Introduction
Rheumatoid arthritis (RA), an autoimmune disease,
presents as a chronic and systemic inflammatory disorder that
affects a number of tissues and organs; however, it frequently
attacks synovial joints and leads to systemic bone loss due to
excessive osteoclast activity (1,2). In
addition, 0.5–1% of the global population suffers from RA (3,4).
Early diagnosis and proper treatment may prevent severe disease
manifestations in patients with RA (5). However, current diagnostic methods
demonstrate various insufficiencies in the early diagnosis of RA,
reflecting the need to investigate novel biomarkers that may
contribute to improved diagnosis and prognosis.
Peptidoglycan recognition protein-1 (PGLYRP-1;
previously termed PGRP-S) is one of the four types of peptidoglycan
recognition proteins in humans that is primarily expressed in
polymorphonuclear leukocyte granules (6,7). In
addition, PGLYRP-1 is a secreted and circulating protein that binds
peptidoglycan and promotes inflammation through the activation of
innate immune mechanisms (8). A
number of studies have demonstrated that an imbalance between the
adaptive and innate immune systems driving excessive immune
responses is involved in the pathogenesis of RA (9–11).
Furthermore, Fodil et al (12) reported that single nucleotide
polymorphisms (SNPs) of Pglyrp1 were associated with RA.
However, limited data are available regarding the potential role of
Pglyrp1/PGLYRP-1 in RA.
In the present study, it was hypothesized that the
plasma levels of PGLYRP-1, reflecting inflammation, may be
associated with RA. This hypothesis was tested by measuring the
expression of PGLYRP-1 in the serum and to determine whether
PGLYRP-1 in the serum may be used as a novel biomarker for RA
diagnosis.
Patients and methods
Patient variables
A total of 62 patients (age, 56.54±11.98 years; 13
males) who fulfilled the revised American College of Rheumatology
criteria for RA (13) were
admitted to The First Affiliated Hospital of Nanchang University
(Nanchang, China) between November 2015 and September 2017. The
Pglyrp1 mRNA expression levels in peripheral blood
mononuclear cells (PBMCs) and the PGLYRP-1 levels in the serum were
simultaneously determined in 22 patients. A further 22 patients
were employed to detect the Pglyrp1 mRNA level in PBMCs
solely because of the limited serum samples and 18 patients were
engaged to identify the PGLYRP-1 level in the serum also solely
because of the limited PBMCs samples. In total, 44 specimens [6
from patients with novel-onset RA (14)] were used to detect the
Pglyrp1 mRNA expression level in PBMCs and 40 specimens (6
cases of novel-onset RA) were used to identify the PGLYRP-1 level
in the serum. The characteristics of the RA patients (44 specimens
for detecting the Pglyrp1 gene and 40 specimens for
detecting the PGLYRP-1 protein) are presented in Table I. The RA disease activity was
measured using the disease activity score 28 (DAS28) (15). In addition, the present study
included 91 healthy controls (HCs; age, 53.76±14.39 years; 14
males, 77 females) who were unrelated to the patients and did not
have inflammatory or autoimmune diseases. Among them, 51 HCs were
employed to detect Pglyrp1 in PBMCs and 40 HCs were engaged
to detect PGLYRP-1 in the serum. RA and HC cases were age- and
sex-matched. As an autoimmune disease control, 41 patients with
systemic lupus erythematosus (SLE; age, 38.61±12.60 years; 3 males,
38 females) who fulfilled the revised American College of
Rheumatology criteria for SLE (16) were also enrolled at The First
Affiliated Hospital of Nanchang University between January 2017 and
September 2017. The study was approved by the Ethics Committee of
The First Affiliated Hospital of Nanchang University (ethical
approval no. 019) and was performed in compliance with the
Declaration of Helsinki. All participants also signed informed
consent forms.
 | Table I.Characteristics of RA cases used for
detecting gene Pglyrp1 and protein PGLYRP-1. |
Table I.
Characteristics of RA cases used for
detecting gene Pglyrp1 and protein PGLYRP-1.
