Introduction
The ST3GAL4 gene encodes for β-galactoside
α-2,3-sialyltransferase 4 (ST3Gal IV), enzyme that transfers sialic
acid to the type 2 precursor (Galβ1,4GlcNAc). It is involved in the
biosynthesis of the sLe(x) and sulfo-sLe(x) epitopes (1–3).
Alteration of sialyltransferase gene expression has
been implicated in carcinogenesis. Specifically, expression of this
gene is altered in different cancer types. ST3GAL4
expression is decreased in renal cell carcinoma and cervical cancer
(4,5), whereas it is increased in gastric
cancer and in premalignant lesions of the cervix (6,7). For
other cancer types, including colorectal cancer, the expression
level of ST3GAL4 does not demonstrate any changes (8).
Sialyltransferase genes are regulated in a tissue
specific manner; that is, gene expression may be regulated by
different promoters, and their alternative use can regulate gene
expression in different tissues. Thus far, five different promoters
have been reported for the ST3GAL4 gene: pA, pB1, pB2, pB3
and pBx (9,10). Complexity of the gene expression
increases since six mRNA isoforms have also been reported: A1, A2,
B1, B2, B3 and B4 (1,11,12)
and these may be produced by combinations of promoter utilization
and alternative splicing.
The transcript isoforms (9,10)
are now listed in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) as variant
transcripts. The expression of these variants has not been
evaluated in cervical cancer cells. To better understand the
mechanisms by which the expression of the ST3GAL4 gene is
regulated in cervical cancer, it is important to determine the
transcript variants present in this tissue and how their expression
is modified during tumour transformation.
Materials and methods
Alignment analysis of transcript
variants of the ST3GAL4 gene
An alignment analysis was performed to compare the
sequences of the transcript variants available in GenBank and the
mRNA B isoforms of the ST3GAL4 gene reported by Kitagawa
et al in 1996 (11). The
gene bank accession numbers for the ST3GAL4 isoforms are: V1
isoform: NM_006278; V2 isoform: NM_001254757; V3 isoform:
NM_001254758 and V4 isoform: NM_001254759.
Cell culture
The human keratinocyte cell line HaCaT was obtained
from the Centro de Investigación Biomédica de Occidente
(Guadalajara, Mexico), and human cervical cell lines (SiHa, HeLa,
C33A) were obtained from the Centro de Investigación Biomédica de
Oriente (Puebla, Mexico). The cells were grown in monolayer culture
in p25 cm2 flasks and were maintained at 37°C in an
atmosphere of CO2 in Dulbecco's modified Eagle's medium
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) with 10 mmol/l
HEPES, which was supplemented with 10% fetal bovine serum, 100
IU/ml penicillin and 100 µg/ml streptomycin (Sigma-Aldrich; Merck
KGaA). The culture medium was replaced every two days. Subconfluent
adherent cells were harvested using a mixture of trypsin (0.025%)
and EDTA (0.02%); (Sigma-Aldrich; Merck KGaA) and were washed with
phosphate buffered saline.
Sample tissue
Cervical specimens were obtained at the Centro
Médico Nacional sXXI (Mexico City, Mexico), between January 2013
and January 2014, from female patients (aged 18–65 years). Samples
of squamous cell carcinoma of the cervix were obtained from women
that underwent radical hysterectomy. Samples exhibiting
premalignant lesions (low and high grade squamous intraepithelial
lesions) were obtained from women who were treated by cervical
conisation. Normal tissue was obtained from women undergoing
hysterectomy for uterine miomatosis. Women with a clinical history
of other cancer types, as well as samples with degraded RNA, were
excluded from the present study. All data, including age and
pathological results, were prospectively recorded. The present
study was approved by the Ethics Committee of Instituto Mexicano
del Seguro Social (Mexico City, Mexico; Number R-2012-785-061). All
patients provided informed consent according to the guidelines of
the Human Ethics Committee. In total, 8 normal samples, 7 samples
of premalignant lesions and 8 samples of cervical cancer were
included.
RNA isolation from cells and
biopsies
Total RNA was extracted from cell lines using a
Nucleo Spin RNA II kit (Macherey-Nagel GmbH, Düren, Germany)
according to the manufacturer's protocols.
