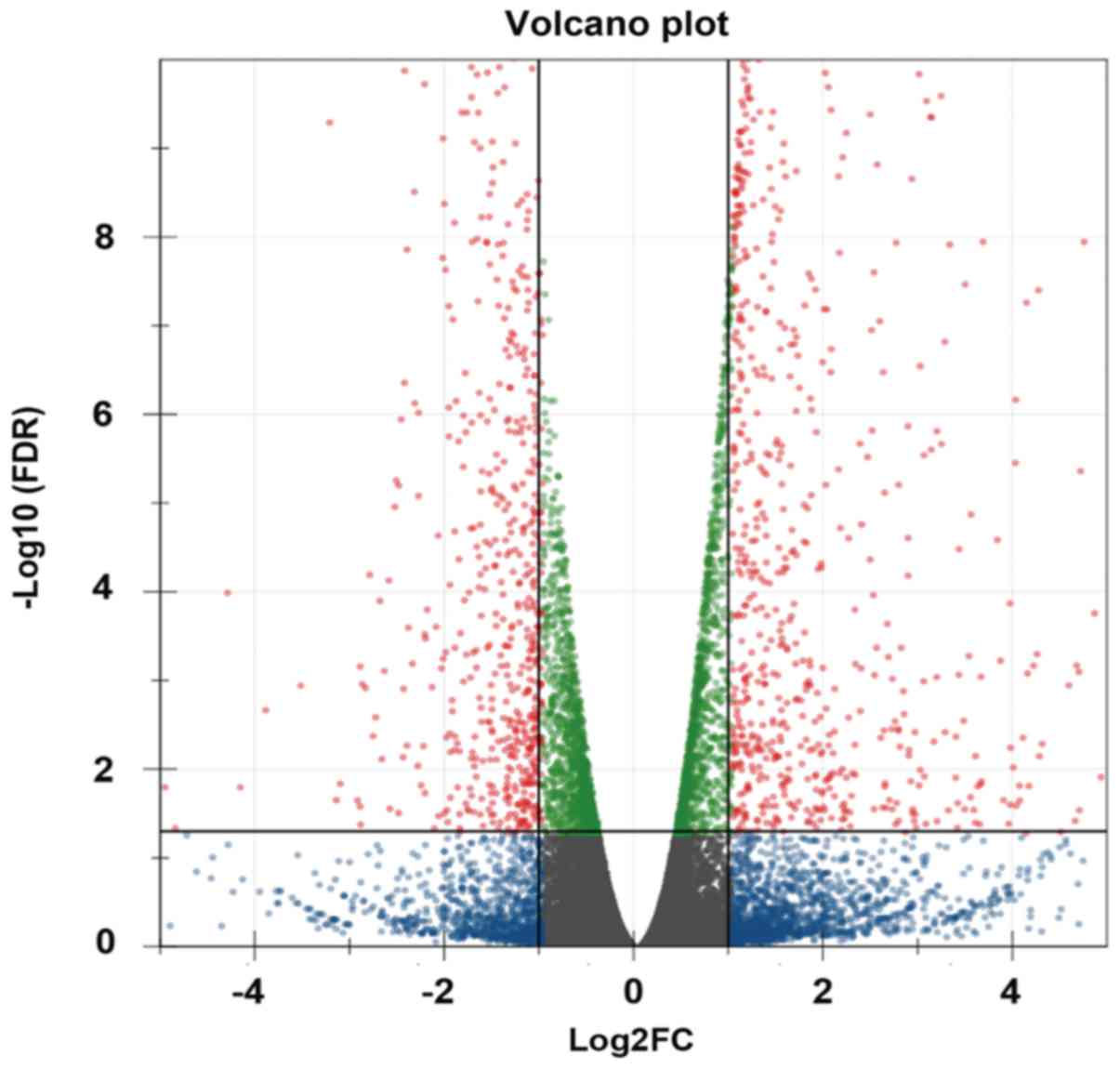
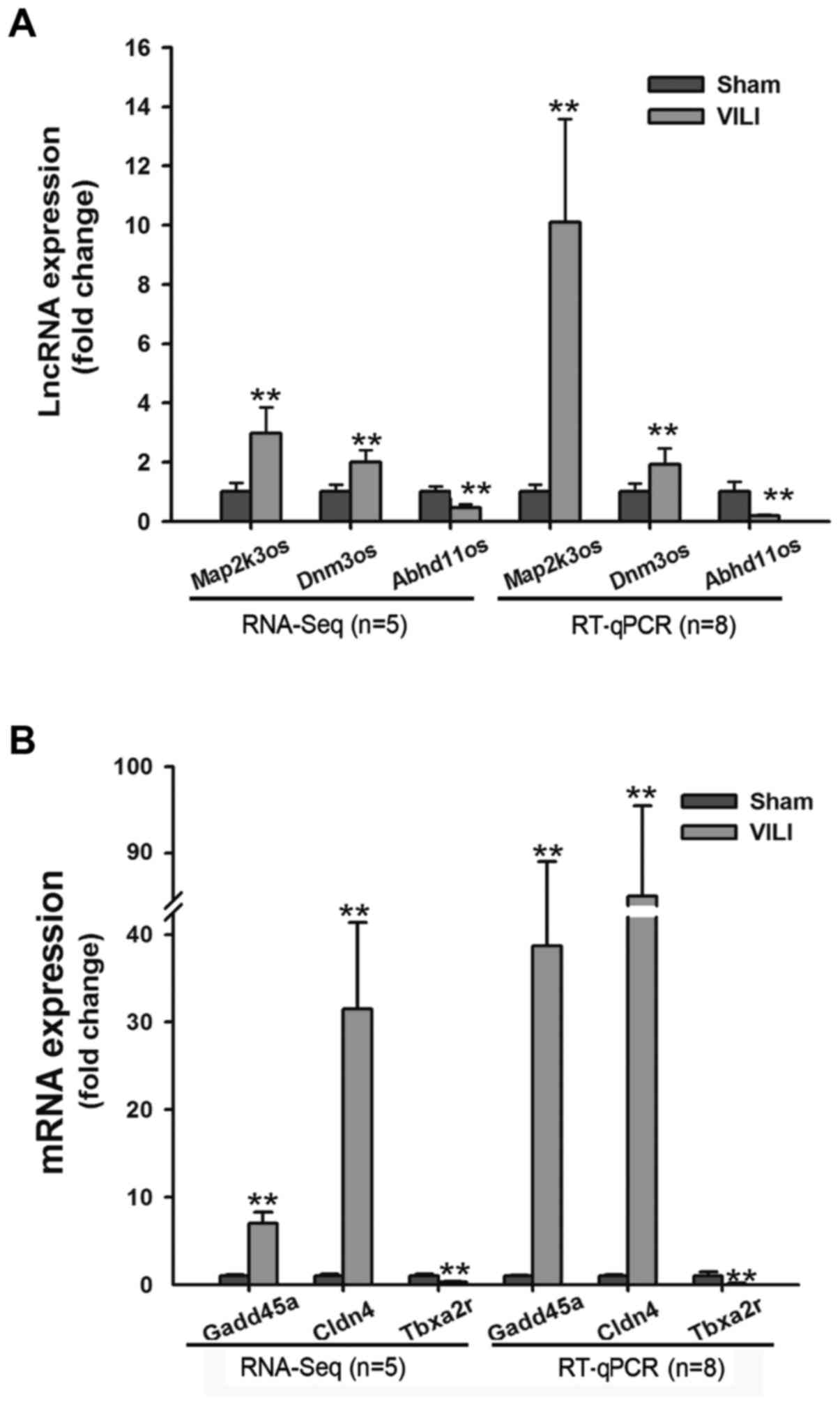