| Characteristic | Detecting gene
Pglyrp1 (n=44) | Detecting protein
PGLYRP-1 (n=40) |
|---|
| Age, years (mean ±
SD) | 54.82±11.83 | 58.43±12.00 |
| Sex
(female/male) | 36/8 | 35/5 |
| DAS28 (mean ±
SD) | 4.91±1.64 | 5.50±1.16 |
| ESR, mm/h (mean ±
SD) | 60.43±33.77 | 68.45±37.13 |
| CRP, mg/l (mean ±
SD) | 49.14±56.51 | 54.25±60.79 |
| RF, IU/ml (mean ±
SD) | 364.47±572.54 | 341.89±538.09 |
| ACPA, RU/ml (mean ±
SD) | 349.75±586.00 | 449.30±656.55 |
| WBC, 109/l
(mean ± SD) | 7.51±2.52 | 6.96±2.08 |
| RBC,
1012/l (mean ± SD) | 3.80±0.58 | 3.74±0.66 |
| HGB, g/l (mean ±
SD) | 106.64±17.44 | 104.08±19.42 |
| HCT, l/l (mean ±
SD) | 0.33±0.07 | 0.32±0.07 |
| PLT, 109/l
(mean ± SD) | 267.37±127.77 | 285.63±122.81 |
| Lymphocytes,
109/l (mean ± SD) | 1.60±0.73 | 1.35±0.66 |
| Lymphocyte, % (mean
± SD) | 22.57±9.22 | 20.62±9.89 |
| Monocytes,
109/l (mean ± SD) | 0.46±0.21 | 0.44±0.21 |
| Monocytes, % (mean
± SD) | 6.37±2.67 | 6.39±3.34 |
| Neutrophils,
109/l (mean ± SD) | 5.29±2.30 | 6.22±7.42 |
| Neutrophils, %
(mean ± SD) | 69.34±12.24 | 70.62±12.88 |
PBMC preparation and total RNA
extraction
PBMCs were isolated from 2 ml EDTA-anticoagulated
blood from each donor using Ficoll-Hypaque density gradients
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 25°C. The cells
were frozen in TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at a concentration of
106/ml and stored at −80°C. Total RNA was extracted from
PBMCs using TRIzol reagent, according to the manufacturer's
protocol. The concentration and quality of the RNA were assessed by
absorbance spectrometry ratios of A260/A280 and A260/A230 using a
NanoDrop ND-1000 spectrophotometer (Agilent Technologies, Inc.,
Santa Clara, CA, USA).
Reverse transcription
quantitative-polymerase chain reaction (RT-qPCR) analysis
Total RNA was reverse-transcribed into cDNA using a
PrimeScript™ RT reagent kit (Takara Bio Inc., Otsu, Japan). RT
assay was set at an initial denaturation step of 37°C for 15 min,
followed by 85°C for 5 sec. The qPCR was performed on an ABI 7500
Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.), using SYBR® Premix Ex Taq™ II (Takara Bio, Inc.)
in 10-µl reactions containing 1X SYBR-Green PCR Master Mix, 0.4 µM
of each specific forward and reverse primer, and 0.5 µl cDNA
template. The PCR assay had an initial denaturation step at 95°C
for 5 min, followed by 40 cycles at 95°C for 15 sec and at 60°C for
1 min, and melt curves were detected to confirm the specificity of
amplification and the lack of primer dimers. The primers used in
RT-qPCR are presented in Table
II. Furthermore, β-actin was used as an internal control.
Following the reactions, the Cq values were determined using the
fixed threshold settings.
 | Table II.Specific gene primers used for
reverse transcription-quantitative polymerase chain reaction
analysis. |
Table II.
Specific gene primers used for
reverse transcription-quantitative polymerase chain reaction
analysis.
| Name | Sequence
(5′→3′) |
|---|
| Pglyrp1 | F:
CACATGAAGACACTGGGCTGGT |
|
| R:
CATGAAGCTGATGCCAATGGAC |
| β-actin | F:
TACTGCCCTGGCTCCTAGCA |
|
| R:
TGGACAGTGAGGCCAGGATAG |
The relative expression levels of Pglyrp1
were calculated using the 2−ΔCq method normalized to the
endogenous control, with
ΔCt=Cttarget-Ctreference (17,18).
ELISA
Stored serum obtained from each categorical subject
was thawed and assayed for PGLYRP1 by using a commercial ELISA kit
(cat. no. SED937Hu; USCN Life Science Inc., Wuhan, China) (19).
Serum C-reactive protein (CRP),
rheumatoid factor (RF), anti-cyclic citrullinated peptide (ACPA),
erythrocyte sedimentation rate (ESR) and routine blood
measurement
The concentrations of serum CRP and RF were
determined by nephelometry methods, according to the manufacturer's
protocol (IMMUNE800; Beckman Coulter, Inc., Brea, CA, USA). ACPA of
immunoglobulin G in serum was measured using a commercial ELISA kit
(cat. no. EC180701; Shanghai Kexin Biotech Co., Ltd., Shanghai,
China). ESR and routine bloods were determined erythrocyte sediment
rate analyzer and hematology analyzer according to the
manufacturer's protocols.
Statistical analysis
Statistical analysis and graphical presentation were
performed using GraphPad Prism version 5.0 (GraphPad Software,
Inc., La Jolla, CA, USA) and SPSS software version 16.0 (SPSS Inc.,
Chicago, IL, USA). A Shapiro-Wilk test was used to assess whether
the data were normally distributed. In addition, a Student's t-test
was used where the normality test passed; otherwise, the
nonparametric Mann-Whitney test was used to analyze the data.
Likewise, the Pearson method or the nonparametric Spearman method
was used for correlation analysis. Receiver operating
characteristic (ROC) curves were constructed to evaluate the
diagnostic value of PGLYRP-1 in the serum of RA patients compared
with the HCs. P<0.05 was considered to indicate a statistically
significant difference.
