Introduction
Helicobacter pylori (H. pylori) is a
Gram-negative, spiral-shaped, microaerophilic bacterium that
colonizes the human gastric mucosa (1). H. pylori infection can cause
chronic gastritis, peptic ulcer disease, gastric carcinoma and
mucosa-associated lymphoid tissue lymphoma (2–4).
More than half of the world's population is infected with H.
pylori (5). However, the
majority of infected individuals remain asymptomatic. There is an
interplay between host genetic susceptibility, environmental
factors and bacterial virulence factors, which influences the
outcome of H. pylori-associated diseases (6–8).
Polymorphisms of several virulence genes, including
cytotoxin-associated gene A (cagA), vacuolating toxin A (vacA),
induced by contact with epithelium gene A (iceA), blood group
antigen binding adhesin and duodenal ulcer promoting gene A are
considered to increase the risk for the development of upper
gastrointestinal diseases (9).
Among these genes, cagA and vacA have been investigated
extensively.
CagA is an oncogenic protein, and is a major
virulence factor associated with gastric cancer (10,11).
CagA is injected into host epithelial cells via the type IV
secretion system encoded by cag pathogenicity island (cagPAI)
following H. pylori infection (12). CagA undergoes phosphorylation at
its EPIYA motif by the Abl and Src family tyrosine kinases
(13). Once tyrosine is
phosphorylated, the cagA EPIYA motifs serve as a recognition site
for Src homology phosphotyrosine phosphatase 2 (SHP2), and activate
an intracellular signal transduction pathway that leads to
cytoskeletal rearrangement and cell elongation, known as the
‘hummingbird’ phenotype (14).
This increases the risk of developing precancerous lesions
(15).
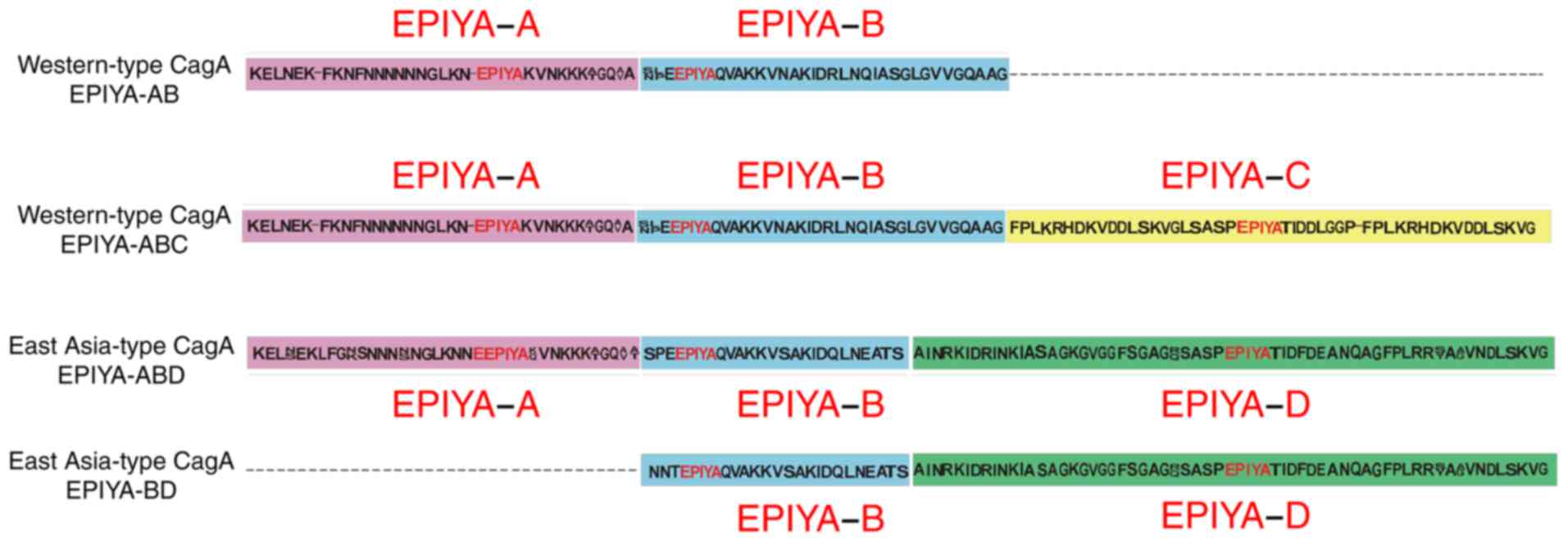
The EPIYA motif is located within the C-terminal of
cagA, and is formed of the conserved amino-acid residues
Glu-Pro-Ile-Tyr-Ala. EPIYA can be further classified into four
types of motif: EPIYA-A, -B, -C and -D, based on the flanking amino
acid sequences (16). H.
pylori is classified as East Asian-type cagA and western-type
cagA, according to the composition of the EPIYA-A, -B, -C and D
motifs. The western-type cagA is mainly cagA-ABC, whereas the East
Asian-type is mainly cagA-ABD (17). The EPIYA-C and EPIYA-D motifs act
as phosphorylation sites for SHP-2 (18). The EPIYA-D segment exhibits a
greater degree of tyrosine phosphorylation and higher binding
affinity to SHP2 than the EPIYA-C segment and shows higher
virulence (2). East Asian-type
cagA is associated with a higher risk of peptic ulcers or gastric
cancer within the same geographical area compared with western-type
cagA (19). Epidemiological
surveys show that ~50–60% of H. pylori strains in western
countries contain the cagA gene, which increases the risk of peptic
ulcers and gastric cancer (20).
Although 90–100% of H. pylori strains in East Asian
countries are cagA-positive, cagA-positive strains may not be
associated with clinical outcomes (21).
VacA is a pore-forming toxin, which has several
effects on epithelial cells. In addition to inducing vacuolation
(22), vacA can induce membrane
channel formation, which leads to the release of cytochrome
c from mitochondria and results in apoptosis (23). Notably, it has immunomodulatory
effects through inhibiting T-cell activation and proliferation
(6,19). There are four sequence diversity
regions of vacA closely associated with H. pylori
vacuolating activity, namely signal region (s-), deletion region
(d-), intermediate region (i-) and middle region (m-) (4). The cytotoxicity of vacA is determined
by variability in the structure of the vacA gene (24). The s- and m- regions of vacA are
the two main polymorphic regions and serve as markers of H.
pylori virulence and the risk of associated diseases (25).
Several studies have shown that iceA has two main
allelic variants, namely iceA1 and iceA2 (26). The expression of iceA1 is
upregulated upon contact between H. pylori and human
epithelial cells. The iceA1 genotype is linked with enhanced mucosa
interleukin-8 expression and acute inflammation (27). Epidemiological data shows that the
geographical distribution of iceA genotypes varies. The iceA1
strains mainly occur in Japan and South Korea, whereas iceA2
strains are predominant in the United States and Columbia (28). A previous meta-analysis showed that
the prevalence of iceA1 was significantly higher in East Asian
countries than in western countries, whereas the prevalence of
iceA2 was higher in western countries than in East Asian countries
(26). Functionally, the iceA1
genotype is associated with peptic ulcers (29) and the iceA2 genotype with the
occurrence of chronic gastritis (28). To date, the majority of studies
have shown that the iceA gene is another virulence gene independent
of cagA and vacA (30).