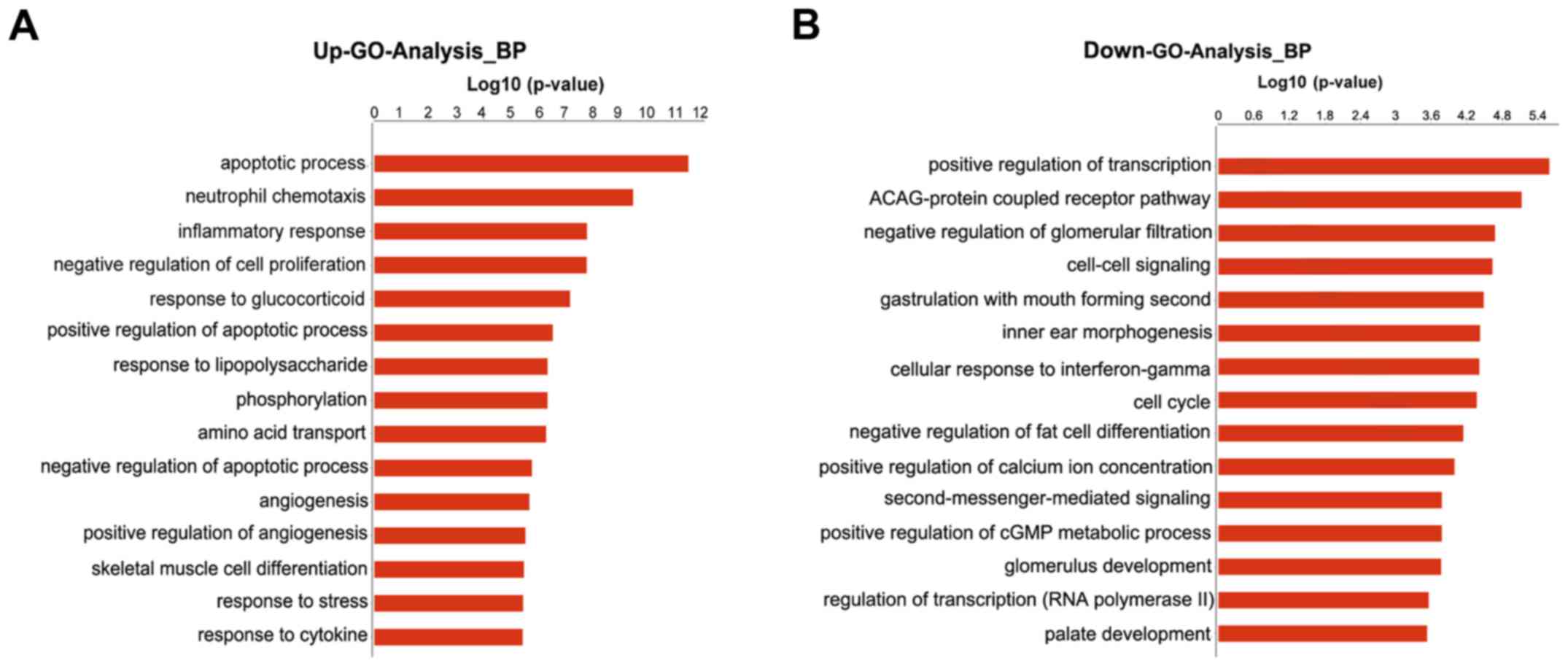
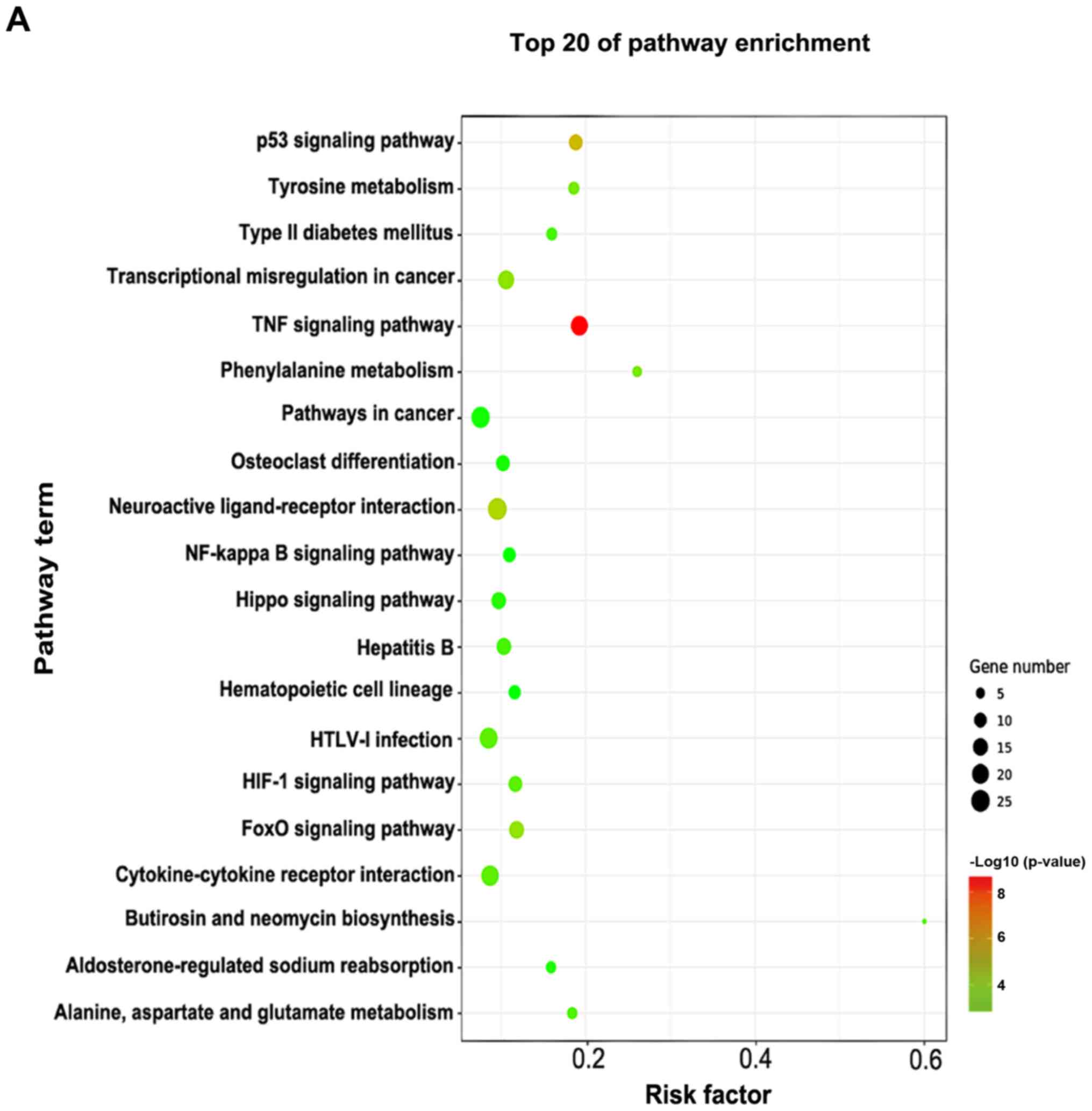
|
1
|
Brower RG and Fessler HE: Mechanical
ventilation in acute lung injury and acute respiratory distress
syndrome. Clin Chest Med. 21(491–510): viii2000.