Results
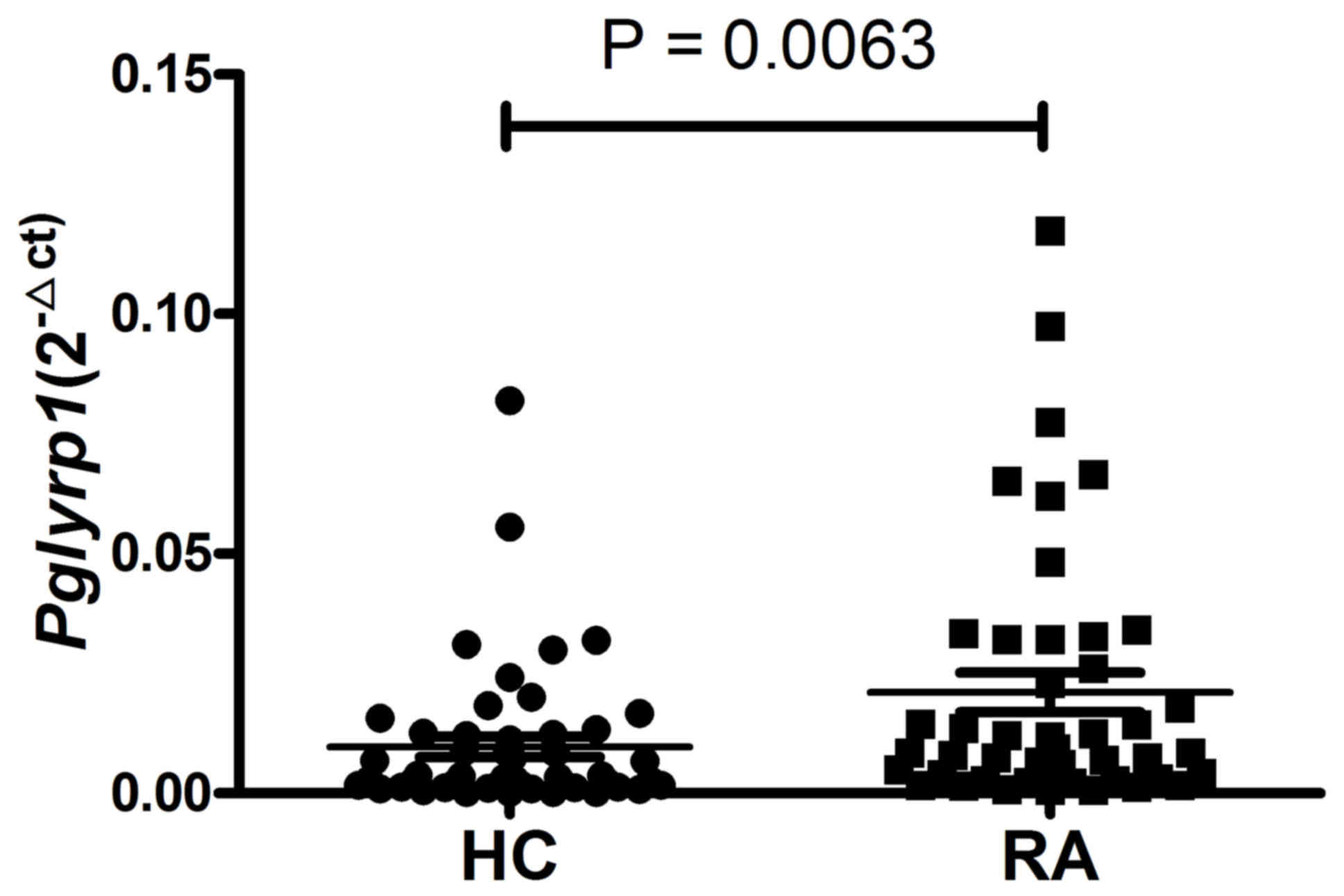
Expression of Pglyrp1 is upregulated
in PBMCs from patients with RA
The expression of Pglyrp1 in PBMCs from 44
patients with RA and 51 HCs was first analyzed using RT-qPCR. As
presented in Fig. 1, compared with
the HCs, the expression level of Pglyrp1 in patients with RA
was significantly upregulated (P=0.0063).
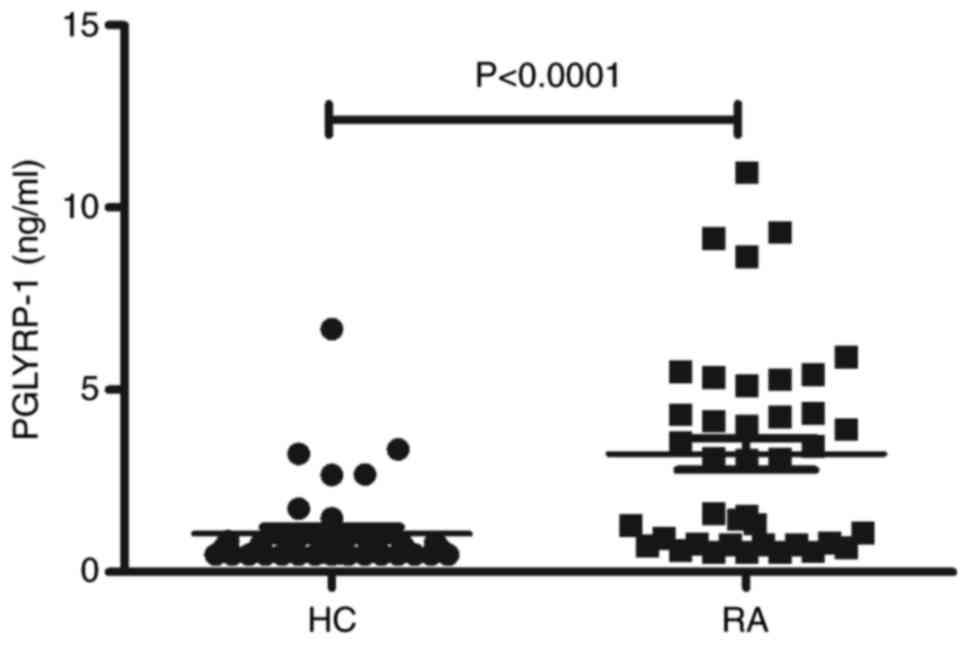
Expression of PGLYRP-1 is increased in
the serum from RA
To determine the expression of the Pglyrp1
gene encoding the PGLYRP-1 protein in serum from RA, additional
serum samples of RA patients (n=40) and HC (n=40) were obtained and
ELISAs were performed. As illustrated in Fig. 2, a significant increase in the
serum levels of PGLYRP-1 in the RA group was demonstrated compared
with the HC group (P<0.0001).