Guizhou, a province located in southwest China, is a
multi-ethnic society. In particular, those in Qiannan, Buyei and
Miao Autonomous Prefecture have a particular lifestyle and are
different from other ethnic groups. The individuals living here
often use herbal medicine to treat diseases, including stomach
disorders. As Xie et al (31) reported, such traditional medicine
may have an effect on bacteria in the stomach, including H.
pylori. Although investigations on the role of H. pylori
virulence cagA and vacA genotypes have been performed worldwide,
the associations between H. pylori virulence genotype and
gastroduodenal diseases, ethnicity or economic conditions in
Guizhou province remain to be fully elucidated. Therefore, the
present study aimed to investigate these associations, which may
facilitate diagnosis and therapeutic strategies for gastroduodenal
diseases.
Materials and methods
Clinical samples
Gastric pylorus mucosa biopsy samples were obtained
by endoscopy from patients with gastric disorders at the First
Affiliated Hospital of Guizhou Medical University (Guizhou, China)
and the People's Hospital of Qiannan Autonomous Prefecture (Duyun,
China) between January and December 2016. Clinicopathological data
were collected, and written informed consent was obtained from all
patients. Patients with the following conditions were excluded from
the present study: A tendency to bleed, lactating or pregnant
women, the inability to undergo surgery to the UGI tract, and
severe cardiovascular or hepatic disease. All protocols were
approved by the Ethical Committee of the First Affiliated hospital
of Guizhou Medical University. All procedures contributing to the
study complied with the Declaration of Helsinki.
H. pylori isolation and bacterial DNA
extract
Gastric mucosa samples were cut into small sections,
homogenized and smeared on the surface of Brain Heart Infusion agar
with 10% sheep blood (Qingdao Hope Biol-Technology Co., Ltd.,
Qingdao, China) and antibiotic supplement (H. pylori
selective supplement, Thermo Fisher Scientific Oxoid, Ltd.,
Basingstoke, UK). The plates were incubated at 37°C for 3–5 days
under microaerophilic conditions. H. pylori was identified
through colonial morphology, Gram staining, a urease test, and
H. pylori-specific 16S rRNA gene fragment polymerase chain
reaction (PCR) amplification. The colony of H. pylori was
smooth and translucent, and the morphology was Gram-negative with
spiral-shaped bacilli. The confirmed colonies were subcultured to
single colonies on fresh medium. Following incubation at 37°C for
3–5 days, the colonies were subjected to DNA extraction using an
Ezup column bacteria genomic DNA purification kit (Sangon Biotech
Co., Ltd., Shanghai, China), according to the manufacturer's
protocol.
PCR amplification and sequencing
PCR assays to amplify cagA and sequence its
C-terminal region were performed according to the report by
Sicinschi et al (14).
Primers (Sangon Biotech Co., Ltd., Shanghai, China) used for
amplification of the cagA, vacA and iceA genes in the present study
are shown in Table I. Following
DNA extraction from pure culture of H. Pylori isolates as
previously mentioned, PCR assays were performed in a volume of 26
µl containing 1 µl forward primer, 1 µl reverse primer, 4 µl
genomic DNA, 13 µl 2X Taq PCR Master Mix (Beijing Solarbio Science
and Technology Co., Ltd., Beijing, China) and 7 µl
ddH2O. Table I
summarizes the expected size of the PCR products and cycling
conditions for cagA gene, vacA subtype gene and iceA allelic genes
(14,32–34).
All runs included one negative (ddH2O) and one positive
(NCTC 11637 or H. pylori 26695) DNA control, and DNA ladder
markers (Tiangen Biotech Co., Ltd., Beijing, China). A total of 6
µl of amplified PCR products was then resolved by electrophoresis
on 2% agarose gels run in acetate EDTA buffer, and stained with
ethidium bromide. The PCR product was visualized under ultraviolet
light. The cagA C-terminal PCR products were sent to Sangon Biotech
Co., Ltd. for Sanger sequencing. The gene sequences were translated
into amino acid sequences using Bioedit software (version 7.1.3.0;
http://www.bioedit.com/). Phylogenetic tree
cluster analysis of cagA carboxyl terminal variable region was
constructed using MEGA software (version 6.0; http://www.megasoftware.net/megamac.php), based on
neighbor joining. The control strain NCTC 11637 was obtained from
the State Key Laboratory of Infectious Disease Prevention and
Control (National Institute for Communicable Disease Control and
Prevention, Beijing, China), and the amino acid sequences of H.
pylori 26695 were obtained from GenBank (https://www.ncbi.nlm.nih.gov/nuccore/CP003904.1).
 | Table I.Primer sequences and reaction
conditions for polymerase chain reaction. |
Table I.
Primer sequences and reaction
conditions for polymerase chain reaction.
