Introduction
Atrial fibrillation (AF) is the most common type of
cardiac arrhythmia in humans, characterized by irregular and rapid
electrical activity of the atria (1,2). In
2010, AF affected ~33.5 million people, causing ~5 million new
cases each year worldwide (3,4). AF
causes 130,000 mortalities/year in the USA (5) and affects 2–3% of the European
population (4,6,7). The
number of patients with AF is predicted to increase rapidly in the
coming decades and to gradually reduce quality of life (8,9). In
Asia, the number of patients with AF and AF-associated stroke is
estimated to reach 72 million and 2.9 million by 2050, respectively
(3,10). Warfarin is commonly applied to
treat nonvalvular persistent atrial fibrillation (NPAF) and
research on NPAF has been extensive (11,12),
although the current treatment options remain inadequate. Thus, it
is necessary to identify a novel biomarker for the prediction,
diagnosis and treatment of NPAF.
Circular RNAs (circRNAs), non-coding RNAs identified
in 1991 (13), regulate gene
expression and act as microRNA (miRNA) ‘sponges’ by competing with
endogenous RNA (ceRNA) networks to suppress specific miRNA activity
(14,15). circRNAs are associated with
numerous diseases and may serve an important role in diagnosis or
pathogenesis (16,17). Previous studies have demonstrated
that circRNA-miRNA-mRNA networks are likely to be involved in
certain diseases (18,19). However, there is little research
regarding the functions of circRNAs in AF, particularly NPAF. The
present study aimed to identify a novel biomarker for diagnosis of
AF.
Materials and methods
Ethics approval statement
Written informed consent was obtained from patients
prior to collection of left atrial appendages (LAAs), which were
abandoned due to surgical techniques. The present study was
conducted in accordance with the Declaration of Helsinki, and all
experimental procedures were approved by the Ethics Committee of
Shanghai East Hospital (approval no. 040–2017) as per relevant
guidelines and regulations (clinical research registration no.
ChiCTR-RRC-17014230).
Clinical specimens
A total of four male patients with NPAF without
valvular disease as an AF group, and four healthy male organ donors
as a control group, were recruited between January 2016 and
December 2017. AF specimens were collected from the LAAs of
patients with NPAF during LAA excision along with surgical atrial
fibrillation ablation. Normal LAAs were collected from healthy
donors. Excised LAAs were stored in RNA later in −20°C prior to RNA
extraction. Characteristics of the four patients are listed in
Table I.
 | Table I.Demographic characteristics of
patients. |
Table I.
Demographic characteristics of
patients.
| No. | Age, years | Sex | NYHA | INR | Coronary
artery | Disease
complications | Duration of AF,
years | Drug
treatments |
|---|
| 1 | 64 | Male | II–III | 3.38 | Negative | Type II
diabetes | 2 | Warfarin,
furosemide, spironolactone and Cordarone |
| 2 | 76 | Male | II–III | 0.92 | Negative | Negative | 3 | Warfarin,
Cordarone, spironolactone, furosemide and Betaloc |
| 3 | 69 | Male | II–III | 2.03 | Negative | Negative | 4 | Warfarin, Betaloc,
furosemide, spironolactone and Cordarone |
| 4 | 60 | Male | II–III | 2.00 | Negative | Negative | 2 | Warfarin,
furosemide and spironolactone |
RNA extraction and qualification
Total RNA was isolated using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocol, and dissolved in
RNase-free water. RNA purity was assessed using a Nanodrop ND-2000
device (Thermo Fisher Scientific, Inc., Wilmington, DE, USA) with
an absorbance (A)260/A280 of 1.8–2.0, and integrity was assessed
using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.,
Santa Clara, CA, USA). The RNA integrity values of the RNAs
obtained from the eight LAA specimens were 7.9, 7.7, 7.1, 7.3, 6.8,
7.2, 8.1 and 7.5, respectively.
RNA sequencing
Sequencing libraries were prepared as recommended by
the VAHTS™ Total RNA-seq (H/M/R) Library Prep kit (Illumina, Inc.,
San Diego, CA, USA). Ribosomal RNA was removed using
target-specific probes, RNase H and DNA polymerase I (Finnzyme;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), and subsequently
fragmented into pieces. Using reverse transcriptase and random
primers, the RNA fragments were copied to the first strand of cDNA:
1 cycle of 25°C for 10 min; and 1 cycle of 50°C for 15 min and 85°C
for 5 min, and the second strand was synthesized using DNA
polymerase I, RNase H and dNTPs (dUTP, dATP, dGTP and dCTP;
Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, single
‘A’ bases were added to the fragments of cDNA and then the adapter
was ligated. To select the appropriate fragment size for
sequencing, the library fragments were selected with VAHTSTM DNA
Clean Beads (Vazyme Biotech Co., Nanjing, China). The second strand
of cDNA was digested using an UDG (uracil-N-glycosylase) enzyme
(Thermo Fisher Scientific, Inc.). Following cluster generation,
150-bp paired-end reads were produced by sequencing the libraries
on the Illumina, Inc. Hiseq X10 platform.
Differential expression analysis
The expression levels of circRNAs were measured by
RNA sequencing and expressed as ‘transcripts per kilobase million’.
Differentially-expressed circRNAs were analyzed using DESeq2 based
on the negative binomial distribution test, and the thresholds were
P<0.05 and fold-change (FC) >2. P-values were calculated
using a statistical algorithm (20).
Validation of circRNAs by reverse
transcription-quantitative PCR (RT-qPCR)
A number of differential circRNAs were randomly
selected to test the accuracy of the RNA sequencing data by
RT-qPCR. Total RNA was reverse transcribed into cDNA using
PrimeScript™ RT Reagent kit (Takara Bio, Inc., Shiga, Japan)
according to standard procedures. A total reaction volume of 20 µl,
including 1,000 ng RNA, 4 µl 5X PrimeScript RT Master Mix and RNase
Free dH2O. The thermocycling conditions were as follows:
37°C for 15 min, then 10 sec at 85°C. RT-qPCR was performed with a
SYBR Green kit (Promega Corporation, Madison, WI, USA) in the
QuantStudio™ 6 Flex system (Applied Biosystems; Thermo Fisher
Scientific, Inc.), The reactions were incubated in 384-well plates
at 50°C for 2 min, and 95°C for 10 min, followed by 40 cycles of
95°C for 15 sec and 60°C for 1 min, followed by a dissociation
curve. The internal reference was 18s rRNA (Applied Biosystems;
Thermo Fisher Scientific, Inc.). Relative expression was quantified
using the 2−ΔΔCq method (21). The primers are listed in Table II.
 | Table II.The list of circRNAs qRT-PCR primers
between NPAF and controls. |
Table II.
The list of circRNAs qRT-PCR primers
between NPAF and controls.
| CircRNA | Forward primer | Reverse primer |
|---|
|
hsa_circRNA011430 |
CGAGCAGTACCCCACAATGG |
TCTGTTGGCATGCTGCTGAA |
|
hsa_circRNA015317 |
ATTAGGCAGACTCTTCAAAACGC |
AAACCCCCACCCACAAAGCA |
|
hsa_circRNA016587 |
TAGGCACAGCTCCTCCAGAT |
TGTGAGATGCTTCACTGCATTC |
|
hsa_circRNA003585 |
GCTGCCCAATGATCTGCTTG |
CCCTGCTTGCAGCTGTAGAAT |
|
hsa_circRNA015019 |
GGAGCCAAAGCCTAATCCGC |
CTGCTGCCAAGGCATACTCA |
|
hsa_circRNA003126 |
ATGGACTGGCGGATCAAGGA |
TGGCCACCAGTCACAAGGTA |
Gene ontology (GO) and kyoto
encyclopedia of genes and genomes (KEGG) enrichment analysis
The differential or target genes of circRNAs were
estimated and determined using GO enrichment analysis at the perl
module (GO: TermFinder, http://search.cpan.org/dist/GO-TermFinder/) The KEGG
enrichment of target genes of differential circRNAs was tested
using R functions (22) (P hyper
and Q value). The significantly-enriched GO terms and KEGG pathways
met the criterion of corrected P<0.05.
circRNA-ceRNA interaction
prediction
The circRNA-miRNA interactions were predicted using
TargetScan 6.0 (http://www.targetscan.org/vert_60/), which identifies
miRNA targets and determines whether or not a given target is
conserved across a given set of species. The sequences of circRNAs
were predicted as miRNA binding seed sequence sites using potential
miRNA response elements.