Biopsies were maintained in RNAlater solution
(Qiagen, Inc., Valencia, CA, USA), and 25 mg of tissue was
disrupted with the TissueLyser II (Qiagen, Inc.) for 2 min.
Subsequently, RNA was extracted with an RNeasy Plus Mini kit
(Qiagen, Inc.) according to the manufacturer's protocols.
cDNA synthesis
First-strand cDNA synthesis was performed with a
RevertAid H Minus First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc., Waltham, MA, US) using the following
concentrations: 1X of reaction buffer, 1 U/µl of Ribolock RNAse
Inhibitor, 1 mM of dNTP mix, 10 U/µl of RevertAid H Minus M-MuLV
Reverse Transcriptase, 5 µM of random primers and 1 µg of total
RNA, in a final volume of 20 µl, according to the manufacturer's
protocols. The reaction was incubated for 5 min at 25°C followed by
incubation for 60 min at 42°C.
Reverse
transcription-semi-quantitative polymerase chain reaction
(RT-sqPCR) analysis
RT-sqPCR was performed on a 1000 Touch thermal
cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA) in a final
volume of 20 µl using the following: 10 µl of 2X PCR Master Mix
(Promega Corporation, Madison, WI, USA), 1 µl of 10 µM forward and
reverse primer, and 2 µl of cDNA. The primer sequences were as
follows: V1 transcript forward 5′-gac tgt gct gga ggt gac ag-3′,
reverse 5′-acc atg ttt ctc agc agg ca-3′; V2 forward 5′-gaa ccg tgc
tgc ccc gcc cc-3′; reverse 5′-ggg act tgc tgac cat gtt t-3′; V3
forward 5′-ttt gta gtg ttt ccc gcc ca-3′, reverse 5′-gct gac cat
gtt tct cag ca-3′. A positive control containing primers for
hypoxanthine-guanine phosphoribosyltransferase (HPRT) (forward
5′-ccc tgg cgt cgt gat tag tga-3′; reverse 5′-agc aag acg ttc agt
cct gtc c-3′) and a negative control (with no template) were added
into each assay. The program cycle was: 95°C for 2 min, 40 cycles
of 95°C for 30 sec, 56°C for 1 min and 72°C for 1 min. A total of
40 cycles were performed to ensure for the successful detection of
RNA variants with a low copy number. The presence of the variants
was confirmed, by performing the reactions twice. The amplification
product was assessed on an agarose gel for low molecular weight
fragments at a 2% concentration. The gels were stained with
ethidium bromide (1 mg/ml), observed in an ultraviolet
transilluminator, and then photographed using a digital camera
(ELP110HS; Canon, Inc., Tokyo, Japan). To confirm that the
amplified product corresponded to the variant transcript, direct
sequencing of the PCR products was performed using the Sanger
method in the Laboratory of Biodiversity and Genomics, CINVESTAV
(Irapuato, Mexico).
RT quantitative (q)PCR validation
To prove that the efficiencies of the ST3Gal
4 gene and HPRT amplification were similar and optimal, RT-qPCR
was performed to validate the endogenous gene and to quantify the
V1 transcription in cell lines and biopsies. Each reaction was
performed in a final volume of 10 µl with the concentrations: 5 µl
of 2X Maxima SYBR Green/Rox qPCR Master Mix (Thermo Fisher
Scientific, Inc.), 1 µl of 10 mM forward and reverse primers for
the V1 isoform of the ST3GAL4 gene or HPRT, and cDNA from
SiHa, HeLa, and C33A cell lines that had been previously
synthesized, using the aforementioned procedure. A standard curve
was determined with the concentrations: 2, 1, 0.5, 0.25, 0.125 and
0.0625 ng/µl. The reactions were performed in triplicate using a
StepOne Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.), and the conditions were as follows: 95°C for 10
min, followed by 40 cycles of 95°C for 10 sec, 30 sec at 60°C, and
70°C for 30 sec.