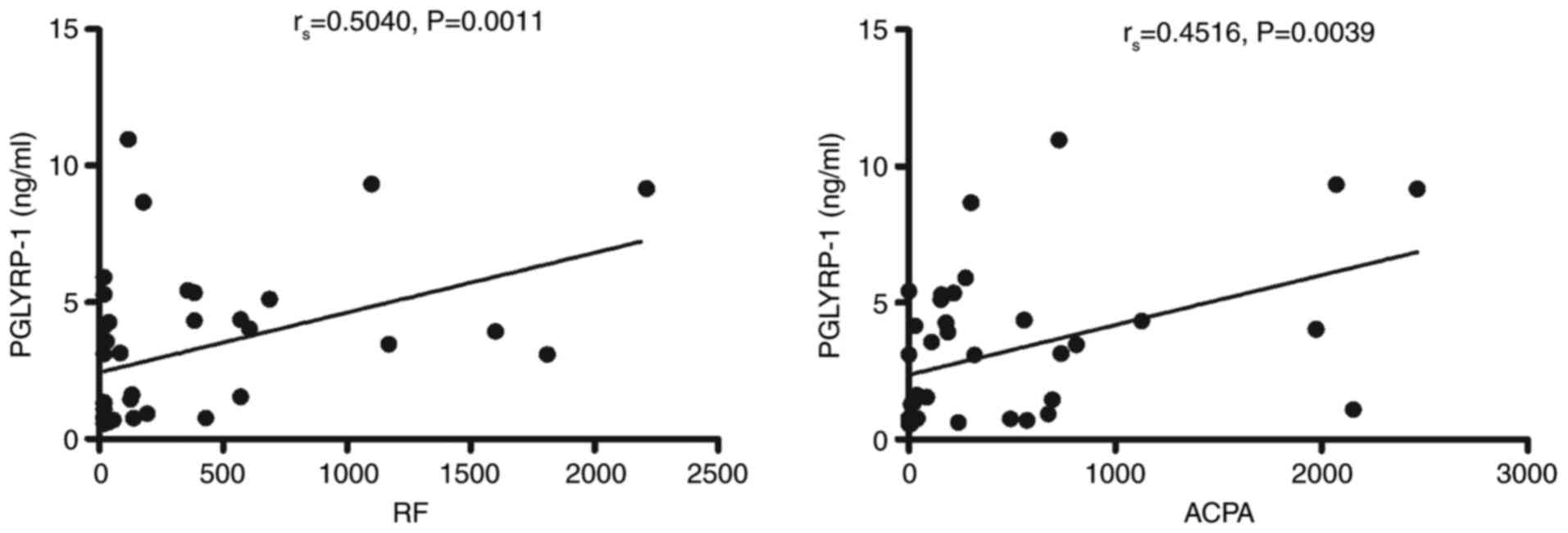
Potential diagnostic values of
PGLYRP-1 in the serum in RA
Next, analysis was conducted to evaluate the
potential diagnostic value of PGLYRP-1 in serum. As presented in
Fig. 3, the PGLYRP-1 level in the
serum from patients with RA was significantly correlated with RF
(r=0.50, P=0.0011) and ACPA (r=0.45, P=0.0039), which are antibody
hallmarks of RA and may reflect the severity of the disease
(20,21). However, the PGLYRP-1 level in the
serum from patients with RA was not correlated with ESR, CRP or
DAS28 (data not shown).