Statistical analysis
Statistical analyses was conducted using SPSS 19.0
software (IBM Corp., Armonk, NY, USA). When comparing two groups of
profile differences, the fold-change (i.e. the ratio of the group
averages) between the groups for each circRNA was computed. The
differential expression of circRNAs was assessed using a Student's
unpaired t-test. Error bars in the figures represent the means ±
standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
Overview of RNA-seq
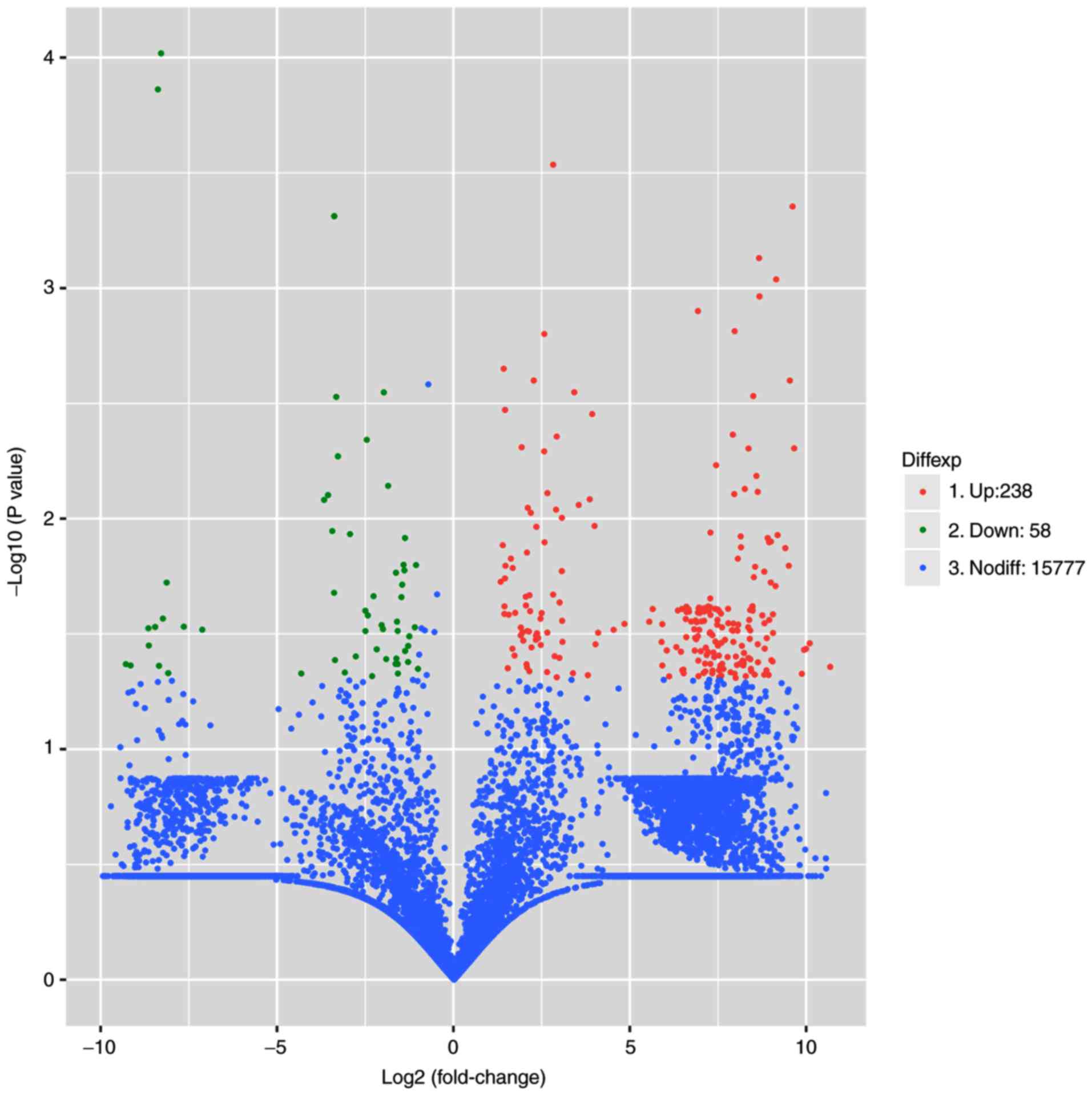
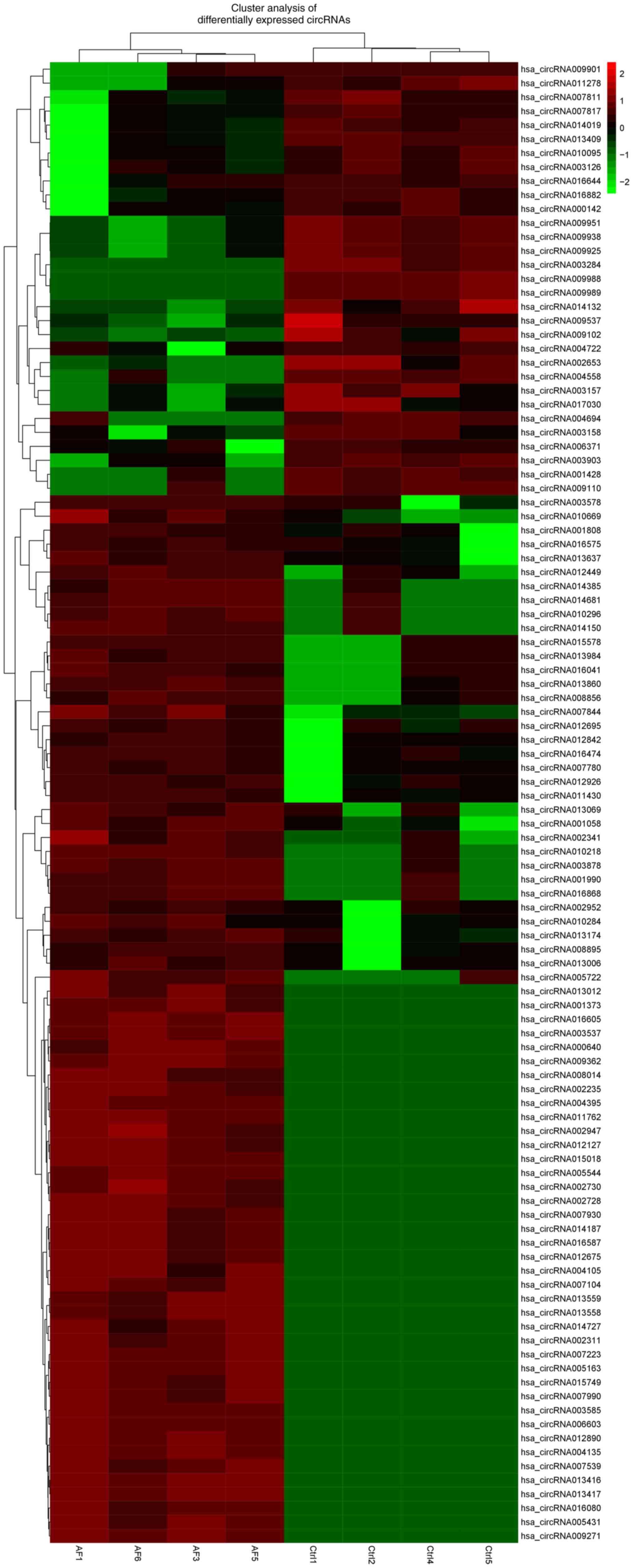
No significant between-group difference was
identified in the expression levels of 15,777 circRNAs. In total,
296 differential circRNAs were identified with FC>2 and
P<0.05. Among them, 238 were upregulated and 58 were
downregulated in NPAF tissues compared with the controls (Figs. 1 and 2). The upregulated circRNAs (data not
shown; available at http://1drv.ms/w/s!Al7sl_MjqbmWgQDki8ZjwTQd2j0H)
with ‘FC >2’ and downregulated circRNAs (data not shown;
available at http://1drv.ms/w/s!Al7sl_MjqbmWgQEKE-v8hCjzTevu)
are listed. A number of the circRNAs were used in the subsequent
analysis.