Relative quantification of the V1
transcript in cell lines and biopsies
Relative quantification was performed using the
comparative CT method with the formula: 2−ΔΔCq (13). The qPCR reaction was performed with
the StepOneReal-Time PCR System (Applied Biosytems; Thermo Fisher
Scientific, Inc.). The final reaction volume of 10 µl included 1 µl
of cDNA template (0.5 ng final concentration in cell lines and 12
ng in biopsies), 5 µl of 2X Maxima SYBR Green/Rox qPCR Master Mix
(Thermo Fisher Scientific, Inc.), 0.5 µl of forward and reverse
primers (0.5 µM final concentration) and 3 µl of RNase free water.
RT-qPCR was performed under the following conditions: 1 cycle at
50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 sec, and
60°C for 1 min. Triplicates of each sample were analysed, and
negative controls with no cDNA template were included in each
assay. Transcript levels were normalized against HPRT
expression.
Statistical analysis
Statistical analysis was performed using the Graph
Pad program (version 7; GraphPad Software, Inc., La Jolla, CA,
USA). A one-way analysis of variance followed by Tukey's post-test
was performed for multiple comparisons. Data are presented as mean
± standard error deviation. P<0.05 was considered to indicate a
statistically significant difference. All experiments were repeated
in triplicate.
Results
Alignment analysis of transcript
variants of the ST3GAL4 gene
Since isoforms A1 and A2 are reported exclusively in
testicle, ovary and placenta these isoforms were excluded from
subsequent analysis. First, an alignment was performed among
isoforms B1, B2, B3 and B4 reported by Kitagawa et al
(11) with those transcript
variants reported in GenBank to determine their correspondence and
their sequence identity. Table I
indicates the B isoforms whose sequence corresponded to the current
nomenclature of transcript variants reported in GenBank and the
protein isoform coded. Analysis demonstrated that B1 isoform
corresponds to V1 variant, BX to V2 and B3 to V4. The B2 isoform
sequence was not available in the GenBank data base, and the V3
transcript was not reported by Kitagawa et al (11) (Access date April 29, 2016).
 | Table I.Current status of ST3GAL4 mRNA
variant nomenclature previously reported by Kitagawa et al
(11). |
Table I.
Current status of ST3GAL4 mRNA
variant nomenclature previously reported by Kitagawa et al
(11).
| Current nomenclature
of mRNA variants and accession numbera | Isoforms B reported
by Kitagawa et al (11) | Transcript size
(bases) | Protein isoform and
accession number |
|---|
| V1 | B1 | 2053 b | Isoform 1 |
| NM_006278 |
|
| NP_006269 |
| V2 | BX | 1854 b | Isoform 2 |
| NM_001254757 |
|
| NP_001241686 |
| V3 | – | 1727 b | Isoform 2 |
| NM_001254758 |
|
| NP_001241687 |
| V4 | B3 | 1871 b | Isoform 3 |
| NM_001254759 |
|
| NP_001241688 |
| – | B2 | – | – |
Analysis of transcript variants in
cervical cell lines
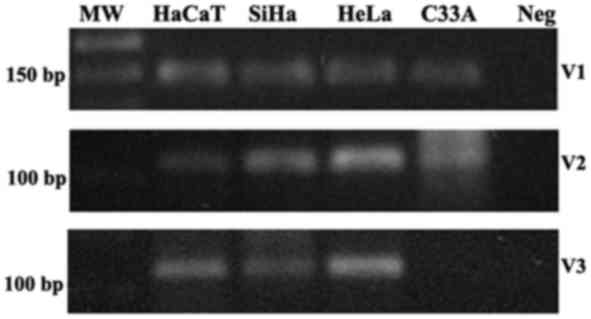
Next, the presence of the transcripts variants V1,
V2 and V3 was evaluated in cervical cell lines. The RT-PCR analysis
demonstrated that V1 and V2 transcripts were present in all the
cervical cell lines, SiHa, HeLa, and C33A, and in the human
keratinocyte cell line HaCaT; V3 transcript was present in SiHa,
HeLa and HaCaT cell lines, however not in C33A (Fig. 1).