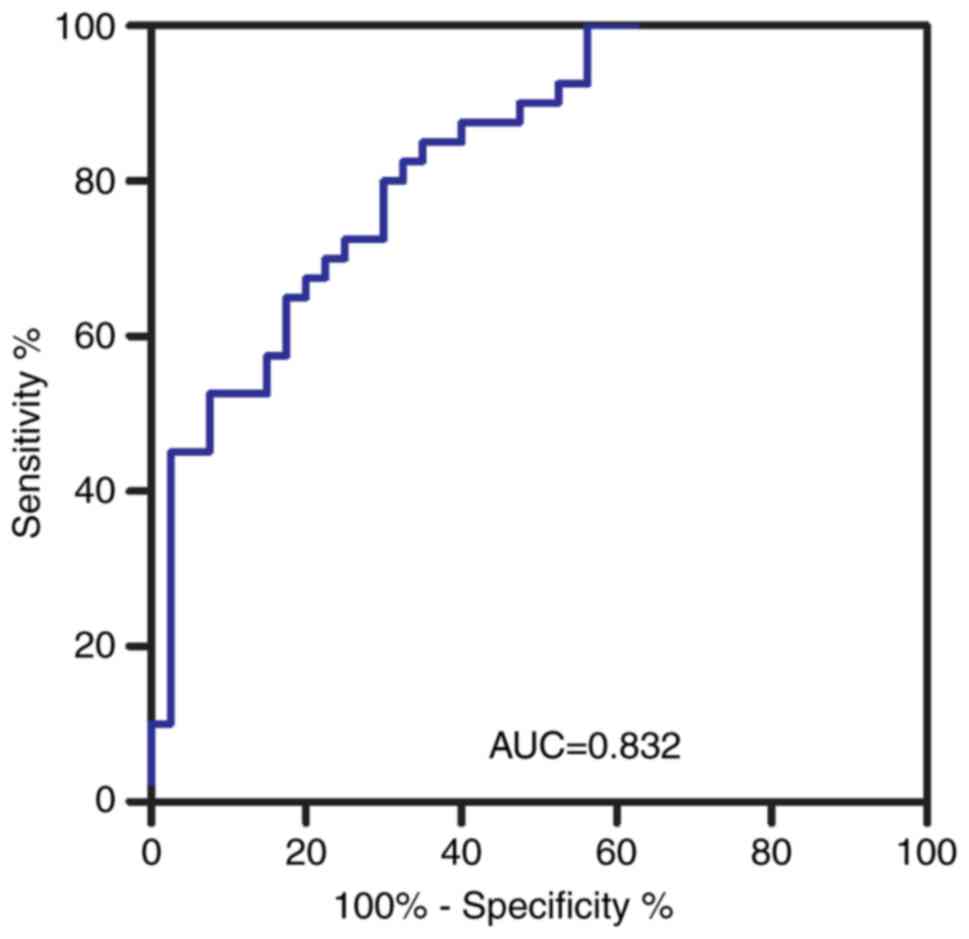
It was hypothesized that the PGLYRP-1 level in the
serum may be used as a biomarker of RA. Therefore, a ROC curve was
constructed. The area under the ROC curve (AUC) was up to 0.832
[95% confidence interval (CI): 0.745–0.919; P<0.0001; Fig. 4] calculated using 0.7306 as the
cutoff point. The sensitivity and specificity were 85.0 and 65.0%,
respectively.
PGLYRP-1 level in the serum in
patients with RA and SLE
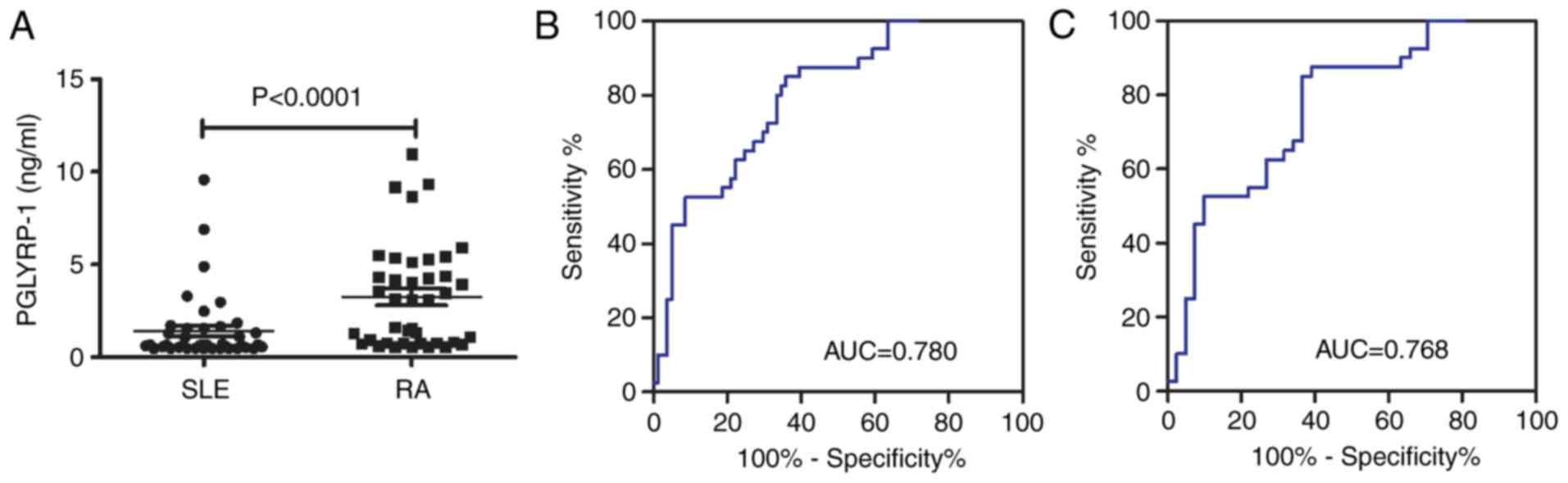
The level of PGLYRP-1 was significantly increased in
patients with RA compared with patients with SLE (P<0.0001;
Fig. 5A). Subsequently, a risk
score based on PGLYRP-1 in serum was further assessed in RA
patients and all controls (HC + SLE). The AUC for the risk score
was 0.780 (95% CI, 0.719–0.880; P<0.0001; Fig. 5B). The sensitivity and specificity
were 85.0 and 64.2%, respectively. The risk score also
significantly differentiated the patients with RA from disease
controls (SLE) and the AUC was 0.768 (95% CI: 0.665–0.871;
P<0.0001; Fig. 5C). The
sensitivity and specificity were 87.5 and 61.0%, respectively.