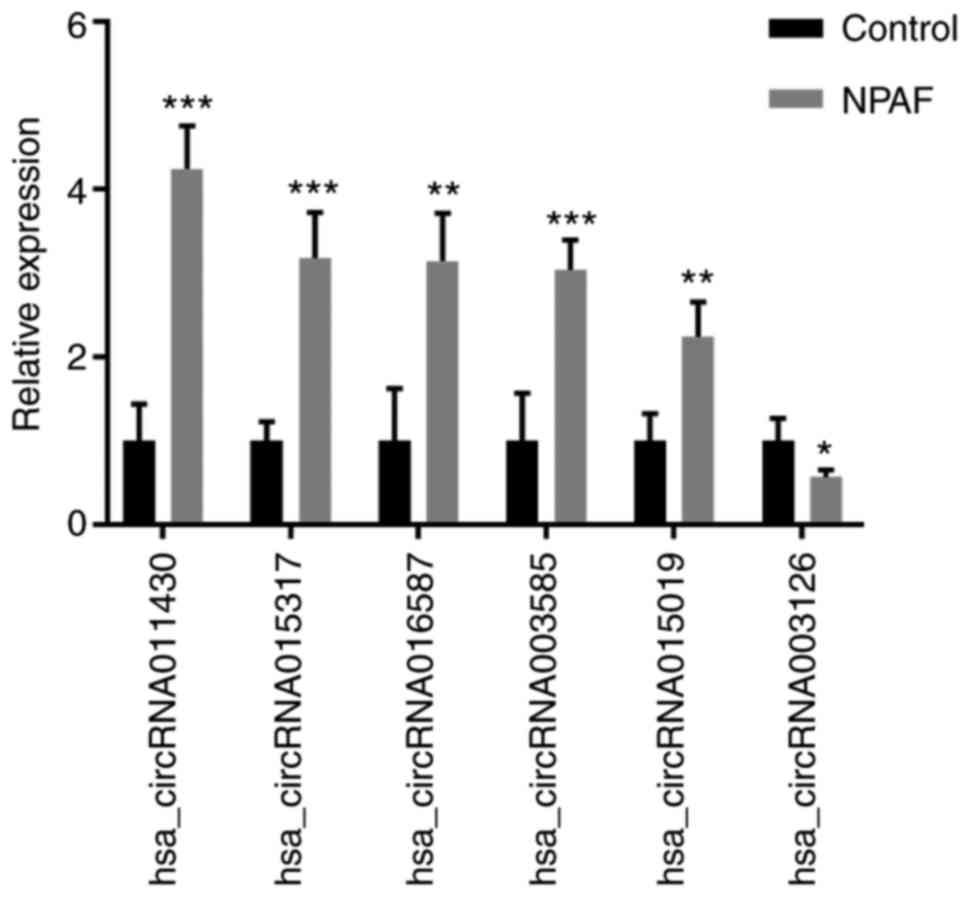
Validation of circRNAs by RT-qPCR
A total of six dysregulated circRNAs were randomly
selected as representatives to validate the RNA sequencing data by
RT-qPCR, and the primers are listed in Table II. All circRNAs were
well-validated by qPCR, and the directions of the changes were
consistent with the RNA-seq data (Fig.
3). The RT-qPCR data suggested the RNA-seq-identified circRNAs
are reliable and require further research.
Functional enrichment analysis: GO and
KEGG pathway analysis
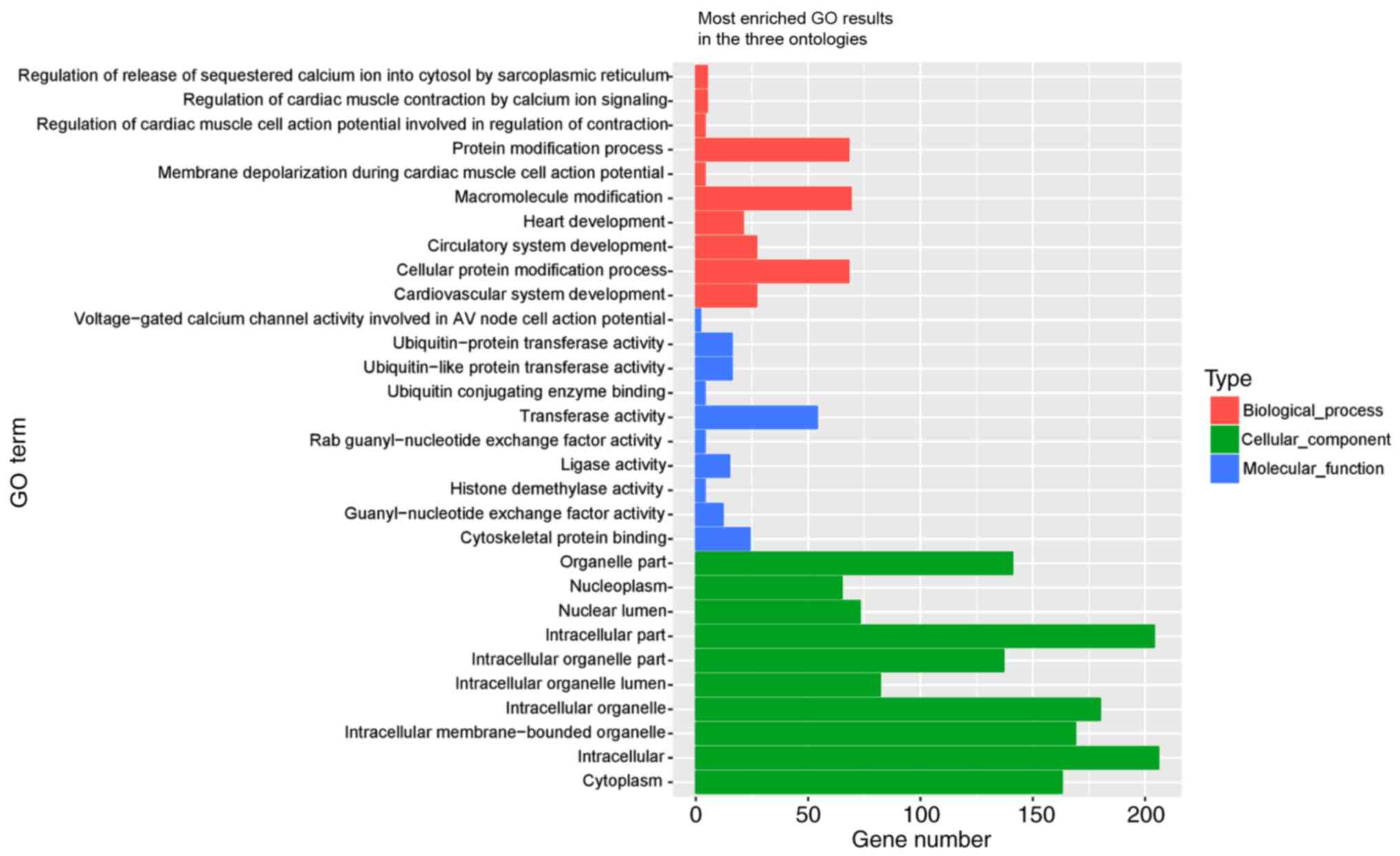
The differential circRNAs were annotated with GO,
and were identified to be involved in the following functions: Cell
component, biological process and molecular function (Fig. 4). GO analysis revealed that a
number of functional pathways were enriched. Some of the top 10
terms were as follows: ‘Voltage-gated calcium channel activity is
involved in AV node cell action potential’ in cell component GO
(no. 0086056); ‘cardiac muscle contraction is regulated by calcium
ion signaling’ in bioprocess GO (no. 0010882); ‘release of
sequestered calcium ion into cytosol is regulated by sarcoplasmic
reticulum’ in bioprocess GO (no. 0010880); and ‘regulation of
cardiac muscle cell action potential is involved in regulation of
contraction’ in bioprocess GO (no. 0098909), which were the most
closely associated with AF. These criteria from the GO analysis
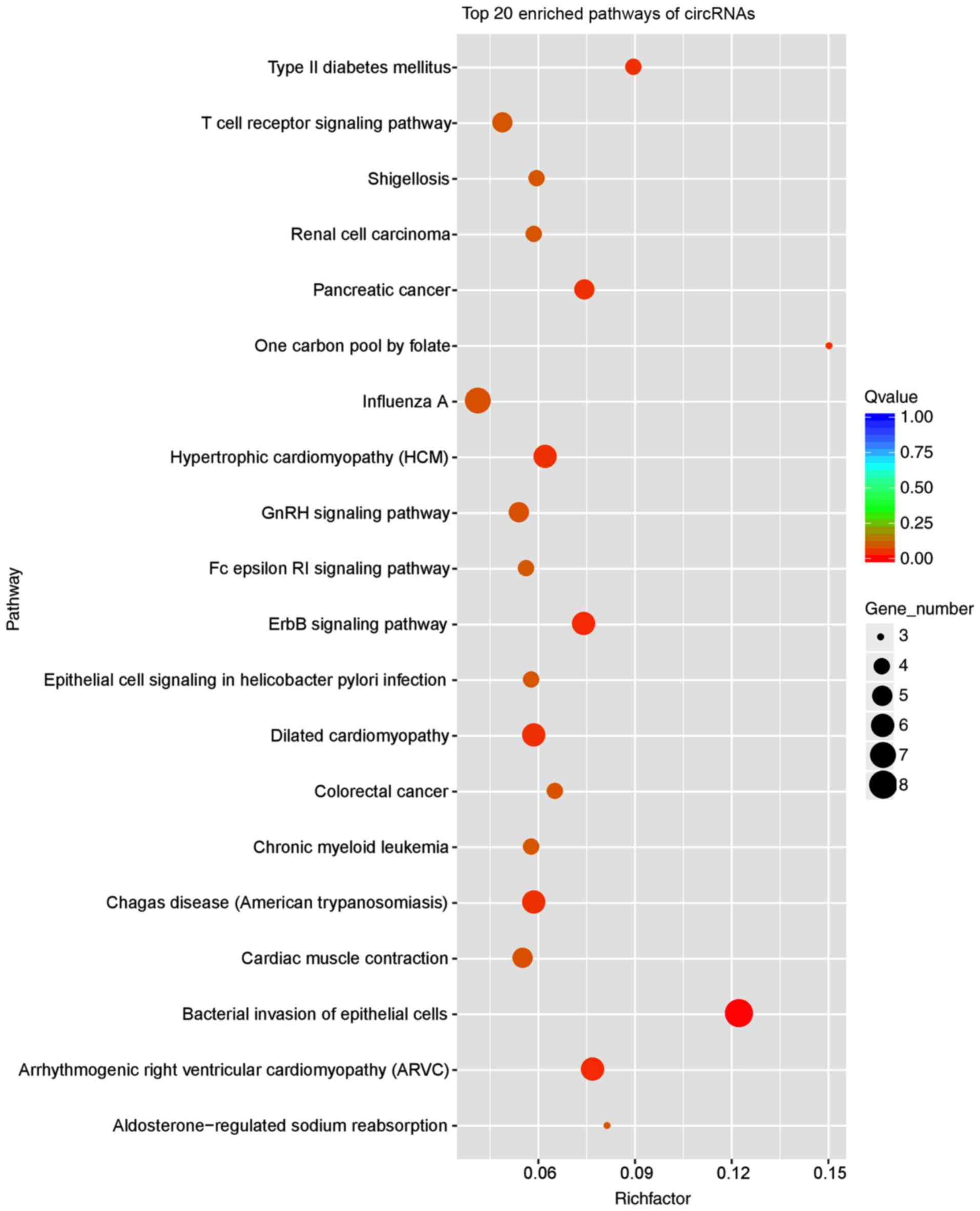
were also applied to the KEGG enrichment analysis. The top 20 KEGG
pathway analysis demonstrated that the most significant pathways
involved in AF were ‘arrhythmogenic right ventricular
cardiomyopathy’ (no. ko05412) and ‘cardiac muscle contraction’ (no.