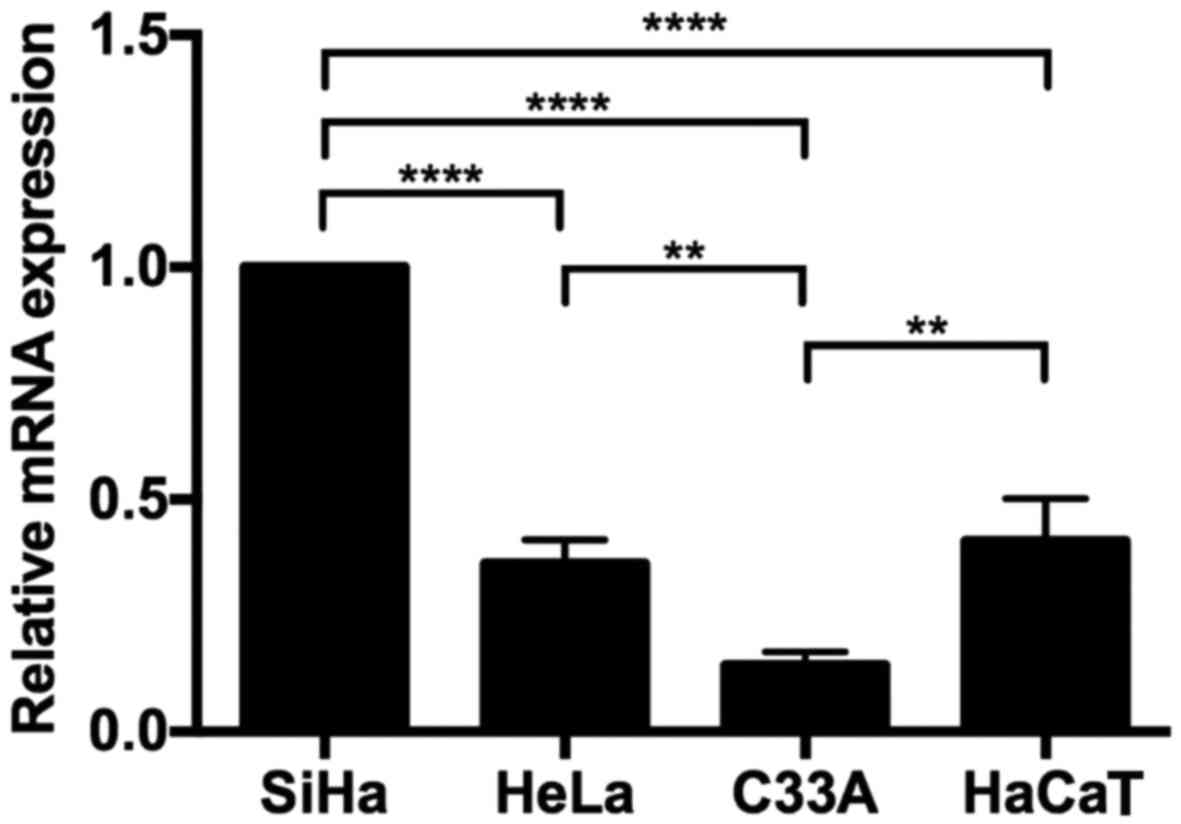
Relative quantification of the V1
variant in cervical cell lines
The expression level of the V1 transcript was
determined in the cervical cell lines SiHa, HeLa and C33A. The ΔΔCq
analysis demonstrated that SiHa cells exhibited the highest
expression (Fig. 2) compared with
HeLa, C33A and HaCaT. In every case, significant differences were
identified.