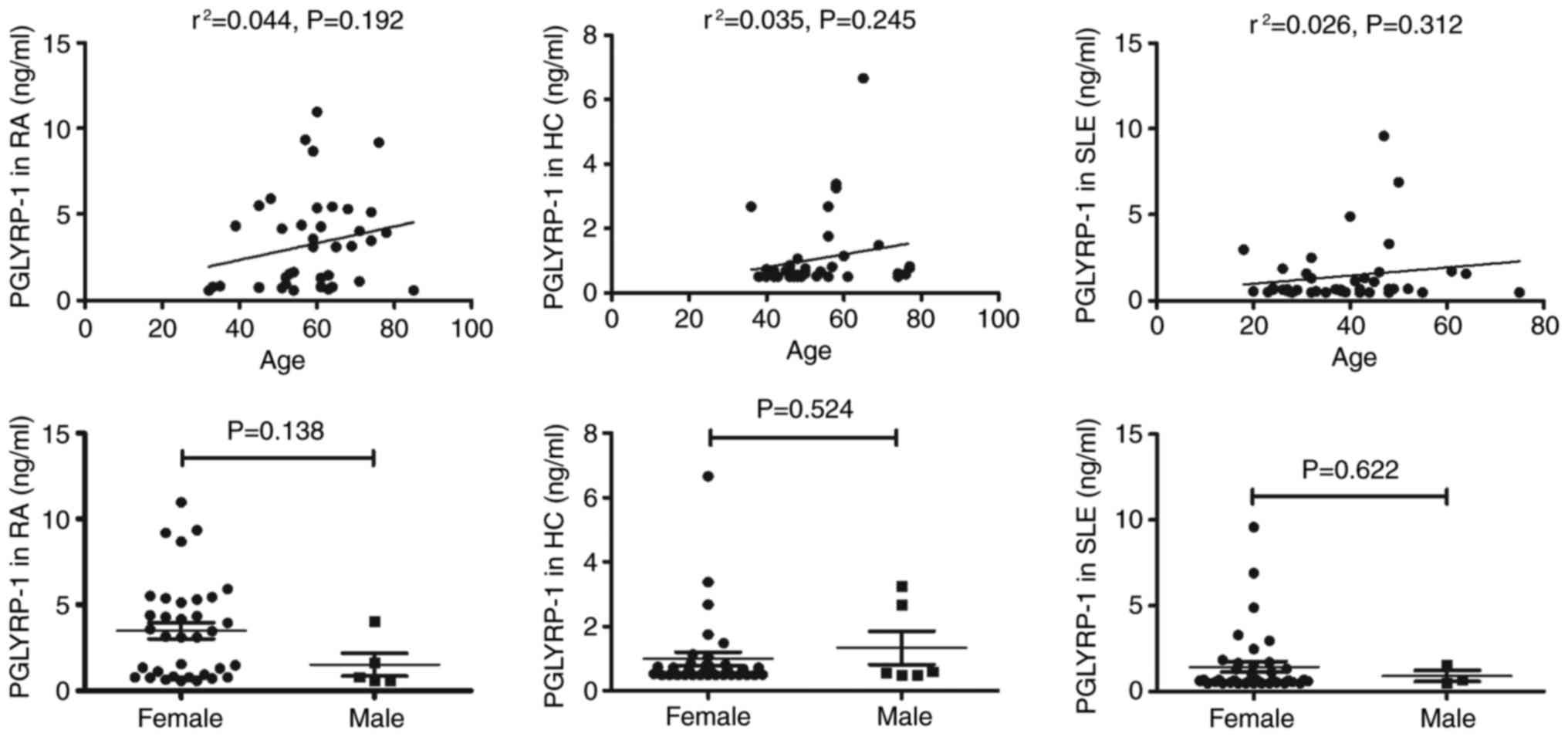
Due to the difference in the ages of high incidence
between RA and SLE (the incidence of RA is high in people of 50–60
years, and the incidence of SLE is high in women of childbearing
age), patients with RA and SLE were not age-matched in the present
study. No correlation between PGLYRP-1 serum levels and age or sex
was observed in SLE, RA and HC (Fig.
6).
Discussion
The Pglyrp1 gene encodes an innate immunity
protein (PGLYRP-1) that directly breaks down the structure of
microbial cell wall peptidoglycan and serves an important role in
antibacterial defenses, and several inflammatory diseases (22). To the best of our knowledge, only
one study has demonstrated that SNPs of Pglyrp1 are
associated with RA (12), which
suggested that the Pglyrp1 gene was correlated with RA.