ko04260; Fig. 5).
Construction of circRNA-ceRNA
interaction network
To analyze the interaction between differential
circRNAs and ceRNAs, a ceRNA network in NPAF was investigated using
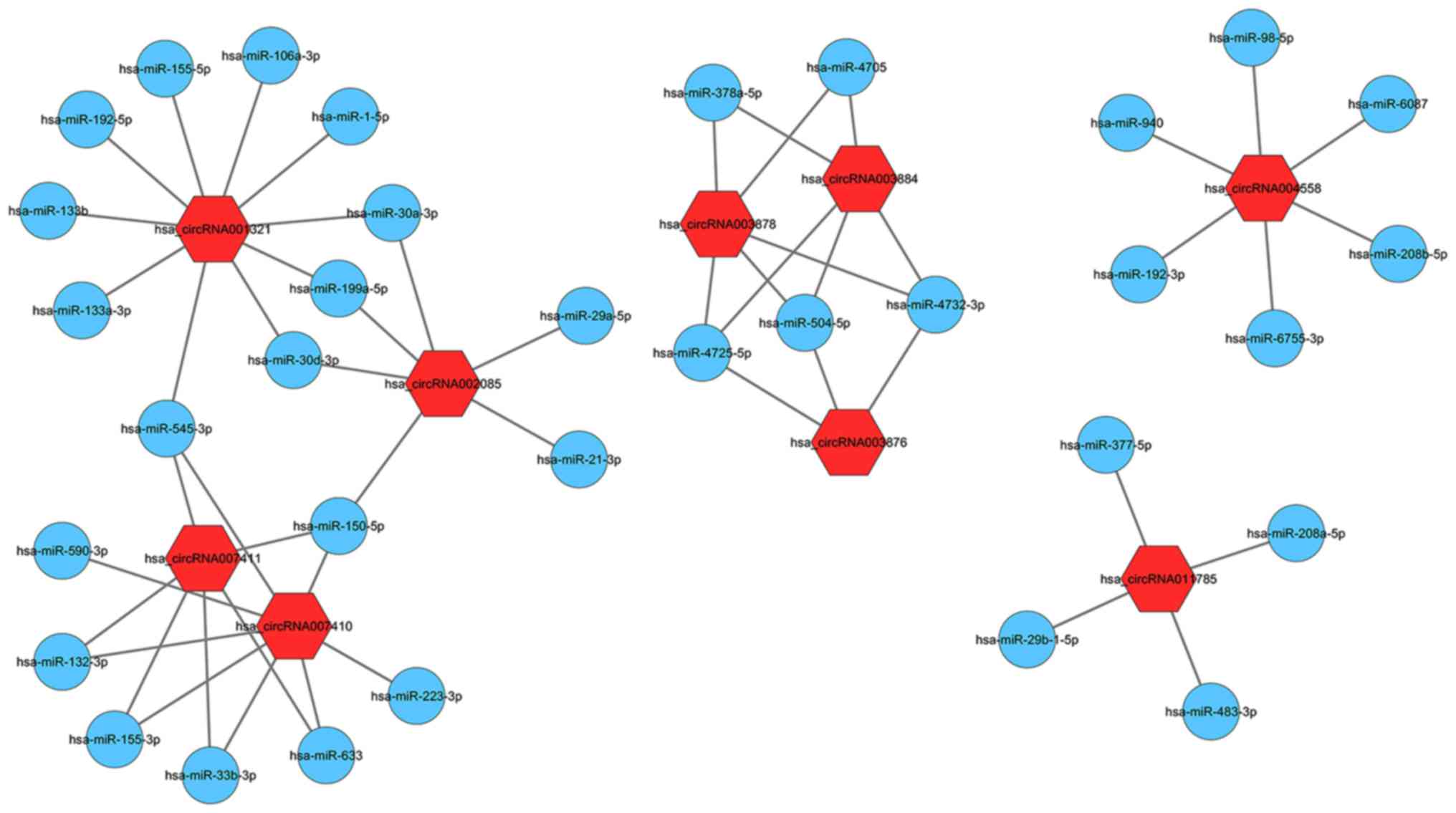
RNA sequencing data. To examine which circRNAs were vital for NPAF
progression, nine differential circRNAs were selected from those
with calcium-associated parental genes. Complete sequences of
hsa_circRNA-011785, −001321, −003878, −002085, −003884, −003876,
−007410, −007411 and −004558 are listed in Table III. Details are provided in
Table IV. A representative
network of circRNAs and miRNAs is presented in Fig. 6. The interactions between
hsa_circRNA002085 and hsa-miRNA (miR)-21, and between
hsa_circRNA001321 and hsa-miR-1, suggest the possibility of an NPAF
regulatory mechanism.
 | Table III.Complete sequence of differentially
expressed circRNAs between the NPAF and control groups. |
Table III.
Complete sequence of differentially
expressed circRNAs between the NPAF and control groups.