| Primer | Gene | Primer
sequence | Size (bp) | Amplification
condition |
|---|
| 16S
rRNA | 16S
rRNA-F |
5′-CTTGCTAGAGTGCTGATTA-3′ | 550 | 35 cycles: 94°C for
30 sec; 55°C for 30 sec; 72°C for 30 sec |
|
| 16S
rRNA-R |
5′-TCCCACACTCTAGAATAGT-3′ |
|
|
| cagA 5′-end
conserved region | cagA F |
5′-GATAACAGGCAAGCTTTTGAGG-3′ | 349 | 30 cycles: 94°C for
1 min; 55°C for 1 min; 72°C for 1 min |
|
| cagA R |
5′-CTGCAAAAGATTGTTTGGCAGA-3′ |
|
|
| cagA 3′-end
variable region | cagA-VF |
5′-ACCCTAGTCGGTAATGGGTTA-3′ | 591–856 | 30 cycles: 94°C for
1 min; 50°C for 1 min; 72°C for 1 min |
|
| cagA-VR |
5′-GTAATTGTCTAGTTTCGC-3′ |
|
|
| cagPAI empty
site | Empty site-F |
5′-ACATTTTGGCTAAATAAACGCTG-3′ | 535 | 30 cycles: 94°C for
1 min; 55°C for 1 min; 72°C for 1 min |
|
| Empty site-R |
5′-GGTTGCACGCATTTTCCCTTAATC-3′ |
|
|
| iceA1 | iceA1-F |
5′-GCTTGTAACGATAAGAAACGCCAGAT-3′ | 297 | 35 cycles: 94°C for
30 sec; 55°C for 30 sec; 72°C for 30 sec |
|
| iceA1-R |
5′-GGAATGAGCTTGTATTTAGAGCCGAT-3′ |
|
|
| iceA2 | iceA2-F |
5′-GTTGGGTATATCACAATTTAT-3′ | 229/334 | 30 cycles: 94°C for
30 sec; 52°C for 30 sec; 72°C for 45 sec |
|
| iceA2-R |
5′-TTRCCCTATTTTCTAGTAGGT-3′ |
|
|
| vacA-s1a | vacA-s1a-F |
5′-CTCTCGCTTTAGTAGGAGC-3′ | 213 | 30 cycles: 94°C for
30 sec; 60°C for 30 sec; 72°C for 45 sec |
|
| vacA-s1a-R |
5′-CTGCTTGAATGCGCCAAAC-3′ |
|
|
| vacA-s1b | vacA-s1b-F |
5′-AGCGCCATACCGCAAGAG-3′ | 187 |
|
|
| vacA-s1b-R |
5′-CTGCTTGAATGCGCCAAAC-3′ |
|
|
| vacA-s1c | vacA-s1c-F |
5′-CTCTCGCTTTAGTGGGGYT-3′ | 213 |
|
|
| vacA-s1c-R |
5′-CTGCTTGAATGCGCCAAAC-3′ |
|
|
| vacA-s2 | vacA-s2-F |
5′-GCTAACACGCCAAATGATCC-3′ | 199 |
|
|
| vacA-s2-R |
5′-CTGCTTGAATGCGCCAAAC-3′ |
|
|
| vacA-m1a | vacA-m1a-F |
5′-GGTCAAAATGCGGTCATGG-3′ | 290 |
|
|
| vacA-m1a-R |
5′-CCATTGGTACCTGTAGAAAC-3′ |
|
|
| vacA-m1b | vacA-m1b-F |
5′-GGCCCCAATGCAGTCATGGAT-3′ | 291 |
|
|
| vacA-m1b-R |
5′-GCTGTTAGTGCCTAAAGAAGCAT-3′ |
|
|
| vacA-m2 | vacA-m2-F |
5′-GGAGCCCCAGGAAACATTG-3′ | 352 |
|
|
| vacA-m2-R |
5′-CATAACTAGCGCCTTGCAC-3′ |
|
|
Statistical analysis
All data were analysed using SPSS 19.0 (IBM SPSS,
Armonk, NY, USA), χ2 test and Fisher's exact test were
used for the analysis of categorical data. P<0.05 was considered
to indicate a statistically significant difference.
Results
Clinical and pathological
information
A total of 73 H. pylori strains were isolated
from patients with upper gastroduodenal disorders. The demographics
of these patients are shown in Table
II. Among the 73 cases, 41 were men with a mean age of
38.85±19.43 years, and range of 3–80 years, and 32 were women with
a mean age of 39.94±18.93 years, and range of 4–66. A total of 60
strains were isolated from Han groups, and 13 from other minority
ethnic groups, including Miao, Dong, Tujia, Buyi, Bai and Yi
groups. A total of 60 strains were isolated from Guiyang city, and
13 from Qiannan autonomous prefecture. A total of 31 cases were
from urban populations, and 42 were from suburban populations.
Finally, 54 strains were isolated from patients with gastritis and
19 from patients with peptic ulcers, the latter comprising eight
with gastric ulcer and 11 with duodenal ulcer. PCR products of 550
bp represented H. pylori-specific 16S rRNA, as shown in
Fig. 1A.
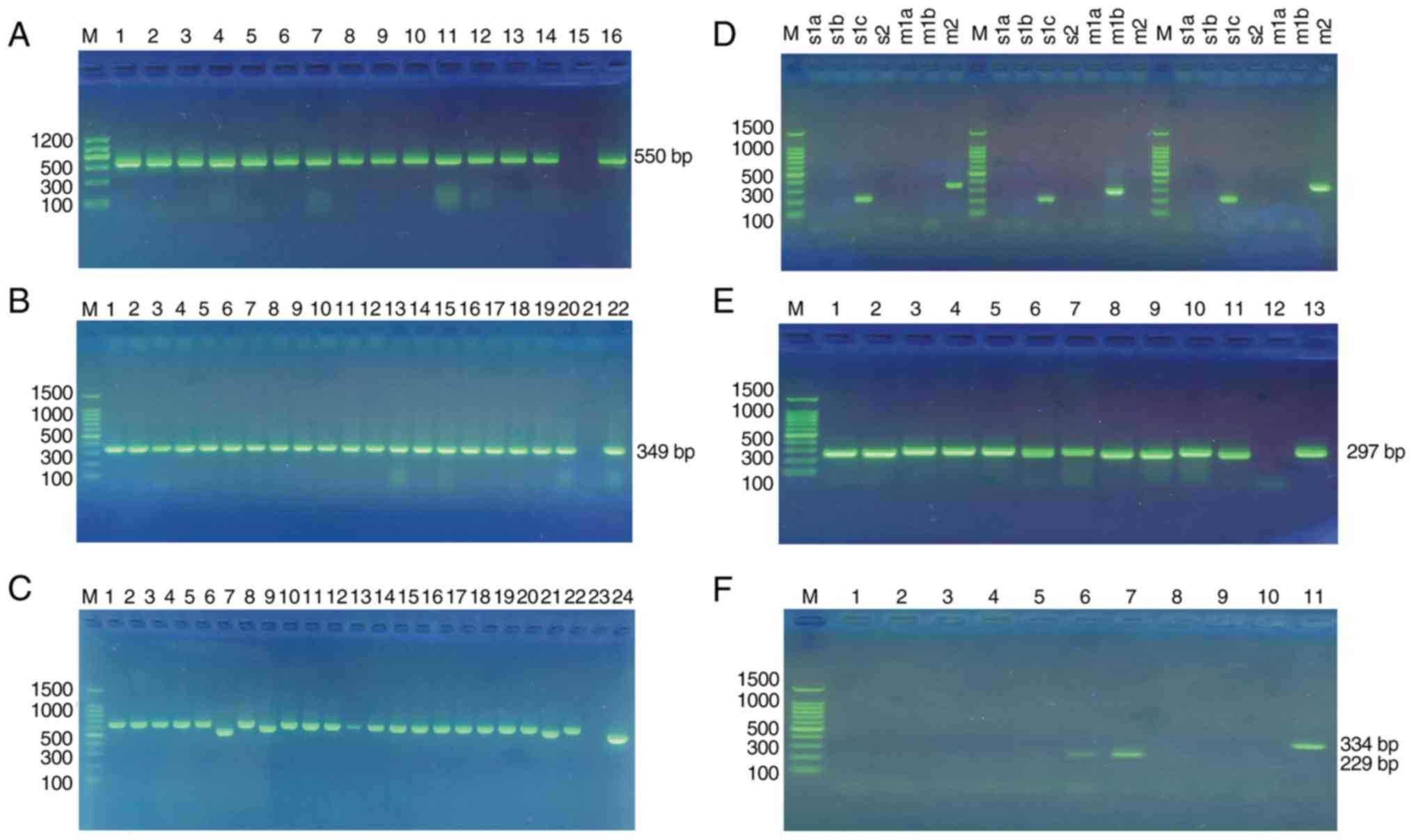 | Figure 1.PCR amplification of H. pylori
cagA, vacA and iceA genes. (A) Gel electrophoresis of
genus-specific 16S rRNA PCR products from H. pylori
isolates. M, Marker II; 1–14, positive sample for 16Sr RNA
PCR; 15, negative control; 16, positive control (NCTC 11637). (B)
Gel electrophoresis of clinical samples with different cagA
N-terminals. M, 100 bp DNA ladder marker; 1–20, clinical sample;
23, negative control; 24, positive control (NCTC 11637). (C) Gel
electrophoresis of clinical samples with different cagA EPIYA
patterns. M, 100 bp DNA ladder marker; 1–22, clinical sample; 23,
negative control; 24, positive control (NCTC 11637). (D) Gel
electrophoresis of H. pylori genotyping of vacA alleles. M,
100 bp DNA ladder marker; s1a, s1b, s1c and s2, vacA signal region;
m1a, m1b and m2, vacA middle region. (E) Gel electrophoresis of
H. pylori genotyping of iceA1 alleles. M, 100 bp DNA ladder
marker; 1–11, clinical sample; 12, negative control; 13, positive
control (H. pylori 26695). (F) Gel electrophoresis of H.