However, there are no reports regarding the assessment of the
Pglyrp1/PGLYRP-1 levels in the peripheral blood from
patients with RA, to the best of our knowledge. The present study
is among the first studies to report the level of Pglyrp1 in
PBMCs, in which the level of PGLYRP-1 in the serum was increased in
patients with RA. These results indicated that
Pglyrp1/PGLYRP-1 may be associated with the pathogenesis of
RA.
RA is a chronic, debilitating systemic autoimmune
disease with unclear etiology. In 2010, the American College of
Rheumatology/European League Against Rheumatism classification
criteria for rheumatoid arthritis, ACPA, RF, CRP and ESR were
adopted to diagnose RA (23).
However, all of these biomarkers suffer from a weak correlation
with the disease severity of RA. Novel biomarkers with a
satisfactory correlation with the disease activity of RA are
required for the evaluation of the curative effect of treatment or
disease development (5). The
present study revealed that the level of PGLYRP-1 was increased and
the regression model from PGLYRP-1 demonstrated that the AUC was
high. Furthermore, the risk score based on PGLYRP-1 indicated that
it may significantly discriminate patients with RA from those with
SLE. The results supported the level of PGLYRP-1 in the serum being
used as a candidate biomarker of RA.
It is well-known that RA is a systemic autoimmune
disease characterized by increased auto-antibodies, including RF
and ACPA (24). Additionally, the
RF/ACPA titer is frequently positively correlated with disease
activity and the severity of joint destruction (25). In the present study, the serum
levels of RF and ACPA were first determined and analyzed due to
their association with the level of PGLYRP-1 in the serum. The
obtained data demonstrated that the level of PGLYRP-1 was
positively correlated with RF and ACPA in the serum of patients
with RA and suggested that the level of PGLYRP-1 in the serum may
be associated with the autoimmune responses of RA and disease
activity.
However, there are a number of limitations to the
present study. The first one is the relatively small sample size of
patients with novel-onset RA, which results in some uncertainty as
to whether the use of corticosteroids or immunosuppressive drugs
affect the expression of PGLYRP-1 in the serum. The second issue is
the relatively small sample size of patients with RA, and the
present study is limited as only patients from one hospital were
included, which may restrict the representativeness of the results.
Third, due to the lack of data on RF and ACPA in HC, the values of
PGLYRP-1 in serum plus RF/ACPA in RA diagnosis were not evaluated.
Fourth, the roles of PGLYRP-1 in RA pathogenesis in this study were
not investigated. However, the increased expression of PGLYRP-1 and
its correlation with RF and ACPA suggest that PGLYRP-1 may serve
specific roles in the pathogenesis of RA, which will be
investigated in the authors' future study.
In conclusion, to the best of our knowledge, the
level of PGLYRP-1 in serum was demonstrated for the first time to
be upregulated in RA patients. The results of the present study
established an association between the level of PGLYRP-1 in the
serum and RA disease activity. Serum PGLYRP-1 is a promising
diagnostic biomarker for RA.
Acknowledgements
The authors would like to acknowledge the advice
received from Dr Rui Wu [affiliated to the Department of
Rheumatology, the First Affiliated Hospital of Nanchang
University].
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81360459), the
Jiangxi Provincial Natural Science Foundation of China (grant nos.
20151BAB215031 and 20171BAB205113), the Science and Technology
Project of Health and Family Planning Commission of Jiangxi
Province of China (grant no. 20165094) and the Foundation for
Distinguished Young Scientists of Jiangxi Province of China (grant
no. 20171BCB23087).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
QL, XL, ZH and JL conceived and designed the
experiments. QL, XL, LZ, FY, ZD, CQ, RS, JX and YG performed the
experiments. QL, XL, ZH and JL analyzed the data. QL, XL, ZH and JL
wrote the manuscript. FY, ZD, CQ, RS, JX and YG contributed
reagents, materials and analytical tools. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the First Affiliated Hospital of Nanchang University (approval no.
019) and was performed in compliance with the Helsinki Declaration.
All participants signed informed consent forms.
Patient consent for publication
All participants signed informed consent forms.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Schett G and Gravallese E: Bone erosion in
rheumatoid arthritis: Mechanisms, diagnosis and treatment. Nat Rev
Rheumatol. 8:656–664. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Crotti TN, Dharmapatni AA, Alias E,
Zannettino AC, Smith MD and Haynes DR: The immunoreceptor
tyrosine-based activation motif (ITAM)-related factors are
increased in synovial tissue and vasculature of rheumatoid
arthritic joints. Arthritis Res Ther. 14:R2452012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wong JB, Ramey DR and Singh G: Long-term
morbidity, mortality, and economics of rheumatoid arthritis.
Arthritis Rheum. 44:2746–2749. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lajas C, Abasolo L, Bellajdel B,
Hernández-García C, Carmona L, Vargas E, Lázaro P and Jover JA:
Costs and predictors of costs in rheumatoid arthritis: A
prevalence-based study. Arthritis Rheum. 49:64–70. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R,
Lou A, Zhu D, Shi GP and Yang M: Microarray expression profile of
circular RNAs in peripheral blood mononuclear cells from rheumatoid
arthritis patients. Cell Physiol Biochem. 42:651–659. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guan R and Mariuzza RA: Peptidoglycan
recognition proteins of the innate immune system. Trends Microbiol.