| circ_ID | Sequence |
|---|
|
hsa_circRNA011785 |
AAAATGTAGGCCGGTTAATCACTCCTGCCAAAAAGCTTGAAGATACAATACGTCTTGCTGAACTAGTCATTGAAGTTCTTCAGCAAAATGAGGAGCACCACGCAGAGCCACATGTTGATAAAGGAGAAGCCTTTGCGTGGTGGTCAGATTTAATGGTGGAGCATGCGGAGACGTTCCTGTCACTCTTTGCAGTAGACATGGATGCAGCCTTAGAGGTGCAACCTCCAGACACATGGGACAGTTTTCCACTATTTCAGCTGCTGAATGATTTTCTCCGTACTGACTATAATTTGTGCAATGGAAAATTTCACAAACACCTGCAAGACCTGTTTGCCCCACTTGTTGTTAGATATGTGGATCTGATGGAGTCCTCAATTGCACAATCCATTCACAGGGGCTTTGAGCGGGAGTCATGGGAACCAGTCAAGAGTTTAACCAGTAACCTACCCAATGTGAACCTACCCAATGTGAACCTTCCCAAAGTACCAAATCTACCAGTTAACATCCCTCTAGGCATCCCACAAATGCCTACTTTTTCGGCACCGTCATGGATGGCTGCTATATATGATGCGGATAATGGGTCAGGCACCTCAGAAGATCTGTTTTGGAAACTTGACGCCCTTCAGACCTTCATTCGGGACCTGCACTGGCCTGAAGAAGAGTTTGGAAAGCACCTGGAACAACGGCTGAAGTTGATGGCAAGTGACATGATCGAATCTTGTGTCAAAAGAACCAGGATTGCATTTGAAGTTAAGCTGCAAAAAACCAGTCGATCAACAGATTTTCGAGTCCCACAGTCAATATGCACCATGTTTAATGTTATGGTTGATGCCAAAGCTCAATCAACAAAACTTTGCAGCATGGAAATGGGCCAAGAGCATCAATACCATTCAAAAATAGACGAACTAATTGAAGAAACTGTTAAAGAAATGATAACACTCTTGGTTGCAAAGTTCGTTACTATCTTGGAAGGAGTGCTGGCAAAATTATCCAGATATGACGAAGGGACTTTGTTTTCTTCTTTTCTGTCATTTACCGTGAAGGCAGCTTCCAAATATGTGGATGTACCTAAACCCGGGATGGACGTGGCCGACGCCTACGTGACTTTCGTCCGCCATTCTCAGGATGTCCTGCGTGATAAGGTCAATGAGGAGATGTACATAGAAAGGTTATTTGAT |
|
hsa_circRNA001321 |
GAGGAATTAGAAGGTCTTCATCTATGTCTTATGTTGATGGCTTCATAGGGACATGGCCCAAAGAGAAAAGATCATCAGTGCATGGCGTATCATTTGATATTTCTTTTGATAAAGAAGATAGTGTACAGAGATCCACTCCAAACCGAGGAATCACTCGTTCTATTAGTAATGAAGGACTTACTCTGAACAACAGTCATGTATCTAAACACATTAGGAAAAATTTGTCCTTCAAGCCAATAAATGGAGAAGAGGAAGCAGAGAGCATTGAAGAAGAACTTAATATAGATTCTCACAGTGACCTCAAATCTTGTGTGCCCCTTAACACAAATGAACTAAATTCTAATGAGAATATTCATTACAAGCTTCCAAATGGAGCTTTACAAAATAGAATACTTCTTGACGAGTTTGGCAATCAGATCGAGACACCAAGCATTGAAGAAGCATTACAAATAATTCATGATACTGAAAAATCTCCTCATACACCTCAGCCAGACCAAATTGCTAATGGCTTCTTTCTTCATAGTCAAGAAATGAGTATCTTAAATTCAAATATCAAGTTAAATCAATCTAGTCCTGATAATGTAACTGATACGAAAGGTGCCTTGAGTCCCATAACTGACAATACTGAAGTAGACACTGGAATTCACGTTCCTTCAGAAGATATTCCTGAAACTATGGACGAAGATTCTTCGTTGAGAGATTATACTGTAAGCTTGGACTCTGACATGGATGATGCATCTAAATTTCTTCAGGATTATGATATTCGAACTGGCAACACCAGGGAAGCTTTGAGTCCTTGTCCAAGTACTGTAAGTACCAAGTCTCAGCCAGGCAGCAGTGCTTCTTCTAGTTCTGGAGTTAAAATGACCAGCTTTGCTGAACAAAAATTCAGGAAACTGAATCATACCGATGGAAAAAGTAGTGGAAGCAGTTCTCAAAAAACTACACCAGAAGGCTCTGAACTTAATATTCCTCATGTGGTTGCTTGGGCACAAATTCCAGAAGAAACAGGGCTTCCACAGGGACGGGACACTACCCAGCTGTTGGCCTCTGAAATGGTGCATCTTAGGATGAAACTAGAAGAAAAGAGGCGTGCTATAGAAGCCCAGAAAAAGAAAATGGAAGCTGCTTTTACCAAACAGAGACAGAAAATGGGAAGGACAGCATTCCTTACTGTAGTGAAAAAGAAAGGGGATGGGATATCTCCTCTACGAGAGGAAGCGGCGGGTGCAGAAGATGAGAAAGTATATACTGATCGAGCAAAAGAAAAGGAATCACAAAAAACTGATGGACAAAGGAGCAAGTCACTGGCAGATATAAAAGAGAGCATGGAGAATCCTCAAGCCAAATGGCTAAAGTCTCCAACTACACCTATTGATCCTGAGAAGCAGTGGAACCTGGCAAGCCCCTCAGAAGAAACTTTAAATGAAGGAGAGATTTTAGAATATACCAAATCCATTGAAAAGTTAAATTCATCCCTGCATTTTCTACAACAAGAAATGCAACGCTTGTCACTTCAGCAGGAGATGTTAATGCAGATGAGAGAGCAACAATCTTGGGTGATTTCACCTCCACAACCCTCTCCACAGAAACAGATTCGAGATTTTAAGCCTTCTAAGCAGGCAGGCCTGTCATCAGCCATTGCACCATTCTCCTCAGACTCCCCTCGTCCTACTCACCCATCTCCACAGTCTTCTAACAGGAAAAGTGCATCTTTTTCTGTTAAAAGTCAAAGGACTCCTAGGCCAAATGAGTTAAAAATAACACCTTTGAATCGAACCTTGACACCTCCTCGGTCTGTGGATAGCCTTCCTCGGTTAAGGAGGTTTTCACCAAGTCAAGTTCCTATTCAAACTAGGTCATTTGTATGTTTTGGGGATGATGGAGAACCTCAGTTAAAGGAATCCAAACCTAAAGAGGAAGTTAAAAAGGAGGAATTGGAATCCAAAGGGACTTTGGAACAGCGTGGACATAATCCAGAAGAAAAGGAAATCAAACCTTTTGAGTCAACAGTCTCTGAAGTCCTATCACTGCCTGTCACAGAGACTGTATGTCTGACACCAAATGAGGACCAATTGAATCAACCCACAGAACCCCCTCCTAAACCCGTTTTCCCACCCACTGCTCCAAAAAATGTTAATCTGATTGAAGTTTCCCTCTCAGATTTGAAACCCCCTGAAAAGGCTGATGTACCTGTTGAAAAATATGATGGAGAAAGTGATAAAGAACAATTTGATGATGACCAGAAAGTATGCTGTGGATTCTTTTTTAAGGATGATCAAAAAGCAGAAAATGATATGGCAATGAAACGGGCAGCTTTGTTGGAGAAAAGATTAAGAAGGGAAAAGGAAACTCAGCTCCGGAAACAACAGTTGGAAGCAGAAATGGAGCATAAGAAGGAGGAAACAAGGCGTAAAACTGAGGAAGAACGTCAGAAGAAAGAAGATGAGAGAGCACGCAGAGAATTTATTAGGCAAGAATATATGAGGCGGAAACAACTGAAATTCTAATGGAAGATATGGATACAGTAATTAAACCCCGTCCTCAAGTAGTAAAACAAAAAAAACAGCGACCAAAATCTATTCACAGAGATCATAGAATCCCCCAAAACACCAATAAAGGGTCCTCCAG |
|
hsa_circRNA003878 |
GTCAGGATCGTGTGATTGTAAACTAGTCAGGTCAGGGTCGTGTGATTGTAAACTAGTCAGGTCAGGGTCGTGTGATTGTAAACTAGTCAG |
|
hsa_circRNA002085 |
TGCTGGGATTACAGGCGTGACCATGGCACCGAGAACAATTTTTTGAAATTTAAGTTTTGGTGTTAAAGGTTTGGGTCAAGTTAAGCAGAGCTTGGGCTTGACACATATGTACACACGTGTGTGTGGGTGCGATGTATGTATTCTGCAACTGGCTATGACACCAATGCCCAGATGTCAAGTGGGAAATTTTCCAAGTTCTTGACAGCATCCAAGCATATATACTTTAGGCAGATACTTATATTCTTTGGTCTGGCAATGAGTGAGGAGAATAATAGATGAGACCTTTGGAAACGATGAAAATGATCATTACAGACCCATCCACACTTGATAGTTGGAGCAGTTGTGCCTGGAGGTGGATTGGGAGAACTTCTCTTTCGAGCTTTCTGAAGTTTTCCTTTAAATCAGTTGAGCTATTGATATAAAAATTGTCTCAGAGTCCTAATATGCTTATTATACATTTTGTAGTTGGTTATAGCTCCAAGTGACTCAGTCACCAGAAAAGTCACTTGAAAGTTGGTGTATCTCTACAAAACCTTGGTGAACATTTAAATCACCTTTTTAAATATTACACTCTTCATGGAACTTTACTTTTAGTAATGTCAAACATCTGCCTACAAAATGGAATGTTATTTTACATGTATACTAACTTGTTCTTCAAAATGCAGCACCTCCTTTCACTATACAGACGGTCCTTGACATACACTGGTTCAACTTAAGATTGTGCAAGTTTATTACAAATTTTTCAGGACATAACCTTATCATAAGTCAAGGTCTGAACTTTACGATGGTTTGACGTCAGACTTTTTACTGGGTCTGTCCACGTGTTAAGTACATTTTGAGTTACCATATTTTCTATTTATGATGGGTTTAGTGAAGTGTAACTCTATCATAAGTTGGGGAGCATCTGTGTTGCCAAAACCACTGGAGGGCTACAATATGGCTGGAGGGAATGGAAGAATTTTTCTGGTATATAGAAAGAAGTGAGGCTGACATTTGCAGGCAGAACTGCTGCTTTATCGCAAGGTGACATAGTCTTTTTCCACTTTAAGAGCCTAGTAGCAGTCCCTAGAAATCACTGAAATCATCTTTGAGATATATAAGAGTGTAAGGTTTATATAGGAATCCCTGCAACTTCACACATTTGAGAAGGAACAGGATAGGCCTCCCTTCCTCTGTGGATTCAGGGGCTCCGTAATTATCAAGGGAAGGTATAAGTTTCACATAATACCTACAGGGCCTTTCATTTTCATTTCATGCATACTATTTTACTTAATTTTGAGCATCTTAGAATTGCTTGCTTCAAGTTATAAGTAGATAGTTTTTGAAATTAATTTGCTTCCTAAAGCAATTCATTTATTGTGCTCAGATTTTGCTCACTTGAAAGTCCTATATTCTGTGTAGAACTTGGAATTCTATATAAACAGAACAATGGTTTGAATTGCAAAAAAAGCCCATGATCAGTATCTTAGTTTGCCAATGGCTGGGCATGGTGGCTCATGCCTGTCATCCCAACA |