pylori genotyping of iceA2 alleles. M, 100 bp DNA ladder
marker; 1–5 and 8–9, negative clinical sample; 6–7, positive
clinical sample (iceA2-229 bp); 10, negative control; 11, positive
control (iceA2-334 bp). H. pylori, Helicobacter pylori;
cagA, cytotoxin associated gene A; vacA, vacuolating cytotoxin A;
iceA induced by contact with epithelium gene A; PCR, polymerase
chain reaction. |
 | Table II.Demographic characteristics of
patients. |
Table II.
Demographic characteristics of
patients.
| Characteristic | Patients (n) | Age
(years)a | Range |
|---|
| Sex |
|
Male | 41 | 38.85±19.43 | 3–80 |
|
Female | 32 | 39.94±18.93 | 4–66 |
| Ethnicity |
|
Han | 60 | 40.68±19.64 | 3–80 |
| Ethnic
minority |
|
Miao | 4 | 25.75±19.45 | 7–44 |
|
Dong | 2 | 23±22.63 | 7–39 |
|
Tujia | 2 | 36.5±16.26 | 25–48 |
|
Buyi | 2 | 42.5±0.71 | 42–43 |
|
Bai | 2 | 36.5±2.12 | 35–38 |
| Yi | 1 | 50 | – |
| Place of
residence |
| Guiyang
city | 60 | 37.97±20.73 | 3–80 |
| Qiannan
autonomous prefecture | 13 | 45.62±4.68 | 38–53 |
|
Urban | 31 | 42.19±21.19 | 3–80 |
|
Suburban | 42 | 36.41±19.49 | 4–72 |
| Clinical
disease |
|
Gastritis | 54 | 38.11±20.34 | 3–80 |
| Peptic
ulcer | 19 | 42.79±14.90 | 9–60 |
Polymorphism of cagA
The cagA N-terminal conserved region (cagA 5′-end)
and C-terminal variable region (cagA 3′-end) were amplified by PCR
using the specific primers (Table
I). PCR products of 349 bp represented the 5′-end fragments, as
shown in Fig. 1B, and were present
in all H. pylori isolates (n=73, 100%). CagPAI empty site
PCR produced negative results in all of these strains, indicating
the absence of cagA-negative strains in the mixture. PCR products
within the range of 450–650 bp represented the 3′-end fragments, as
shown in Fig. 1C. All H.
pylori isolates were found to contain the 3′-end variable
region expressing various EPIYA motifs. Notably, four distinct PCR
products (500, 600, 610 and 550 bp amplicons) were obtained from
the various H. pylori isolates (Fig. 1C).
Polymorphism of vacA
The vacA gene polymorphism was also analyzed. The
vacA gene has variation regions including signal (s-) and middle
(m-) regions (24). Usually,
alleles are further divided into sub-alleles, including s1a, s1b,
s1c, s2, m1a, m1b and m2 (34).
The H. pylori isolates were screened for all sub-alleles by
the PCR assay, as shown in Fig.
1D. vacA s1c/m1b and s1c/m2 genotypes were identified in 24.66
and 65.75%, respectively, whereas the s1c/m1b/m2 mixed genotype was
only identified in 9.59%, as shown in Table III. The distribution of vacA
genotypes in Guizhou province is shown in Table III. No significant association
between the vacA genotypes and clinical outcomes was identified
(P=1.000). There was also no significant association between the
vacA genotypes and age, place of residence (Guiyang city vs.
Qiannan autonomous prefecture, urban vs. suburban), or ethnic group
(P=0.605, P=0.400, P=0.718 and P=0.210, respectively), as shown in
Tables IV and V.
 | Table III.Distribution of cagA genotypes in
Guizhou province. |
Table III.
Distribution of cagA genotypes in
Guizhou province.
| Genotype | n (%) | Guiyang city
strains (n) | Qiannan autonomous
prefecture strains (n) |
|---|
| Western-type
cagA-AB | 5 (6.85) | 5 | 0 |
| Western-type
cagA-ABC | 3 (4.11) | 3 | 0 |
| East Asia-type
cagA-ABD | 63 (86.30) | 50 | 13 |
| East Asia-type
cagA-BD | 2 (2.74) | 2 | 0 |
| vacA s1c/m1b | 18 (24.66) | 13 | 5 |
| vacA s1c/m2 | 48 (65.75) | 41 | 7 |
| vacA
s1c/m1b/m2 | 7 (9.59) | 6 | 1 |
| iceA1 | 58 (79.45) | 49 | 9 |
| iceA2 | 2 (2.74) | 1 | 1 |
| iceA1+iceA2 | 13 (17.81) | 10 | 3 |
 | Table IV.Distribution of virulence genotypes
by clinical disease and age of patients. |
Table IV.
Distribution of virulence genotypes
by clinical disease and age of patients.
| Genotype | Gastritis
(n=54) | Peptic ulcer
(n=19) | P-value | Age <18 years
(n=16) | Age >18 years
(n=57) | P-value |
|---|
| cagA-AB | 5 | 0 | 0.417 | 0 | 4 | 0.180 |
| cagA-ABC | 3 | 0 |
| 2 | 1 |
|
| cagA-ABD | 44 | 19 |
| 14 | 50 |
|
| cagA-BD | 2 | 0 |
| 0 | 2 |
|
| vacA s1c/m1b | 13 | 5 | 1.000 | 5 | 13 | 0.605 |
| vacA s1c/m2 | 36 | 12 |
| 9 | 39 |
|
| vacA
s1c/m1b/m2 | 5 | 2 |
| 2 | 5 |
|
| iceA1 | 41 | 17 | 0.146 | 12 | 46 | 0.679 |
| iceA2 | 1 | 1 |
| 0 | 2 |
|
| iceA1+iceA2 | 12 | 1 |
| 4 | 9 |
|
 | Table V.Distribution of cagA genotypes by
place of residence and ethnicity of patients. |
Table V.