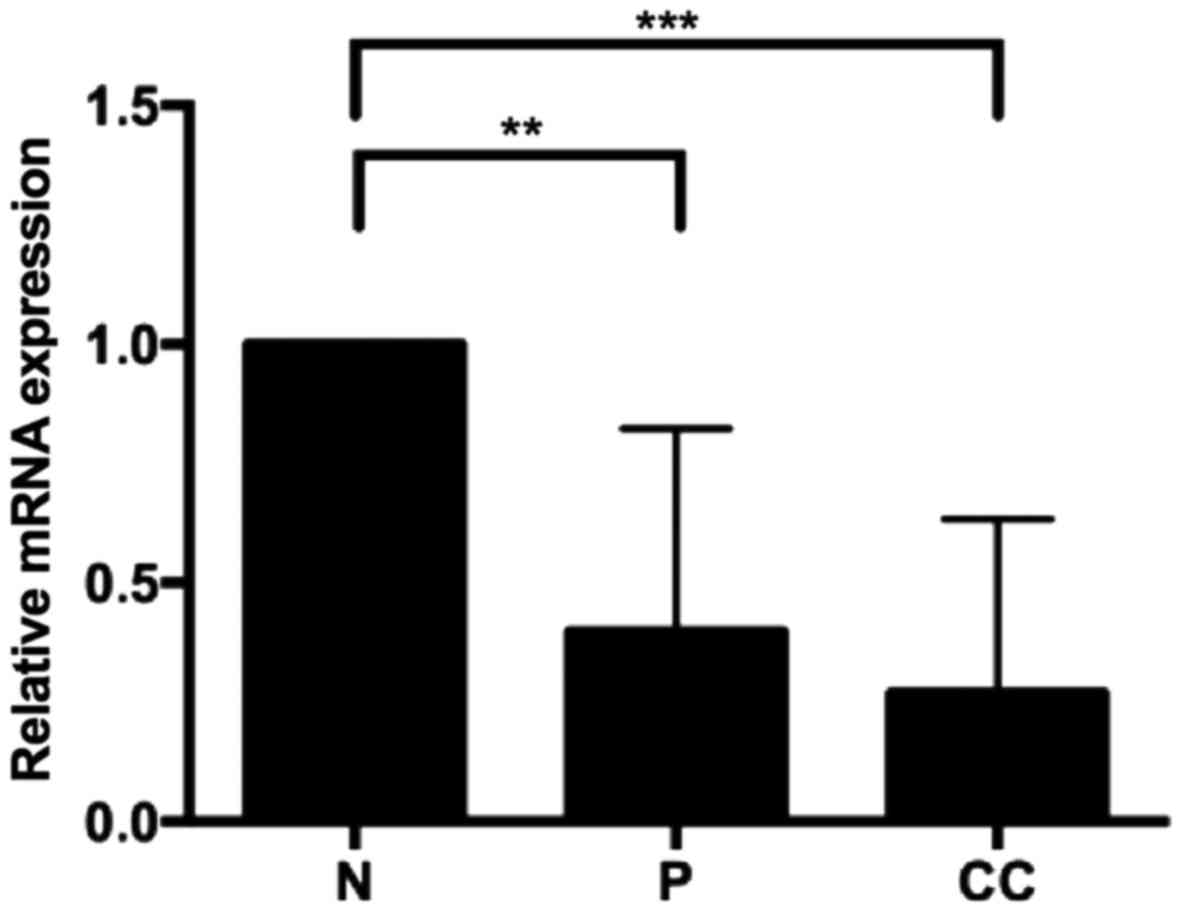
Quantification of transcript variant 1
in patients
Next, the V1 transcript levels in biopsies were
analysed to determine whether the V1 transcript modifies its
expression in relation to cervical neoplasia grade. Relative
quantification was performed by RT-qPCR using 8 samples of normal
tissue, 7 samples of squamous intraepithelial lesions (low and high
grade) and 8 samples of cervical cancer. Premalignant lesions and
cervical cancer demonstrated a decreased level of V1 transcript
compared with normal tissue (Fig.
3). In every case, significant differences were identified.
Discussion
The ST3GAL4 gene encodes sialyltransferase
ST3Gal IV. It produces at least 5 transcripts by alternative
promoter utilization in combination with alternative splicing
(1). This gene has been reported
to have increased expression in pancreatic cancer (14). In contrast, ST3GAL4 has
decreased expression in renal cancer and is associated with
malignant progression (4).
Decreased expression has also been reported in cervical cancer
(5). The present study detected
the presence of the variants V1, V2 and V3 in cervical cell lines.
The expression level of the V1 transcript in cervical cell lines
and in cervical tissue with different neoplasia grades was
analysed. The V1 transcript represents the longest transcript and
encodes protein isoform 1. This transcript demonstrated decreased
expression in premalignant and malignant tissues. These results
demonstrated that glycosylation changes occur in previous stages of
cancer, as has been reported for sialic acid and sialyltransferase
expression (7,15,16).