15:127–134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Royet J and Dziarski R: Peptidoglycan
recognition proteins: Pleiotropic sensors and effectors of
antimicrobial defences. Nat Rev Microbiol. 5:264–277. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dziarski R and Gupta D: Review: Mammalian
peptidoglycan recognition proteins (PGRPs) in innate immunity.
Innate Immun. 16:168–174. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo Q, Zeng L, Mei H, Huang Z, Luo X, Ye
J, Li X, Guo Y and Li J: PD-L1-expressing neutrophils as a novel
indicator to assess disease activity of rheumatoid arthritis. Int J
Clin Exp Med. 10:7716–7724. 2017.
|
|
10
|
Villanueva-Romero R, Gutiérrez-Cañas I,
Carrión M, Pérez-García S, Seoane IV, Martínez C, Gomariz RP and
Juarranz Y: The anti-inflammatory mediator, vasoactive intestinal
peptide, modulates the differentiation and function of th subsets
in rheumatoid arthritis. J Immunol Res. 2018:60437102018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calabresi E, Petrelli F, Bonifacio AF,
Puxeddu I and Alunno A: One year in review 2018: Pathogenesis of
rheumatoid arthritis. Clin Exp Rheumatol. 36:175–184.
2018.PubMed/NCBI
|
|
12
|
Fodil M, Teixeira VH, Chaudru V, Hilliquin
P, Bombardieri S, Balsa A, Westhovens R, Barrera P, Alves H,
Migliorin P, et al: Relationship between SNPs and expression level
for candidate genes in rheumatoid arthritis. Scand J Rheumatol.
44:2–7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Arnett FC, Edworthy SM, Bloch DA, McShane
DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS,
et al: The American Rheumatism Association 1987 revised criteria
for the classification of rheumatoid arthritis. Arthritis Rheum.
31:315–324. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang J, Shan Y, Jiang Z, Feng J, Li C, Ma
L and Jiang Y: High frequencies of activated B cells and T
follicular helper cells are correlated with disease activity in
patients with new-onset rheumatoid arthritis. Clin Exp Immunol.
174:212–220. 2013.PubMed/NCBI
|
|
15
|
Prevoo ML, van't Hof MA, Kuper HH, van
Leeuwen MA, van de Putte LB and van Riel PL: Modified disease
activity scores that include twenty-eight-joint counts. Development
and validation in a prospective longitudinal study of patients with
rheumatoid arthritis. Arthritis Rheum. 38:44–48. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hochberg MC: Updating the American College
of rheumatology revised criteria for the classification of systemic
lupus erythematosus. Arthritis Rheum. 40:17251997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luo Q, Zhang L, Li X, Fu B, Deng Z, Qing
C, Su R, Xu J, Guo Y, Huang Z and Li J: Identification of circular
RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for
rheumatoid arthritis. Clin Exp Immunol. 194:118–124. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang F, Wu L, Qian J, Qu B, Xia S, La T,
Wu Y, Ma J, Zeng J, Guo Q, et al: Identification of the long
noncoding RNA NEAT1 as a novel inflammatory regulator acting
through MAPK pathway in human lupus. J Autoimmun. 75:96–104. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park HJ, Noh JH, Eun JW, Koh YS, Seo SM,
Park WS, Lee JY, Chang K, Seung KB, Kim PJ and Nam SW: Assessment
and diagnostic relevance of novel serum biomarkers for early
decision of ST-elevation myocardial infarction. Oncotarget.
6:12970–12983. 2015.PubMed/NCBI
|
|
20
|
Alemao E, Guo Z, Frits ML, Iannaccone CK,
Shadick NA and Weinblatt ME: Association of anti-cyclic
citrullinated protein antibodies, erosions, and rheumatoid factor
with disease activity and work productivity: A patient registry
study. Semin Arthritis Rheum. 47:630–638. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Haschka J, Englbrecht M, Hueber AJ, Manger
B, Kleyer A, Reiser M, Finzel S, Tony HP, Kleinert S,
Feuchtenberger M, et al: Relapse rates in patients with rheumatoid
arthritis in stable remission tapering or stopping anti-rheumatic
therapy: Interim results from the prospective randomized controlled
RETRO study. Ann Rheum Dis. 75:45–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tydell CC, Yuan J, Tran P and Selsted ME:
Bovine peptidoglycan recognition protein-S: antimicrobial activity,
localization, secretion, and binding properties. J Immunol.
176:1154–1162. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kedar MP, Acharya RV and Prakashini K:
Performance of the 2010 American College of Rheumatology/European
League against Rheumatism (ACR/EULAR) criteria for classification
of rheumatoid arthritis in an Indian population: An observational
study in a single centre. Indian J Med Res. 144:288–292. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Falkenburg WJJ, Kempers AC, Dekkers G,
Ooijevaar-de Heer P, Bentlage AEH, Vidarsson G, van Schaardenburg
D, Toes REM, Scherer HU and Rispens T: Rheumatoid factors do not
preferentially bind to ACPA-IgG or IgG with altered
galactosylation. Rheumatology (Oxford). 56:2025–2030. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schett G: The role of ACPAs in at-risk
individuals: Early targeting of the bone and joints. Best Pract Res
Clin Rheumatol. 31:53–58. 2017. View Article : Google Scholar : PubMed/NCBI
|