Distribution of cagA genotypes by
place of residence and ethnicity of patients.
|
| Place of
residence | Ethnic group |
|---|
|
|
|
|
|---|
| Genotype | Guiyang city | Qiannan autonomous
prefecture | P-value | Urban | Suburban | P-value | Han | Minority | P-value |
|---|
| CagA-AB | 5 | 0 | 0.845 | 3 | 2 | 0.602 | 4 | 1 | 1.000 |
| CagA-ABC | 3 | 0 |
| 2 | 1 |
| 3 | 0 |
|
| cagA-ABD | 50 | 13 |
| 25 | 38 |
| 51 | 12 |
|
| CagA-BD | 2 | 0 |
| 1 | 1 |
| 2 | 0 |
|
| vacA s1c/m1b | 13 | 5 | 0.400 | 7 | 11 | 0.718 | 13 | 5 | 0.210 |
| vacA s1c/m2 | 41 | 7 |
| 22 | 26 |
| 42 | 6 |
|
| vacA
s1c/m1b/m2 | 6 | 1 |
| 2 | 5 |
| 5 | 2 |
|
| iceA1 | 49 | 9 | 0.304 | 24 | 34 | 0.884 | 46 | 12 | 0.621 |
| iceA2 | 1 | 1 |
| 1 | 1 |
| 2 | 0 |
|
| iceA1/iceA2 | 10 | 3 |
| 6 | 7 |
| 12 | 1 |
|
Polymorphism of iceA
The polymorphism of iceA genes was also assessed.
Gel electrophoresis genotyping of H. pylori iceA1 and iceA2
alleles is shown in Fig. 1E and F,
respectively. Among the 73 cases, the iceA1 and iceA2 genotypes
were present in 79.45% (58/73) and 2.74% (2/73), respectively. The
iceA1/iceA2 mixed genotype occurred in 17.81% (13/73) of cases. The
distribution of iceA genotypes in Guizhou province is shown in
Table III. No significant
association was found between iceA genotypes and disease outcomes,
age, place of residence (Guiyang city, vs. Qiannan autonomous
prefecture, urban vs. suburban), or ethnic group (P=0.146, P=0.679,
P=0.304, P=0.884 and P=0.621, respectively), as shown in Tables IV and V.
EPIYA motifs of cagA
As described above, the EPIYA motif patterns were
determined for each strain. A comparison of the results indicated
that the EPIYA genotypes of the 500, 600, 610 and 550 bp amplicons
were these of the cagA-AB, -ABC, -ABD and -BD genotypes,
respectively. The structural polymorphism of the cagA amino acid
sequence is shown in Fig. 2. It
was noted that cagA EPIYA-ABD (63/73, 86.30%) was the predominant
genotype in the present study, followed by cagA-AB (5/73, 6.85%),
cagA-ABC (3/73, 4.11%) and cagA-BD (2/73, 2.74%). The distribution
of cagA genotypes in Guizhou province is shown in Table III. However, statistical analysis
revealed no significant correlation between the disease outcome and
the cagA genotypes (P=0.417). There was also no significant
correlation between the cagA genotypes and age, place of residence
(Guiyang city, vs. Qiannan autonomous prefecture, urban vs.
suburban), or ethnic group (P=0.180, P=0.845, P=0.602 and P=1.000,
respectively), as shown in Tables
IV and V.
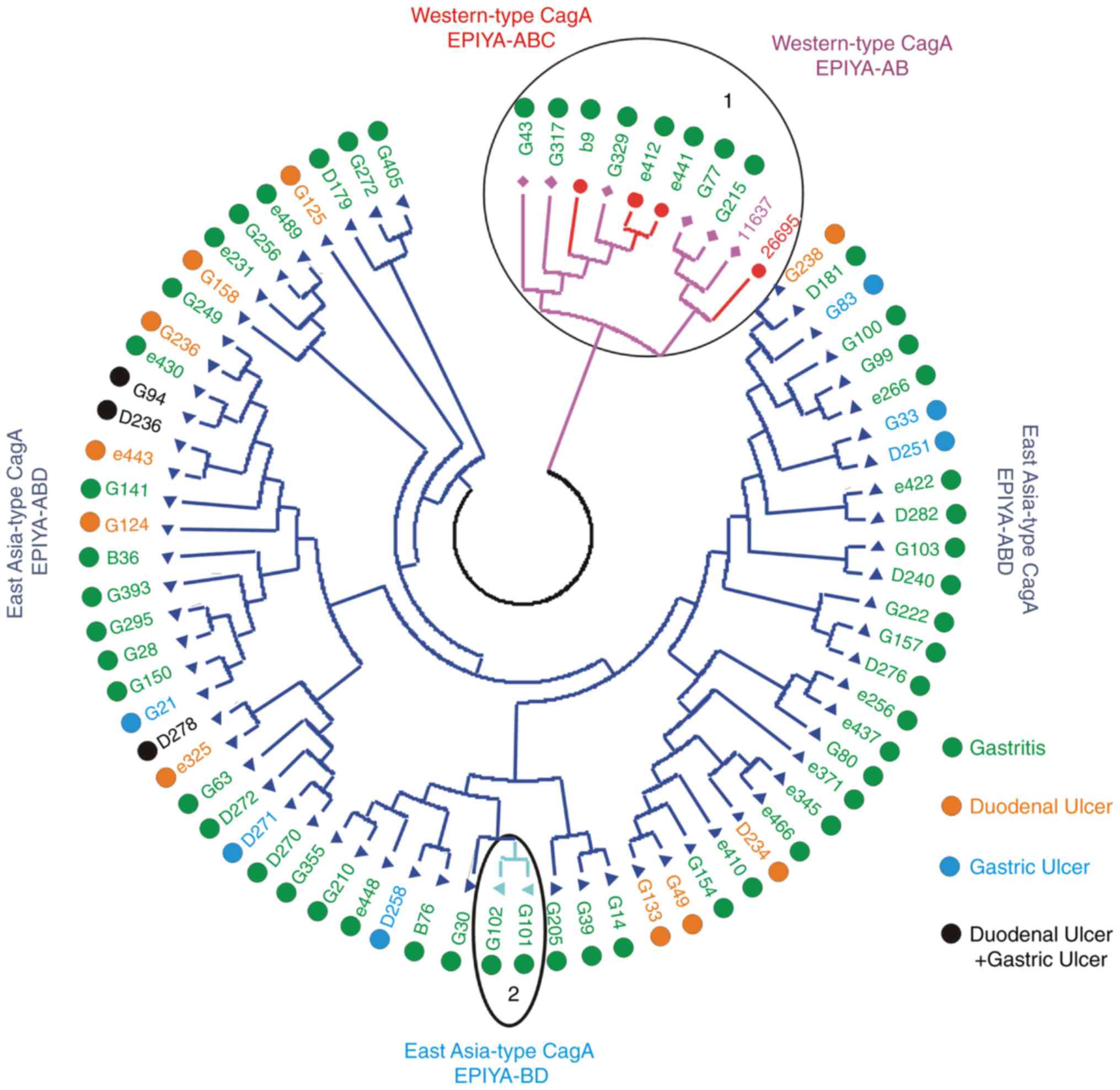
The results of phylogenetic tree cluster analysis of
the cagA carboxyl terminal variable region is shown in Fig. 3. There was a clustering association
between western-type cagA-AB and cagA-ABC; there was also a
clustering association between East Asian-type cagA-ABD and
cagA-BD. All five strains with western-type cagA-AB and three
strains with western-type cagA-ABC were isolated from patients with
chronic gastritis; 19 strains with East Asian-type cagA-ABD were
isolated from patients with peptic ulcers, and a further 46 with
East Asia-type cagA were isolated from patients with chronic
gastritis. As mentioned above, it appeared that East Asia-type cagA
strains may exhibit higher virulence than western-type cagA
strains.