|
hsa_circRNA003884 |
GTGTGATTGTAAACTAGTAAGGTCAGGATCGTGTGATTGTAAAGTAGTCAGGTCAGGATCGTGTGATTGTAAACTAGTCAGGTCAGGGTC |
|
hsa_circRNA003876 | GTCAGGGTCGTGTGA
TTGTAAACTAGTCAG |
|
hsa_circRNA007410 |
GCAGAGGAAAATATTGACTTATTAGATGATGGCTCTAATTCTTTTGCAACTGACTTGTCATCAGGAACTATTAACCACAAGAAATACATCAAGTTTTCTAAAACAATAGAGAAGGAAATTTCACCGGAAATTAGGAGTTTGAGCCCAGAATATAAAAAAATATTTGAAACATCAATAATCTTTTGTGGAGAAGAAAAGTCCTCTGATTTTTCAGGAGAAAAAAAAGTTGGGAGAAAGAGTTTACAAGTACAACAGCACAGTAAAAGAACTGAGATTATCCCTCCTTTTCTGAAGCTGTCAAAGGAGAAGGTGACAAGGAAAGAAAACTCTTTATGCAAGTTGCCGAATCAGTACAGCGTTCACAAGACTTCATCACCTCTTTGTACATCTTCTGCAATTACTCGGGAAAAGGAAATGCTGTCTAACCTCTACATGACATTATATGATGAAGTAACCCATGGATATTTACACTCAAAAGAATTAAGTGCACTTCATAAAGCCTGTAAAATTTTTAGTAAAATTCGAAGTGGTAAGATTTATGTGAATGATCTTCCAGTGATCCTTTGCATCTTGAGAATTTCTATAAGTGATTTAGAAATGCGACAGGCACTAAAGACTGTTGATATTGATGCATTCCAGGATGCCTTGAAGATTTTCTGTAGGATAAAAGGTGGTCGAGTTTCAACTGATGACGTGTTTGCTGTTTTGGATAGCATGGGTATCCCTATAAACCGTGAAATTTTAGAAGAAGTGACAAAACATACCTATATTGACA |
|
hsa_circRNA007411 |
AGGAAAATATTGACTTATTAGATGATGGCTCTAATTCTTTTGCAACTGACTTGTCATCAGGAACTATTAACCACAAGAAATACATCAAGTTTTCTAAAACAATAGAGAAGGAAATTTCACCGGAAATTAGGAGTTTGAGCCCAGAATATAAAAAAATATTTGAAACATCAATAATCTTTTGTGGAGAAGAAAAGTCCTCTGATTTTTCAGGAGAAAAAAAAGTTGGGAGAAAGAGTTTACAAGTACAACAGCACAGTAAAAGAACTGAGATTATCCCTCCTTTTCTGAAGCTGTCAAAGGAGAAGGTGACAAGGAAAGAAAACTCTTTATGCAAGTTGCCGAATCAGTACAGCGTTCACAAGACTTCATCACCTCTTTGTACATCTTCTGCAATTACTCGGGAAAAGGAAATGCTGTCTAACCTCTACATGACATTATATGATGAAGTAACCCATGGATATTTACACTCAAAAGAATTAAGTGCACTTCATAAAGCCTGTAAAATTTTTAGTAAAATTCGAAGTGGTAAGATTTATGTGAATGATCTTCCAGTGATCCTTTGCATCTTGAGAATTTCTATAAGTGATTTAGAAATGCGACAGGCACTAAAGACTGTTGATATTGATGCATTCCAGGATGCCTTGAAGATTTTCTGTAGGATAAAAGGTGGTCGAGTTTCAACTGATGACGTGTTTGCTGTTTTGGATAGCATGGGTATCCCTATAAACCGTGAAATTTTAGAAGAAGTGACAAAACATACCTATATTGACAGTGA |
|
hsa_circRNA004558 |
ACCAATCTGACCTTCGTTGGCTGCGTGGGCATGCTGGATCCTCCGAGAATCGAGGTGGCCTCCTCCGTGAAGCTGTGCCGGCAAGCAGGCATCCGGGTCATCATGATCACTGGGGACAACAAGGGCACTGCTGTGGCCATCTGTCGCCGCATCGGCATCTTCGGGCAGGATGAGGACGTGACGTCAAAAGCTTTCACAGGCCGGGAGTTTGATGAACTCAACCCCTCCGCCCAGCGAGACGCCTGCCTGAACGCCCGCTGTTTTGCTCGAGTTGAACCCTCCCACAAGTCTAAAATCGTAGAATTTCTTCAGTCTTTTGATGAGATTACAGCTATGACTGGCGATGGCGTGAACGATGCTCCTGCTCTGAAGAAAGCCGAGATTGGCATTGCTATGGGCTCTGGCACTGCGGTGGCTAAAACCGCCTCTGAGATGGTCCTGGCGGATGACAACTTCTCCACCATTGTGGCTGCCGTTGAGGAGGGGCGGGCAATCTACAACAACATGAAACAGTTCATCCGCTACCTCATCTCGTCCAACGTCGGGGAAGTTGTCTG |
 | Table IV.Parental genes corresponding to the
differentially expressed circRNAs. |
Table IV.
Parental genes corresponding to the
differentially expressed circRNAs.
| A, upregulated
circRNAs |
|---|
|
|---|
| circ_ID | P-value | FC | Chr | Start | End | Str | Loci | Gene list | Description |
|---|
|
hsa_circRNA003878 | 0.003525 |
15.10051 | chr12 |
2255950 |
2256039 | – | Exon | CACNA1C | Calcium
voltage-gated channel subunit α1C |
|
hsa_circRNA003884 | 0.027306 | 94.9909 | chr12 |
2256001 |
2256090 | – | Exon | CACNA1C | Calcium
voltage-gated channel subunit α1C |
|
hsa_circRNA003876 | 0.033396 | 116.6174 | chr12 |
2255950 |
2255979 | – | Exon | CACNA1C | Calcium
voltage-gated channel subunit α1C |
|
hsa_circRNA007411 | 0.042442 | 295.9099 | chr17 |
47335200 |
47361525 | + | Exon | EFCAB13 | EF-hand calcium
binding domain 13 |
|
hsa_circRNA007410 | 0.041746 | 522.6585 | chr17 |
47335196 |
47361521 | + | Exon | EFCAB13 | EF-hand calcium
binding domain 13 |
|
hsa_circRNA002085 | 0.025361 | 101.3261 | chr10 |
18508039 |
18509552 | – | Exon | CACNB2 | Calcium
voltage-gated channel auxiliary subunit β2 |
|
hsa_circRNA011785 | 0.026138 | 2.935789 | chr3 |
62438104 |
62499268 | – | Exon | CADPS | Ca2+
dependent secretion activator |
|
hsa_circRNA001321 | 0.012686 | 5.92702 | chr1 | 200847640 | 200853495 | + | Exon | CAMSAP2 | Calmodulin
regulated spectrin associated protein family member 2 |
|
| B, downregulated
circRNAs |
|
| circ_ID | P-value | FC | Chr | Start | End | Str | Loci |
GeneList |
Description |
|
|
hsa_circRNA004558 | 0.007921 | 0.08415 | chr12 | 110340659 | 110342448 | + | Exon | ATP2A2 | ATPase
sarcoplasmic/endoplasmic reticulum Ca2+ transporting
2 |
Discussion
In total, 296 differential circRNAs were identified
between NPAF tissues and the controls (FC>2; P<0.05),
including 238 upregulated circRNAs and 58 downregulated circRNAs.