|
|
2
|
Belperio JA, Keane MP, Lynch JP III and
Strieter RM: The role of cytokines during the pathogenesis of
ventilator-associated and ventilator-induced lung injury. Semin
Respir Crit Care Med. 27:350–364. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Neto AS, Simonis FD, Barbas CS, Biehl M,
Determann RM, Elmer J, Friedman G, Gajic O, Goldstein JN, Linko R,
et al: Lung-protective ventilation with low tidal volumes and the
occurrence of pulmonary complications in patients without acute
respiratory distress syndrome: A systematic review and individual
patient data analysis. Crit Care Med. 43:2155–2163. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Neto Serpa A, Hemmes SN, Barbas CS,
Beiderlinden M, Biehl M, Binnekade JM, Canet J,
Fernandez-Bustamante A, Futier E, Gajic O, et al: Protective versus
conventional ventilation for surgery: A systematic review and
individual patient data meta-analysis. Anesthesiology. 123:66–78.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choi WI, Quinn DA, Park KM, Moufarrej RK,
Jafari B, Syrkina O, Bonventre JV and Hales CA: Systemic
microvascular leak in an in vivo rat model of ventilator-induced
lung injury. Am J Respir Crit Care Med. 167:1627–1632. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Slutsky AS and Ranieri VM:
Ventilator-induced lung injury. N Engl J Med. 369:2126–2136. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Q, Ge YL, Li M, Fang XZ, Yuan YP, Liang
L and Huang SQ: miR-127 contributes to ventilator-induced lung
injury. Mol Med Rep. 16:4119–4126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brower RG, Lanken PN, MacIntyre N, Matthay
MA, Morris A, Ancukiewicz M, Schoenfeld D and Thompson BT: National
Heart, Lung, and Blood Institute ARDS Clinical Trials Network:
Higher versus lower positive end-expiratory pressures in patients
with the acute respiratory distress syndrome. N Engl J Med.
351:327–336. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su JC and Hu XF: Long non-coding RNA
HOXA11-AS promotes cell proliferation and metastasis in human
breast cancer. Mol Med Rep. 16:4887–4894. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang N, Chen J, Zhang H, Wang X, Yao H,
Peng Y and Zhang W: LncRNA OIP5-AS1 loss-induced microRNA-410
accumulation regulates cell proliferation and apoptosis by
targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple
myeloma. Cell Death Dis. 8:e29752017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Delás MJ and Hannon GJ: lncRNAs in
development and disease: From functions to mechanisms. Open Biol.
7:pii: 170121. 2017. View Article : Google Scholar
|
|
13
|
Ren C, Deng M, Fan Y, Yang H, Zhang G,
Feng X, Li F, Wang D, Wang F and Zhang Y: Genome-wide analysis
reveals extensive changes in LncRNAs during skeletal muscle
development in Hu sheep. Genes (Basel). 8:pii: E191. 2017.
View Article : Google Scholar
|
|
14
|
Zhang HJ, Wei QF, Wang SJ, Zhang HJ, Zhang
XY, Geng Q, Cui YH and Wang XH: LncRNA HOTAIR alleviates rheumatoid
arthritis by targeting miR-138 and inactivating NF-κB pathway. Int
Immunopharmacol. 50:283–290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yi H, Peng R, Zhang LY, Sun Y, Peng HM,
Liu HD, Yu LJ, Li AL, Zhang YJ, Jiang WH and Zhang Z:
LincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3
inflammasome-mediated inflammation in diabetic nephropathy. Cell
Death Dis. 8:e25832017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie
Y, Wang K, Jia W, Chu WM and Sun B: The long noncoding RNA lnc-EGFR
stimulates T-regulatory cells differentiation thus promoting
hepatocellular carcinoma immune evasion. Nat Commun. 8:151292017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mumtaz PT, Bhat SA, Ahmad SM, Dar MA,
Ahmed R, Urwat U, Ayaz A, Shrivastava D, Shah RA and Ganai NA:
LncRNAs and immunity: Watchdogs for host pathogen interactions.