The frequencies of the four EPIYA or EPIYA-like
motifs are shown in Table VI. In
total, 212 EPIYA motifs were obtained from 73 cagA sequences. The
most frequent EPIYA motif was EPIYA (197/212, 92.92%), followed by
EPIYT (8/212, 3.77%), ESIYA (6/212, 2.83%) and ESIYT (1/212,
0.47%). The EPIYA-B motif had a high degree of variation in the
five amino acids (EPIYA, EPIYT, ESIYA or ESIYT). In addition, 15
EPIYA-like motifs (EPIYT, ESIYA, ESIYT) were isolated from patients
with gastritis.
 | Table VI.Frequencies of EPIYA motifs. |
Table VI.
Frequencies of EPIYA motifs.
| Motif type | EPIYA | EPIYT | ESIYA | ESIYT | Total |
|---|
| All cagA type |
|
|
|
|
|
| All
motifs | 197 | 8 | 6 | 1 | 212 |
| A
motifs | 71 | 0 | 0 | 0 |
|
| B
motifs | 59 | 7 | 6 | 1 |
|
| C
motifs | 2 | 1 | 0 | 0 |
|
| D
motifs | 65 | 0 | 0 | 0 |
|
|
Western-type-ABC |
|
|
|
|
|
| All
motifs | 5 | 4 | 0 | 0 | 9 |
| A
motifs | 3 | 0 | 0 | 0 |
|
| B
motifs | 0 | 3 | 0 | 0 |
|
| C
motifs | 2 | 1 | 0 | 0 |
|
|
Eastern-type-ABD |
|
|
|
|
|
| All
motifs | 181 | 2 | 5 | 1 | 189 |
| A
motifs | 63 | 0 | 0 | 0 |
|
| B
motifs | 55 | 2 | 5 | 1 |
|
| D
motifs | 63 | 0 | 0 | 0 |
|
|
Western-type-AB |
|
|
|
|
|
| All
motifs | 7 | 2 | 1 |
| 10 |
| A
motifs | 5 | 0 | 0 |
|
|
| B
motifs | 2 | 2 | 1 |
|
|
|
Eastern-type-BD |
|
|
|
|
|
| All
motifs | 4 | 0 | 0 |
| 4 |
| B
motifs | 2 | 0 | 0 |
|
|
| D
motifs | 2 | 0 | 0 |
|
|
Discussion
H. pylori is well known for its genetic
diversity and geographical differences, which may be associated
with compound clinical disease outcomes (7). In the present study, the
epidemiological characteristics of major virulence genes of H.
pylori were investigated in Guizhou province. H. pylori
was cultured from clinical samples, and virulence genotypes of
cagA, vacA and iceA were examined by PCR assays. The results showed
the prevalent strains of H. pylori in Guizhou province were
cagA-positive, vacA s1c/m2-positive and iceA-positive. The results
describe the prevalence of H. pylori strains in Guizhou
province between January 2015 and December 2016, and provides an
experimental basis for molecular epidemiological investigations of
H. pylori in this region.
Epidemiological investigations have shown that the
cagA genotype varies markedly worldwide. The prevalence of cagA
among H. pylori in different regions varies between 50 and
60% in certain western countries to almost 100% in East Asia
(19). In the present study, it
was shown that cagA genotypes were detected in all strains in
Guizhou province. In another study in this region, Zhou et
al reported that 30 H. pylori strains isolated from
gastric cancer were cagA-positive, with cagA having a positive rate
of 100% (35), which was
consistent with the results of the present study. The vacA
genotypes also vary geographically. The vacA s1a genotype is common
in South Asia, the s1b genotype is common in Latin America and
Africa, and s1c is common in East Asia (24,36,37).
The vacA m1 genotype is common in North Asian countries, including
Japan and South Korea, whereas the m2 genotype is predominant in
Southeast Asia, including Taiwan, China and Vietnam (20). The results of the present study
showed that the predominant vacA genotype was s1c/m2 in Guizhou
province. Notably, the iceA genotype also varies among different
regions. A previous meta-analysis showed that the prevalence of
iceA1 was significantly higher in East Asian countries than in
western countries, whereas the prevalence of iceA2 was higher in
western countries than in East Asian countries (26). Consistent with this finding, the
iceA1 genotype was predominant in Guizhou province in the present
study.
Although Guizhou province is a multi-ethnic society,
there was no ethnic specificity of H. pylori infection. This
differs from Malaysia, which has three major ethnic groups, namely,
Malay, Chinese and Indian; vacA s1a/m2 has been detected in all of
these ethnic groups, whereas vacA s1c/m2 was only isolated from
Chinese patients (38). Therefore,
vacA genotypes were associated with ethnic groups in Malaysia. In
the present study, as no predictive value of these H. pylori
virulence genes was identified in Guizhou province, there may be no
need to classify patients according to ethnicity during treatment,
with patients of different ethnicities offered the same diagnosis
and treatment strategies.
H. pylori infection can result in compound
gastroduodenal diseases, including gastritis, peptic ulcer and
gastric carcinoma. The present study did not find an association
between H. pylori virulence diversity and clinical disease
outcomes in the Guizhou region. Zhou et al reported that
H. pylori infection may induce the demethylation of lactate
dehydrogenase, dihydrolipoamide dehydrogenase and calmodulin genes,
and increase methylation of the Ran-specific GTPase-activating
protein gene, which leads to dysfunctional gene expression in
gastric cancer tissues and cells (35). A number of reports have shown that
infection with a higher number of cagA EPIYA-C motif strains was
associated with increased gastric inflammation and atrophy in
western countries (39–41). However, the genetic diversity of
H. pylori virulence genes is not associated with disease
progression in certain eastern countries (9,16).
Matsunari et al reported that, in the Bhutan region, 209
isolate strains, in which >50% of the strains had multiple EPIYA
motifs repeats in East Asian-type cagA-ABD, are associated with
atrophic gastritis and gastric cancer (42). An association between vacA
genotypes and gastric precancerous or gastric carcinoma was
observed in Brazil (39). However,
no correlation has been found between vacA genotypes and diseases
in Japan or Sweden (9,41). Additionally, an association has
been reported between the iceA1 genotype and peptic ulcers, and the
iceA2 genotype and gastritis (26). Among the 73 isolates in the present
study, no association was found between the iceA1 or iceA2
genotypes and clinical disease outcomes.
