedgire SS and Harisinghani MG: Prediction of 5-year survival in
advanced-stage ovarian cancer patients based on computed tomography
peritoneal carcinomatosis index. Abdom Radiol (NY). 41:2196–2202.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pignata S, Cannella L, Leopardo D, Pisano
C, Bruni GS and Facchini G: Chemotherapy in epithelial ovarian
cancer. Cancer Lett. 303:73–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kurman RJ and Shih IeM: The origin and
pathogenesis of epithelial ovarian cancer: A proposed unifying
theory. Am J Surg Pathol. 34:433–443. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vaughan S, Coward JI, Bast RC Jr, Berchuck
A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R,
Etemadmoghadam D, et al: Rethinking ovarian cancer: Recommendations
for improving outcomes. Nat Rev Cancer. 11:719–725. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huarte M and Rinn JL: Large non-coding
RNAs: Missing links in cancer? Hum Mol Genet. 19:R152–R161. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J, Li Z, Liu L, Wang Q, Li S, Chen
D, Hu Z, Yu T, Ding J, Li J, et al: Long noncoding RNA TSLNC8 is a
tumor suppressor that inactivates the interleukin-6/STAT3 signaling
pathway. Hepatology. 67:171–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shih JW, Chiang WF, Wu ATH, Wu MH, Wang
LY, Yu YL, Hung YW, Wang WC, Chu CY, Hung CL, et al: Long noncoding
RNA LncHIFCAR/MIR31HG is a HIF-1α co-activator driving oral cancer
progression. Nat Commun. 8:158742017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP
and Gao YF: LncSox4 promotes the self-renewal of liver
tumour-initiating cells through Stat3-mediated Sox4 expression. Nat
Commun. 7:125982016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Martini P, Paracchini L, Caratti G,
Mello-Grand M, Fruscio R, Beltrame L, Calura E, Sales G, Ravaggi A,
Bignotti E, et al: lncRNAs as novel indicators of patients'
prognosis in stage I epithelial ovarian cancer: A retrospective and
multicentric study. Clin Cancer Res. 23:2356–2366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jayson GC, Kohn EC, Kitchener HC and
Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fujiwara K, McAlpine JN, Lheureux S,
Matsumura N and Oza AM: Paradigm shift in the management strategy
for epithelial ovarian cancer. Am Soc Clin Oncol Educ Book.
35:e247–e257. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Montagnana M, Benati M and Danese E:
Circulating biomarkers in epithelial ovarian cancer diagnosis: From
present to future perspective. Ann Transl Med. 5:2762017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Holschneider CH and Berek JS: Ovarian
cancer: Epidemiology, biology, and prognostic factors. Semin Surg
Oncol. 19:3–10e. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun X, Zhang W, Li H, Niu C, Ou Y, Song L
and Zhang Y: Stonin 2 overexpression is correlated with unfavorable
prognosis and tumor invasion in epithelial ovarian cancer. Int J
Mol Sci. 18:pii: E1653. 2017. View Article : Google Scholar
|
|
21
|
Yang X, Liang L, Zhang XF, Jia HL, Qin Y,
Zhu XC, Gao XM, Qiao P, Zheng Y, Sheng YY, et al: MicroRNA-26a
suppresses tumor growth and metastasis of human hepatocellular
carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology.
58:158–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bagnoli M, Canevari S, Califano D, Losito
S, Maio MD, Raspagliesi F, Carcangiu ML, Toffoli G, Cecchin E,
Sorio R, et al: Development and validation of a microRNA-based
signature (MiROvaR) to predict early relapse or progression of
epithelial ovarian cancer: A cohort study. Lancet Oncol.
17:1137–1146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Walsh CS, Ogawa S, Karahashi H, Scoles DR,
Pavelka JC, Tran H, Miller CW, Kawamata N, Ginther C, Dering J, et
al: ERCC5 is a novel biomarker of ovarian cancer prognosis. J Clin
Oncol. 26:2952–2958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gagnon A and Ye B: Discovery and
application of protein biomarkers for ovarian cancer. Curr Opin
Obstet Gynecol. 20:9–13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xing Z, Lin A, Li C, Liang K, Wang S, Liu
Y, Park PK, Qin L, Wei Y, Hawke DH, et al: lncRNA directs
cooperative epigenetic regulation downstream of chemokine signals.
Cell. 159:1110–1125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Trimarchi T, Bilal E, Ntziachristos P,
Fabbri G, Dalla-Favera R, Tsirigos A and Aifantis I: Genome-wide
mapping and characterization of Notch-regulated long noncoding RNAs
in acute leukemia. Cell. 158:593–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Abraham JM and Meltzer SJ: Long noncoding
RNAs in the pathogenesis of barrett's esophagus and esophageal
carcinoma. Gastroenterology. 153:27–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hua F, Li CH, Chen XG and Liu XP: Long
noncoding RNA CCAT2 knockdown suppresses tumorous progression by
sponging miR-424 in epithelial ovarian cancer. Oncol Res.
26:241–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiu YL, Sun KX, Chen X, Chen S, Zhao Y,
Guo QG and Zong ZH: Upregulation of the lncRNA Meg3 induces
autophagy to inhibit tumorigenesis and progression of epithelial
ovarian carcinoma by regulating activity of ATG3. Oncotarget.
8:31714–31725. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Scaletta G, Plotti F, Luvero D,
Capriglione S, Montera R, Miranda A, Lopez S, Terranova C, De Cicco
Nardone C and Angioli R: The role of novel biomarker HE4 in the
diagnosis, prognosis and follow-up of ovarian cancer: A systematic
review. Expert Rev Anticancer Ther. 17:827–839. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Resnick KE, Alder H, Hagan JP, Richardson
DL, Croce CM and Cohn DE: The detection of differentially expressed
microRNAs from the serum of ovarian cancer patients using a novel
real-time PCR platform. Gynecol Oncol. 112:55–59. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kan CW, Hahn MA, Gard GB, Maidens J, Huh
JY, Marsh DJ and Howell VM: Elevated levels of circulating
microRNA-200 family members correlate with serous epithelial
ovarian cancer. BMC Cancer. 12:6272012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo F, Tian J, Lin Y, Jin Y, Wang L and
Cui M: Serum microRNA-92 expression in patients with ovarian
epithelial carcinoma. J Int Med Res. 41:1456–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX,
Ye C, Yang N, Zhou WP, Li WL, Li W and Sun SH: Oncofetal long
noncoding RNA PVT1 promotes proliferation and stem cell-like
property of hepatocellular carcinoma cells by stabilizing NOP2.
Hepatology. 60:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|