To validate the RNA sequencing data, six representative
dysregulated circRNAs (hsa_circRNA-011430, −015317, −016587,
−003585, −015019 and −003126) were selected. The RT-qPCR results
suggested that the RNA sequencing-identified circRNAs are reliable
and merit further research. The significantly dysregulated circRNAs
in patients with NPAF may serve a regulatory role in the mechanism
of AF progression.
Unlike linear RNAs, circRNAs are covalently linked
to form closed-loop structures without 5′ caps or 3′ tails
(18,23–25).
In circRNAs, a downstream splice donor is joined by ‘back-splicing’
to the upstream splice acceptor (17,26,27).
circRNAs are associated with, and serve important roles in the
diagnosis and pathogenesis of, numerous diseases (16,17),
including colorectal cancer (28),
breast cancer (29) and gastric
cancer (30). Furthermore,
circRNAs are able to upregulate the expression levels of
fibrosis-associated genes in cardiac fibroblasts (31). Other studies have reported that a
circRNA-ceRNA network may be present in certain diseases (18,19).
However, the circRNAs and circRNA-ceRNA network leading to AF
remain to be elucidated.
AF remains a common cause of stroke, heart failure
and cardiovascular mortality worldwide. AF may occur
idiopathically, which is associated with familial inherent specific
genetic mutations (32). Certain
mechanisms leading to AF are primarily associated with the
remodeling of ion channel functions, including those of
K+ channels, and cellular Ca2+ handling and
release (33–36). Other mechanisms of AF include
mutations and the abnormal expression of genes encoding cardiac ion
channels, including potassium voltage-gated channel subfamily Q
member 1, potassium voltage-gated channel subfamily E regulatory
subunit 2, potassium voltage-gated channel subfamily J member 2
(KCNJ2) and sarcoplasmic/endoplasmic reticulum calcium ATPase 2A
(SERCA2) (36).
For instance, calcium gene expression was identified
to be abnormal in AF (37), and
was hypothesized to contribute to the propensity for structural
remodeling in AF. The calcium signaling pathway is essential in the
electrical remodeling of AF and may induce the recurrence of AF
(37). Abnormalities in
intracellular Ca2+ handling are crucially involved in
AF-initiated focal activity and perpetuation through rapidly firing
foci and reentry (37).
GO analysis performed using differential circRNAs
between the NPAF and control groups demonstrated that a number of
functional pathways were enriched. The primary cell component was
GO (no. 0086056) ‘voltage-gated calcium channel activity is
involved in AV node cell action potential’. The main bioprocesses
were GO (no. 0010882) ‘cardiac muscle contraction is regulated by
calcium ion signaling’, GO (no. 0010880) ‘release of sequestered
calcium ion into cytosol is regulated by sarcoplasmic reticulum’
and GO (no. 0098909) ‘regulation of cardiac muscle cell action
potential is involved in regulation of contraction’. The specific
genomes were ‘CACNB2, CACNA1C’ (GO:0086056); ‘SLC8A1, ATP2A2,
CACNA1C, ANK2, RYR2’ (GO:0010882); ‘DHRS7C, CACNA1C, SLC8A1, RYR2,
ANK2’ (GO:0010880); ‘CACNA1C, ATP2A2, RYR2, ANK2’ (GO:0098909).
These genes are primarily involved in Ca2+ channels and
may be the most closely associated with AF. A total of two of the
most significant top 20 KEGG pathways were ‘arrhythmogenic right
ventricular cardiomyopathy’ (no. ko05412) and ‘cardiac muscle
contraction’ (no. ko04260), which were also associated with AF.
Differential expression of circRNAs may be affected by AF and
atrial remodeling. The present study identified a number of
differential circRNAs in AF.
The circRNA-ceRNA interactions were constructed to
examine which circRNAs serve important roles in NPAF progression. A
total of nine differential circRNAs (hsa_circRNA-011785, −001321,
−003878, −002085, −003884, −003876, −007410, −007411 and −004558)
from calcium-associated parental genes were selected. miRNAs serve
multiple roles in atrial fibrillation, including regulating
electrical remodeling by targeting the genes involved in different
ion channels, and regulating structural remodeling in cardiac
tissues by increasing cardiac fibrosis or apoptosis. Different
miRNAs have been demonstrated to be upregulated or downregulated in
patients with AF (21), and may
target different genes to regulate cardiac function. Different
miRNAs may target single genes and serve similar roles, including
repressing IK1 (potassium current) by targeting KCNJ2
via miR-1 and miR-26 (22,38). In the present study, certain
circRNAs were also dysregulated in AF, which might indicate an
association. The interactions between hsa_circRNA004558 and
miR-208b, between hsa_circRNA002085 and hsa-miR-21, and between
hsa_circRNA001321 and hsa-miR-1 may suggest a possible regulatory
mechanism in AF. For instance, miR-208b was demonstrated to be
upregulated in patients with AF and an ovine model. Furthermore, a
high miR-208b level was demonstrated to increase MYH7 expression
and alter the subcellular localization of connexin43 (39). miR-208b has been reported to reduce
the expression levels of CaV1.2 and SERCA2, which further reduce
L-type Ca2+ current density and sarcoplasmic reticulum
Ca2+ load/release, respectively (39). These alterations are hallmarks of
atrial remodeling during AF (40).
miR-21 is reportedly associated with atrial fibrosis regulation in
AF, which inhibits the proliferation of cardiac fibroblasts by
inactivating the transforming growth factor (TGF)-β1/mothers
against decapentaplegic homolog (Smad)2 signaling pathway (41).
miR-21-3p may regulate sepsis-associated cardiac
dysfunction and the development of cardiac hypertrophy. When
miR-21-3p is inhibited, such diseases may be treated via a
protective strategy (42,43). The upregulated inward rectifier
currents (IK1) associated with the electrical remodeling
of AF are required to maintain AF. Additionally, miR-1 expression
is downregulated in patients with AF, which may increase the levels
of inwardly rectifying potassium channel (Kir)2.1 subunits and
IK1 (44,45). miR-4732-3p is targeted by three
circRNAs (hsa_circRNA-003876, −003878 and −003884), and represses
TGF-β signaling by targeting Smad2 and Smad4, and promoting cell
proliferation (46). The TGF-β and
Smad signaling pathways serve essential roles in atrial
fibrosis.
The results of the present study provide a potential
novel insight into the molecular mechanisms and therapeutic
implications of AF. The circRNA-ceRNA interactions identified may
act as biosignatures for AF, and provide evidence for identifying a
novel agent for the diagnosis and gene-targeted therapy of AF.
The current study has a few limitations. Firstly,
though the effects of clinical patient characteristics (e.g.
comorbidities, duration of AF and use of drugs) on the analysis of
the RNA network were considered during patient selection, it is not
possible to disregard these effects. Secondly, the circRNA data for
NPAF and normal tissues were obtained from RNA sequencing, although
further validation is required, including through the use of
knockdown and overexpression experiments on target circRNAs and
miRNAs.
Acknowledgements
Not applicable.
Funding
The present study was funded by the General Program
of the National Natural Science Foundation of China (grant nos.
81500252 and 81770267, to DL), and the Outstanding Young Talent
Training Program of Shanghai Municipal Commission of Health and
Family Planning (grant no. 2017YQ045, to DL).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CG and YLu conceived and designed the experiments.
YZ, XK and JL performed the experiments. XM, YLi, DL and LW
analyzed the data. YZ, CG and YLu wrote the paper. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent from patients was obtained
prior to collection of the left atrial appendages, which were
abandoned due to the surgical technique. The study was approved by
the ethics committees (no. 040-2017) in accordance with the
relevant guidelines and regulations (clinical research registration
no. ChiCTR-RRC-17014230). The patients provided written informed
consent.
Patient consent for publication
The patients provided written informed consent for
publication.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
circRNA
|
circular RNA
|
|
ceRNA
|
competing endogenous RNA
|
|
NPAF
|
nonvalvular persistent atrial
fibrillation
|
|
LAA
|
left atrial appendages
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
AF
|
atrial fibrillation
|
|
FC
|
fold-change
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
TGF-β1
|
transforming growth factor-β1
|
References
|
1
|
Nattel S: New ideas about atrial
fibrillation 50 years on. Nature. 415:219–226. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gudbjartsson DF, Arnar DO, Helgadottir A,
Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A,
Thorleifsson G, Kristjansson K, et al: Variants conferring risk of
atrial fibrillation on chromosome 4q25. Nature. 448:353–357. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Du X, Dong J and Ma C: Is atrial
fibrillation a preventable disease? J Am Coll Cardiol.