It is important to mention that ST3GAL4 encodes the
sialyltransferase that is involved in the synthesis of the sLe(x)
antigen, which has been reported to increase in different cancer
types including cervical cancer and premalignant lesions of the
cervix (17,18). This suggests that other
sialyltransferases could participate in the synthesis of the sLe(x)
antigen, including ST3Gal III (19). The expression level of sLe(x) does
not appear to be associated with the mRNA level reported in
cervical cancer samples where a diminished expression of
ST3GAL4 has been detected. The increased level of sLewis(x)
could be the result of an increased expression of ST3GAL3
that may participate in the synthesis of these antigens, as has
been reported previously (20).
Increased expression of the ST3GAL3 gene has been reported
in cervical cancer and premalignant lesions, and this could explain
the increased expression of the sLe(x) antigen (5,7,20,21).
Transcript variants of this and other glycogenes may
be regulated in a different manner as a result of promoter
utilization. Analysis of the transcript variants could help to
elucidate the regulation mechanisms that could be participating in
malignant transformation.
In conclusion, the V1 transcript of the
ST3GAL4 gene has decreased expression in premalignant and
malignant tissues. The expression level analysis of this transcript
may be utilized to develop diagnostic methods for the early
detection of cervical tissue transformation.
Acknowledgements
The authors would like thank Miss Jocelyn Serna
Villalobos from the Universidad Politécnica Metropolitana de Puebla
(Puebla, Mexico) for her technical support.
Funding
The present study was supported by Instituto
Mexicano del Seguro Social (grant no. FIS/IMSS/PROT/G14/1293),
Consejo Nacional de Ciencia y Tecnología (CONACYT); Fondo Sectorial
de Investigación en Salud (grant no. SALUD-2012-01-180219); Fondo
Redes Temáticas de Investigación-CONACYT (grant no. 253596). IM was
supported by an MD fellowship from CONACYT (grant no. 444914) and
IMSS (grant no. 98225300).
Availability of data and materials
All data generated or analysed during this study are
included in this published article.
Authors' contributions
CGR, CGF, CRA and TAG were responsible for the
recruitment of patients. LMF, NRM, COM, RMS, GSL, LRC, PMM, IMC,
AAL and LJS performed the experiments. AAL, LJS, GSL JRL and VVR
contributed to the study design, data analysis and the writing of
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Instituto Mexicano del Seguro Social (Mexico City,
Mexico; Number R-2012-785-061). All patients provided informed
consent according to the guidelines of the Ethics Committee of
Instituto Mexicano del Seguro Social.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kitagawa H and Paulson JC: Cloning of a
novel alpha 2,3-sialyltransferase that sialylates glycoprotein and
glycolipid carbohydrate groups. J Biol Chem. 269:1394–1401.
1994.PubMed/NCBI
|
|
2
|
Higai K, Miyazaki N, Azuma Y and Matsumoto
K: Interleukin-1beta induces sialyl Lewis X on hepatocellular
carcinoma HuH-7 cells via enhanced expression of ST3Gal IV and FUT
VI gene. FEBS Lett. 580:6069–6075. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Colomb F, Krzewinski-Recchi MA, Machour F,
Mensier E, Jaillard S, Steenackers A, Harduin-Lepers A, Lafitte JJ,
Delannoy P and Groux-Degroote S: TNF regulates sialyl-Lewisx and
6-sulfo-sialyl-Lewisx expression in human lung through
up-regulation of ST3GAL4 transcript isoform BX. Biochimie.
94:2045–2053. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saito S, Yamashita S, Endoh M, Yamato T,
Hoshi S, Ohyama C, Watanabe R, Ito A, Satoh M, Wada T, et al:
Clinical significance of ST3Gal IV expression in human renal cell
carcinoma. Oncol Rep. 9:1251–1255. 2002.PubMed/NCBI
|
|
5
|
Wang PH, Li YF, Juang CM, Lee YR, Chao HT,
Tsai YC and Yuan CC: Altered mRNA expression of sialyltransferase
in squamous cell carcinomas of the cervix. Gynecol Oncol.