Biol Proced Online. 19:32017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Misawa A, Takayama KI and Inoue S: Long
non-coding RNAs and prostate cancer. Cancer Sci. 108:2107–2114.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Haemmig S, Simion V, Yang D, Deng Y and
Feinberg MW: Long noncoding RNAs in cardiovascular disease,
diagnosis, and therapy. Curr Opin Cardiol. 32:776–783. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhuang YT, Xu DY, Wang GY, Sun JL, Huang Y
and Wang SZ: IL-6 induced lncRNA MALAT1 enhances TNF-α expression
in LPS-induced septic cardiomyocytes via activation of SAA3. Eur
Rev Med Pharmacol Sci. 21:302–309. 2017.PubMed/NCBI
|
|
21
|
Wahlestedt C: Targeting long non-coding
RNA to therapeutically upregulate gene expression. Nat Rev Drug
Discov. 12:433–446. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li H, Su X, Yan X, Wasserloos K, Chao W,
Kaynar AM, Liu ZQ, Leikauf GD, Pitt BR and Zhang LM: Toll-like
receptor 4-myeloid differentiation factor 88 signaling contributes
to ventilator-induced lung injury in mice. Anesthesiology.
113:619–629. 2010.PubMed/NCBI
|
|
23
|
Li LF, Yang CT, Huang CC, Liu YY, Kao KC
and Lin HC: Low-molecular-weight heparin reduces
hyperoxia-augmented ventilator-induced lung injury via
serine/threonine kinase-protein kinase B. Respir Res. 12:902011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang K, Singh D, Zeng Z, Coleman SJ, Huang
Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al:
MapSplice: Accurate mapping of RNA-seq reads for splice junction
discovery. Nucleic Acids Res. 38:e1782010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mortazavi A, Williams BA, McCue K,
Schaeffer L and Wold B: Mapping and quantifying mammalian
transcriptomes by RNA-Seq. Nat Methods. 5:621–628. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanehisa M, Goto S, Kawashima S, Okuno Y
and Hattori M: The KEGG resource for deciphering the genome.
Nucleic Acids Res. 32:(Database Issue). D277–D280. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang GT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pujana MA, Han JD, Starita LM, Stevens KN,
Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al:
Network modeling links breast cancer susceptibility and centrosome
dysfunction. Nat Genet. 39:1338–1349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ravasz E, Somera AL, Mongru DA, Oltvai ZN
and Barabási AL: Hierarchical organization of modularity in
metabolic networks. Science. 297:1551–1555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun D, Wang J, Yang N and Ma H: Matrine
suppresses airway inflammation by downregulating SOCS3 expression
via inhibition of NF-κB signaling in airway epithelial cells and
asthmatic mice. Biochem Biophys Res Commun. 477:83–90. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Xu CF, Liu YJ, Mao YF, Lv Z, Li
SY, Zhu XY and Jiang L: Salidroside attenuates ventilation induced
lung injury via SIRT1-dependent inhibition of NLRP3 inflammasome.
Cell Physiol Biochem. 42:34–43. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang W, Meng M, Zhang Y, Wei C, Xie Y,
Jiang L, Wang C, Yang F, Tang W, Jin X, et al: Global
transcriptome-wide analysis of CIK cells identify distinct roles of
IL-2 and IL-15 in acquisition of cytotoxic capacity against tumor.
BMC Med Genomics. 7:492014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lin ZW, Gu J, Liu RH, Liu XM, Xu FK, Zhao
GY, Lu CL and Ge D: Genome-wide screening and co-expression network
analysis identify recurrence-specific biomarkers of esophageal
squamous cell carcinoma. Tumour Biol. 35:10959–10968. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc Series B. 57:pp289–300. 1995.
|
|
38
|
Pawitan Y, Michiels S, Koscielny S,
Gusnanto A and Ploner A: False discovery rate, sensitivity and
sample size for microarray studies. Bioinformatics. 21:3017–3024.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ma SF, Grigoryev DN, Taylor AD, Nonas S,
Sammani S, Ye SQ and Garcia JG: Bioinformatic identification of
novel early stress response genes in rodent models of lung injury.
Am J Physiol Lung Cell Mol Physiol. 289:L468–L477. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dolinay T, Kim YS, Howrylak J, Hunninghake
GM, An CH, Fredenburgh L, Massaro AF, Rogers A, Gazourian L,
Nakahira K, et al: Inflammasome-regulated cytokines are critical
mediators of acute lung injury. Am J Respir Crit Care Med.