Among the 73 patients, none were diagnosed with
gastric cancer; only 14 gastric biopsy samples were obtained from
patients with gastric cancer at the First Affiliated Hospital of
Guizhou Medical University and the People's Hospital of Qiannan
Autonomous Prefecture between January and December 2016, however,
H. pylori isolation was negative. This may be associated
with reduced Helicobacter abundance and overrepresentation
of bacterial genera.
Current knowledge of bacteria other than H.
pylori in the human stomach remains limited. The investigation
of the gastric microbiota in healthy individuals or disease states
is lacking due to limitations of culture conditions and the
collection of gastric mucosal specimens. With the development of
molecular biology and bacterial 16S rRNA gene identification
technology, the constitution and diversity of gastric flora has
been gradually identified. Notably, You et al reported a
microarray used to compare the genomic profiles of strains isolated
from patients with gastroduodenal diseases in the Heilongjiang
province of China, and their findings may provide insight into
novel biomarkers for the prediction of gastric diseases (43). Novel genetic variations may be
examined in our future investigations using similar approaches. The
gastric flora may be important roles in human health and disease,
which remain to be elucidated. The composition of the gastric flora
is dynamic and affected by several factors, including H.
pylori infection and combination therapy with antibiotics and
proton pump inhibitors. The gastric flora may also affect the
development of gastric diseases following H. pylori
colonization. H. pylori infection in Guizhou province may
lead to various diseases without specificity. This may be explained
by gastric flora in the stomach of local residents. Finally, the
clarification of genotypes of gastric flora from different areas
may facilitate clinical therapeutics.
Acknowledgements
The authors would like to thank Professor Jie Yang,
Professor Yonghong Zhang, and Dr Chen Pan from the Department of
Gastrointestinal medicine (The First Affiliated Hospital of Guizhou
Medical University) for case selection.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81460314).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZC conceived, designed and supervised the
experiments. LY, FL, CG and QW performed the experiments. KP, LX,
YX and YC collected the samples. ZC and LY performed the data
analysis and wrote the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All protocols were approved by the Ethical Committee
of the First Affiliated hospital of Guizhou Medical University. All
procedures contributing to the study complied with the Declaration
of Helsinki. Written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
H. pylori
|
Helicobacter pylori
|
|
cagA
|
cytotoxin associated gene A
|
|
vacA
|
vacuolating cytotoxin A
|
|
iceA
|
induced by contact with epithelium
gene A
|
|
PCR
|
polymerase chain reaction
|
References
|
1
|
Pereira WN, Ferraz MA, Zabaglia LM, de
Labio RW, Orcini WA, Bianchi Ximenez JP, Neto AC, Payão SL and
Rasmussen LT: Association among H. pylori virulence markers dupA,
cagA and vacA in Brazilian patients. J Venom Anim Toxins Incl Trop
Dis. 20:12014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang HP, Zhu YL and Shao W: Role of
Helicobacter pylori virulence factor cytotoxin-associated gene A in
gastric mucosa-associated lymphoid tissue lymphoma. World J
Gastroenterol. 19:8219–8226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krisch LM, Posselt G, Hammerl P and
Wessler S: CagA phosphorylation in Helicobacter pylori-infected B
cells is mediated by the non-receptor tyrosine kinases of the Src
and Abl families. Infect Immun. 84:2671–2680. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hashinaga M, Suzuki R, Akada J, Matsumoto
T, Kido Y, Okimoto T, Kodama M, Murakami K and Yamaoka Y:
Differences in amino acid frequency in CagA and VacA sequences of
Helicobacter pylori distinguish gastric cancer from gastric MALT
lymphoma. Gut Pathog. 8:542006. View Article : Google Scholar
|
|
5
|
Zhang M, Zhou YZ, Li XY, Tang Z, Zhu HM,
Yang Y and Chhetri JK: Seroepidemiology of Helicobacter pylori
infection in elderly people in the Beijing region, China. World J
Gastroenterol. 20:3635–3639. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ahn HJ and Lee DS: Helicobacter pylori in
gastric carcinogenesis. World J Gastrointest Oncol. 7:455–465.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Datta De D and Roychoudhury S: To be or
not to be: The host genetic factor and beyond in Helicobacter
pylori mediated gastro-duodenal diseases. World J Gastroenterol.
21:2883–2895. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Myint T, Shiota S, Vilaichone RK, Ni N,
Aye TT, Matsuda M, Tran TT, Uchida T, Mahachai V and Yamaoka Y:
Prevalence of Helicobacter pylori infection and atrophic gastritis
in patients with dyspeptic symptoms in Myanmar. World J
Gastroenterol. 21:629–636. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kita M, Yokota K, Okada H, Take S,
Takenaka R, Kawahara Y, Oguma K, Matsushita O and Yamamoto K: The
genetic diversity of Helicobacter pylori virulence genes is not
associated with gastric atrophy progression. Acta Med Okayama.
67:93–98. 2013.PubMed/NCBI
|
|
10
|
Figura N, Marano L, Moretti E and Ponzetto
A: Helicobacter pylori infection and gastric carcinoma: Not all the
strains and patients are alike. World J Gastrointest Oncol.
8:40–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiménez-Soto LF and Haas R: The CagA toxin
of Helicobacter pylori Abundant production but relatively low
amount translocated. Sci Rep. 6:232272016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Patra R, Chattopadhyay S, De R, Datta S,
Chowdhury A, Ramamurthy T, Nair GB, Berg DE and Mukhopadhyay AK:
Intact cag pathogenicity island of Helicobacter pylori without
disease association in Kolkata, India. Int J Med Microbiol.
301:293–302. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Senda Y, Murata-Kamiya N and Hatakeyama M:
C-terninal Src kinase-mediated EPIYA phosphorylation of pragmin
creates a feed-forward C-terminal Src kinase activation loop that
promotes cell motility. Cancer Sci. 107:972–980. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sicinschi LA, Correa P, Peek RM, Camargo
MC, Piazuelo MB, Romero-Gallo J, Hobbs SS, Krishna U, Delgado A,
Mera R, et al: CagA C-terminal variations in Helicobacter pylori
strains from Colombian patients with gastric precancerous lesions.