83:121–127. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jun L, Yuanshu W, Yanying X, Zhongfa X,
Jian Y, Fengling W, Xianjun Q, Kokudo N, Wei T, Weixia Z and
Shuxiang C: Altered mRNA expressions of sialyltransferases in human
gastric cancer tissues. Med Oncol. 29:84–90. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
López-Morales D, Velázquez-Márquez N,
Valenzuela O, Santos-López G, Reyes-Leyva J and Vallejo-Ruiz V:
Enhanced sialyltransferases transcription in cervical
intraepithelial neoplasia. Invest Clin. 50:45–53. 2009.PubMed/NCBI
|
|
8
|
Petretti T, Kemmner W, Schulze B and
Schlag PM: Altered mRNA expression of glycosyltransferases in human
colorectal carcinomas and liver metastases. Gut. 46:359–366. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taniguchi A, Morishima T, Tsujita Y,
Matsumoto Y and Matsumoto K: Genomic structure, expression, and
transcriptional regulation of human Gal beta 1,3 GalNAc alpha
2,3-sialyltransferase gene. Biochem Biophys Res Commun.
300:570–576. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taniguchi A: Promoter structure and
transcriptional regulation of human beta-galactoside alpha2,
3-sialyltransferase genes. Curr Drug Targets. 9:310–316. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kitagawa H, Mattei MG and Paulson JC:
Genomic organization and chromosomal mapping of the Gal beta
1,3GalNAc/Gal beta 1,4GlcNAc alpha 2,3-sialyltransferase. J Biol
Chem. 271:931–938. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Taniguchi A and Matsumoto K:
Down-regulation of human sialyltransferase gene expression during
in vitro human keratinocyte cell line differentiation. Biochem
Biophys Res Commun. 243:177–183. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pérez-Garay M, Arteta B, Llop E, Cobler L,
Pagès L, Ortiz R, Ferri MJ, de Bolós C, Figueras J, de Llorens R,
et al: α2,3-Sialyltransferase ST3Gal IV promotes migration and
metastasis in pancreatic adenocarcinoma cells and tends to be
highly expressed in pancreatic adenocarcinoma tissues. Int J
Biochem Cell Biol. 45:1748–1757. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Roy A and Chakraborty S: Detection of
cancer cervix by estimation of sialic acid. J Indian Med Assoc.
103:589–590. 2005.PubMed/NCBI
|
|
16
|
López-Morales D, Reyes-Leyva J,
Santos-López G, Zenteno E and Vallejo-Ruiz V: Increased expression
of sialic acid in cervical biopsies with squamous intraepithelial
lesions. Diagn Pathol. 5:742010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Velázquez-Márquez N, Santos-López G,
Jiménez-Aranda L, Reyes-Leyva J and Vallejo-Ruiz V: Sialyl Lewis ×
expression in cervical scrapes of premalignant lesions. J Biosci.
37:999–1004. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Engelstaedter V, Fluegel B, Kunze S, Mayr
D, Friese K, Jeschke U and Bergauer F: Expression of the
carbohydrate tumour marker Sialyl Lewis A, Sialyl Lewis X, Lewis Y
and Thomsen-Friedenreich antigen in normal squamous epithelium of
the uterine cervix, cervical dysplasia and cervical cancer. Histol
Histopathol. 27:507–514. 2012.PubMed/NCBI
|
|
19
|
Grahn A, Barkhordar GS and Larson G:
Cloning and sequencing of nineteen transcript isoforms of the human
alpha2,3-sialyltransferase gene, ST3Gal III; its genomic
organization and expression in human tissues. Glycoconj J.
19:197–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui HX, Wang H, Wang Y, Song J, Tian H,
Xia C and Shen Y: ST3Gal III modulates breast cancer cell adhesion
and invasion by altering the expression of invasion-related
molecules. Oncol Rep. 36:3317–3324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang PH, Li YF, Juang CM, Lee YR, Chao HT,
Ng HT, Tsai YC and Yuan CC: Expression of sialyltransferase family
members in cervix squamous cell carcinoma correlates with lymph
node metastasis. Gynecol Oncol. 86:45–52. 2002. View Article : Google Scholar : PubMed/NCBI
|