185:1225–1234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mirzapoiazova T, Moitra J, Moreno-Vinasco
L, Sammani S, Turner JR, Chiang ET, Evenoski C, Wang T, Singleton
PA, Huang Y, et al: Non-muscle myosin light chain kinase isoform is
a viable molecular target in acute inflammatory lung injury. Am J
Respir Cell Mol Biol. 44:40–52. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Uhlig U, Fehrenbach H, Lachmann RA,
Goldmann T, Lachmann B, Vollmer E and Uhlig S: Phosphoinositide
3-OH kinase inhibition prevents ventilation-induced lung cell
activation. Am J Respir Crit Care Med. 169:201–208. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vanderbilt JN, Mager EM, Allen L, Sawa T,
Wiener-Kronish J, Gonzalez R and Dobbs LG: CXC chemokines and their
receptors are expressed in type II cells and upregulated following
lung injury. Am J Respir Cell Mol Biol. 29:661–668. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sakashita A, Nishimura Y, Nishiuma T,
Takenaka K, Kobayashi K, Kotani Y and Yokoyama M: Neutrophil
elastase inhibitor (sivelestat) attenuates subsequent
ventilator-induced lung injury in mice. Eur J Pharmacol. 571:62–71.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hammerschmidt S, Kuhn H, Grasenack T,
Gessner C and Wirtz H: Apoptosis and necrosis induced by cyclic
mechanical stretching in alveolar type II cells. Am J Respir Cell
Mol Biol. 30:396–402. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gao J, Huang T, Zhou LJ, Ge YL, Lin SY and
Dai Y: Preconditioning effects of physiological cyclic stretch on
pathologically mechanical stretch-induced alveolar epithelial cell
apoptosis and barrier dysfunction. Biochem Biophys Res Commun.
448:342–348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chess PR, Benson RP, Maniscalco WM, Wright
TW, O'Reilly MA and Johnston CJ: Murine mechanical ventilation
stimulates alveolar epithelial cell proliferation. Exp Lung Res.
36:331–341. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mukhopadhyay S, Hoidal JR and Mukherjee
TK: Role of TNFalpha in pulmonary pathophysiology. Respir Res.
7:1252006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wilson MR and Takata M: Inflammatory
mechanisms of ventilator-induced lung injury: A time to stop and
think? Anaesthesia. 68:175–178. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wilson MR, Wakabayashi K, Bertok S, Oakley
CM, Patel BV, O'Dea KP, Cordy JC, Morley PJ, Bayliffe AI and Takata
M: Inhibition of TNF receptor p55 by a domain antibody attenuates
the initial phase of acid-induced lung injury in mice. Front
Immunol. 8:1282017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Eckle T, Kewley EM, Brodsky KS, Tak E,
Bonney S, Gobel M, Anderson D, Glover LE, Riegel AK, Colgan SP and
Eltzschig HK: Identification of hypoxia-inducible factor HIF-1A as
transcriptional regulator of the A2B adenosine receptor during
acute lung injury. J Immunol. 192:1249–1256. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Eckle T, Grenz A, Laucher S and Eltzschig
HK: A2B adenosine receptor signaling attenuates acute lung injury
by enhancing alveolar fluid clearance in mice. J Clin Invest.
118:3301–3315. 2008.PubMed/NCBI
|
|
53
|
Wang T, Gross C, Desai AA, Zemskov E, Wu
X, Garcia AN, Jacobson JR, Yuan JX, Garcia JG and Black SM:
Endothelial cell signaling and ventilator-induced lung injury:
Molecular mechanisms, genomic analyses, and therapeutic targets. Am
J Physiol Lung Cell Mol Physiol. 312:L452–L476. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hert DG, Fredlake CP and Barron AE:
Advantages and limitations of next-generation sequencing
technologies: A comparison of electrophoresis and
non-electrophoresis methods. Electrophoresis. 29:4618–4626. 2008.
View Article : Google Scholar : PubMed/NCBI
|