Clin Microbiol Infect. 16:369–378. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Furuta Y, Yahara K, Hatakeyama M and
Kobayashi I: Evolution of cagA oncogene of Helicobacter pylori
through recombination. PLoS One. 6:e234992011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang CC, Kuo WS, Chen YC, Perng CL, Lin
HJ and Ou YH: Fragmentation of CagA reduces hummingbird phenotype
induction by Helicobactor pylori. PLoS One. 11:e01500612016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jones KR, Joo YM, Jang S, Yoo YJ, Lee HS,
Chung IS, Olsen CH, Whitmire JM, Merrell DS and Cha JH:
Polymorphism in the CagA EPIYA motif impacts development of gastric
cancer. J Clin Microbiol. 47:959–968. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Braga LL, Oliveira MA, Gonçalves MH,
Chaves FK, Benigno TG, Gomes AD, Silva CI, Anacleto C, Batista Sde
A and Queiroz DM: CagA phosphorylation EPIYA-C motifs and the vacA
i genotype in Helicobacter pylori strains of asymptomatic children
from a high-risk gastric cancer area in northeastern Brazil. Mem
Inst Oswaldo Cruz. 109:1045–1049. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yamaoka Y: Mechanisms of disease:
Helicobacter pylori virulence factors. Nat Rev Gastroenterol
Hepatol. 7:629–641. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shiota S, Suzuki R and Yamaoka Y: The
significance of virulence factors in Helicobacter pylori. J Dig
Dis. 14:341–349. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wen S and Moss SF: Helicobacter pylori
virulence factors in gastric carcinogenesis. Cancer Lett. 282:1–8.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Utsch C and Haas R: VacA's induction of
VacA-containing vacuoles (VCVs) and their immunomodulatory
activities on human T cells. Toxins (Basel). 8:E1902016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Amilon KR, Letley DP, Winter JA, Robinson
K and Atherton JC: Expression of the Helicobacter pylori virulence
factor vacuolating cytotoxin A(vacA) is influenced by apotential
stem-loop structure in the 5′untranslated region of the transcript.
Mol Microbiol. 98:831–846. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamaoka Y and Graham DY: Helicobacter
pylori virulence and cancer pathogenesis. Future Oncol.
10:1487–1500. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thi Huyen Trang T, Thanh Binh T and
Yamaoka Y: Relationship between vacA Tyeps and development of
gastroduodenal diseases. Toxins (Basel). 8:E1822016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shiota S, Watada M, Matsunari O, Iwatani
S, Suzuki R and Yamaoka Y: Helicobacter pylori iceA, clinical
outcomes, and correlation with cagA: A meta-analysis. PLoS One.
7:e303542012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Roesler BM, Rabelo-Goncalves EM and
Zeitune JM: Virulence factors of Helicobacter pylori A review. Clin
Med Insights Gastroenterol. 7:9–17. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ben Mansour K, Fendri C, Zribi M, Masmoudi
A, Labbene M, Fillali A, Ben Mami N, Najjar T, Meherzi A, Sfar R
and Burucoa C: Prevalence of Helicobacter pylori vacA, cagA, iceA
and oipA genotypes in Tunisian patients. Ann Clin Microbiol
Antimicrob. 9:102010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Miftahussurur M, Syam AF, Makmun D, Nusi
IA, Zein LH, Akil F, Uswan WB, Simanjuntak D, Uchida T, et al:
Helicobacter pylori virulence genes in the five largest islands of
Indonesia. Gut Pathog. 7:262015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
da Coata DM, Pereira Edos S and Rabenhorst
SH: What exists beyond cagA and vacA? Helicobacter pylori genes in
gastric diseases. World J Gastroenterol. 21:10563–10572. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xie JH, Chen YL, Wu QH, Wu J, Su JY, Cao
HY, Li YC, Li YS, Liao JB, Lai XP, et al: Gastroprotective and
anti-Helicobacter pylori potential of herbal formula HZJW: Safety
and efficacy assessment. BMC Complement Altern Med. 13:1192013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Secka O, Antonio M, Berg DE, Tapgun M,
Bottomley C, Thomas V, Walton R, Corrah T, Thomas JE and Adegbola
RA: Mixed infection with cagA positive and cagA negative strains of
Helicobacter pylori lowers disease burden in The Gambia. PLoS One.
6:e279542011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Secka O, Antonio M, Tapgun M, Berg DE,
Bottomley C, Thomas V, Walton R, Corrah T, Adegbola RA and Thomas
JE: PCR-based genotyping of Helicobacter pylori of Gambian children
and adults directly from biopsy specimens and bacterial cultures.
Gut Pathog. 3:52011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yamazaki S, Yamakawa A, Okuda T, Ohtani M,
Suto H, Ito Y, Yamazaki Y, Keida Y, Higashi H, Hatakeyama M and
Azuma T: Distinct diversity of vacA, cagA, and cagE genes of
Helicobacter pylori associated with peptic ulcer in Japan. J Clin
Microbiol. 43:3906–3916. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou J, Wang W, Xie Y, Zhao Y, Chen X, Xu
W, Wang Y and Guan Z: Proteomics-based identification and analysis
of proteins associated with Helicobacter pylori in gastric cancer.
PLoS One. 11:e01465212016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dabiri H, Maleknejad P, Yamaoka Y,
Feizabadi MM, Jafari F, Rezadehbashi M, Nakhjavani FA, Mirsalehian
A and Zali MR: Distribution of Helicobacter pylori cagA, cagE, oipA
and vacA in different major ethnic groups in Tehran, Iran. J
Gastroenterol Hepatol. 24:1380–1386. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Camorlinga-Ponce M, Perez-Perez G,
Gonzalez-Valencia G, Mendoza I, Peñaloza-Espinosa R, Ramos I,
Kersulyte D, Reyes-Leon A, Romo C, Granados J, et al: Helicobacter
pylori genotyping from American indigenous groups shows novel
Amerindian vacA and cagA alleles and Asian, African and European
admixture. PLoS One. 6:e272122011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Alfizah H, Rukman AH, Norazah A, Hamizah R
and Ramelah M: Ethnicity association of Helicobacter pylori
virulence genotype and metronidazole susceptibility. World J
Gastroenterol. 19:1283–1291. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Queiroz DM, Silva CI, Goncalves MH,
Braga-Neto MB, Fialho AB, Fialho AM, Rocha GA, Rocha AM, Batista
SA, Guerrant RL, et al: Higher frequency of cagA EPIYA-C
phosphorylation sites in H. pylori strains from first-degree
relatives of gastric cancer patients. BMC Gastroenterol.
12:1072012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Batista SA, Rocha GA, Rocha AM, Saraiva
IE, Cabral MM, Oliveira RC and Queiroz DM: Higher number of
Helicobacter pylori CagA EPIYA C phosphorylation sites increases
the risk of gastric cancer, but not duodenal ulcer. BMC Microbiol.
11:612011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Karlsson A, Ryberg A, Dehnoei MN, Borch K
and Monstein HJ: Association between cagA and vacA genotypes and
pathogenesis in a Helicobacter pylori infected population from
South-eastern Sweden. BMC Microbiol. 12:1292012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Matsunari O, Miftahussurur M, Shiota S,
Suzuki R, Vilaichone RK, Uchida T, Ratanachu-ek T, Tshering L,
Mahachai V and Yamaoka Y: Rare Helicobacter pylori virulence
genotypes in Bhutan. Sci Rep. 6:225842016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
You Y, He L, Zhang M, Fu J, Gu Y, Zhang B,
Tao X and Zhang J: Comparative genomics of Helicobacter pylori
strains of China associated with different clinical outcome. PLoS
One. 7:e385282012. View Article : Google Scholar : PubMed/NCBI
|