69:1968–1982. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chugh SS, Havmoeller R, Narayanan K, Singh
D, Rienstra M, Benjamin EJ, Gillum RF, Kim YH, McAnulty JH Jr,
Zheng ZJ, et al: Worldwide epidemiology of atrial fibrillation: A
Global Burden of Disease 2010 Study. Circulation. 129:837–847.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hijazi Z, Oldgren J, Lindbäck J, Alexander
JH, Connolly SJ, Eikelboom JW, Ezekowitz MD, Held C, Hylek EM,
Lopes RD, et al: A biomarker-based risk score to predict death in
patients with atrial fibrillation: The ABC (age, biomarkers,
clinical history) death risk score. Eur Heart J. 39:477–485. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schnabel RB, Yin X, Gona P, Larson MG,
Beiser AS, McManus DD, Newton-Cheh C, Lubitz SA, Magnani JW,
Ellinor PT, et al: 50 year trends in atrial fibrillation
prevalence, incidence, risk factors, and mortality in the
Framingham Heart Study: A cohort study. Lancet. 386:154–162. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kirchhof P: The future of atrial
fibrillation management: Integrated care and stratified therapy.
Lancet. 390:1873–1887. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kotecha D, Calvert M, Deeks JJ, Griffith
M, Kirchhof P, Lip GY, Mehta S, Slinn G, Stanbury M, Steeds RP and
Townend JN: A review of rate control in atrial fibrillation, and
the rationale and protocol for the RATE-AF trial. BMJ Open.
7:e0150992017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kirchhof P, Benussi S, Kotecha D, Ahlsson
A, Atar D, Casadei B, Castella M, Diener HC, Heidbuchel H, Hendriks
J, et al: 2016 ESC guidelines for the management of atrial
fibrillation developed in collaboration with EACTS. Europace.
18:1609–1678. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chiang CE, Wang KL and Lip GY: Stroke
prevention in atrial fibrillation: An Asian perspective. Thromb
Haemost. 111:789–797. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fauchier L, Philippart R, Clementy N,
Bourguignon T, Angoulvant D, Ivanes F, Babuty D and Bernard A: How
to define valvular atrial fibrillation? Arch Cardiovasc Dis.
108:530–539. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Boriani G, Cimaglia P, Fantecchi E,
Mantovani V, Ziacchi M, Valzania C, Martignani C, Biffi M and
Diemberger I: Non-valvular atrial fibrillation: Potential clinical
implications of the heterogeneous definitions used in trials on new
oral anticoagulants. J Cardiovasc Med (Hagerstown). 16:491–496.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nigro JM, Cho KR, Fearon ER, Kern SE,
Ruppert JM, Oliner JD, Kinzler KW and Vogelstein B: Scrambled
exons. Cell. 64:607–613. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cortés-López M and Miura P: Emerging
functions of circular RNAs. Yale J Biol Med. 89:527–537.
2016.PubMed/NCBI
|
|
17
|
He L, Zhang A, Xiong L, Li Y, Huang R,
Liao L, Zhu Z and Wang AY: Deep circular RNA sequencing provides
insights into the mechanism underlying grass carp reovirus
infection. Int J Mol Sci. 18(pii): E19772017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang S, Zhu DN, Li H, Li H, Feng C and
Zhang W: Characterization of circRNA-associated-ceRNA networks in a
senescence-accelerated mouse prone 8 brain. Mol Ther. 25:2053–2061.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nan A, Chen LJ, Zhang N, Liu Z, Yang T,
Wang Z, Yang C and Jiang Y: A novel regulatory network among
LncRpa, CircRar1, MiR-671 and apoptotic genes promotes lead-induced
neuronal cell apoptosis. Arch Toxicol. 91:1671–1684. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Audic S and Claverie JM:
Self-identification of protein-coding regions in microbial genomes.
Proc Natl Acad Sci USA. 95:10026–10031. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Su H, Lin F, Deng X, Shen L, Fang Y, Fei
Z, Zhao L, Zhang X, Pan H, Xie D, et al: Profiling and
bioinformatics analyses reveal differential circular RNA expression
in radioresistant esophageal cancer cells. J Transl Med.
14:2252016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Starke S, Jost I, Rossbach O, Schneider T,
Schreiner S, Hung LH and Bindereif A: Exon circularization requires
canonical splice signals. Cell Rep. 10:103–111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo JN, Li J, Zhu CL, Feng WT, Shao JX,
Wan L, Huang MD and He JD: Comprehensive profile of differentially
expressed circular RNAs reveals that hsa_circ_0000069 is
upregulated and promotes cell proliferation, migration, and
invasion in colorectal cancer. Onco Targets Ther. 9:7451–7458.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Galasso M, Costantino G, Pasquali L,
Minotti L, Baldassari F, Corrà F, Agnoletto C and Volinia S:
Profiling of the predicted circular RNAs in ductal in situ and
invasive breast cancer: A pilot study. Int J Genomics.
2016:45038402016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN,
Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, et al: CircRNA_000203
enhances the expression of fibrosis-associated genes by
derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac
fibroblasts. Sci Rep. 7:403422017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Woods CE and Olgin J: Atrial fibrillation
therapy now and in the future: Drugs, biologicals, and ablation.
Circ Res. 114:1532–1546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brundel BJ, Van Gelder IC, Henning RH,
Tuinenburg AE, Wietses M, Grandjean JG, Wilde AA, Van Gilst WH and
Crijns HJ: Alterations in potassium channel gene expression in
atria of patients with persistent and paroxysmal atrial
fibrillation: Differential regulation of protein and mRNA levels
for K+ channels. J Am Coll Cardiol. 37:926–932. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Santulli G, D'ascia SL and D'ascia C:
Development of atrial fibrillation in recipients of cardiac
resynchronization therapy: The role of atrial reverse remodelling.
Can J Cardiol. 28:245.e17–e18. 2012. View Article : Google Scholar
|
|
35
|
Xie W, Santulli G, Guo X, Gao M, Chen BX
and Marks AR: Imaging atrial arrhythmic intracellular calcium in
intact heart. J Mol Cell Cardiol. 64:120–123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Andrade J, Khairy P, Dobrev D and Nattel
S: The clinical profile and pathophysiology of atrial fibrillation:
Relationships among clinical features, epidemiology, and
mechanisms. Circ Res. 114:1453–1468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dobrev D and Nattel S: Calcium handling
abnormalities in atrial fibrillation as a target for innovative
therapeutics. J Cardiovasc Pharmacol. 52:293–299. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Luo X, Pan Z, Shan H, Xiao J, Sun X, Wang
N, Lin H, Xiao L, Maguy A, Qi XY, et al: MicroRNA-26 governs
profibrillatory inward-rectifier potassium current changes in
atrial fibrillation. J Clin Invest. 123:1939–1951. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cañón S, Caballero R, Herraiz-Martinez A,
Pérez-Hernández M, López B, Atienza F, Jalife J, Hove-Madsen L,
Delpón E and Bernad A: miR-208b upregulation interferes with
calcium handling in HL-1 atrial myocytes: Implications in human
chronic atrial fibrillation. J Mol Cell Cardiol. 99:162–173. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lai LP, Su MJ, Lin JL, Lin FY, Tsai CH,
Chen YS, Huang SK, Tseng YZ and Lien WP: Down-regulation of L-type
calcium channel and sarcoplasmic reticular Ca(2+)-ATPase mRNA in
human atrial fibrillation without significant change in the mRNA of
ryanodine receptor, calsequestrin and phospholamban: An insight
into the mechanism of atrial electrical remodeling. J Am Coll
Cardiol. 33:1231–1237. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tao H, Zhang M, Yang JJ and Shi KH:
MicroRNA-21 via dysregulation of WW domain-containing protein 1
regulate atrial fibrosis in atrial fibrillation. Heart Lung Circ.
27:104–113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang H, Bei Y, Shen S, Huang P, Shi J,
Zhang J, Sun Q, Chen Y, Yang Y, Xu T, et al: miR-21-3p controls
sepsis-associated cardiac dysfunction via regulating SORBS2. J Mol
Cell Cardiol. 94:43–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yan MW, Chen C, Gong W, Yin Z, Zhou L,
Chaugai S and Wang DW: miR-21-3p regulates cardiac hypertrophic
response by targeting histone deacetylase-8. Cardiovasc Res.
105:340–352. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Girmatsion Z, Biliczki P, Bonauer A,
Wimmer-Greinecker G, Scherer M, Moritz A, Bukowska A, Goette A,
Nattel S, Hohnloser SH and Ehrlich JR: Changes in microRNA-1
expression and I-K1 up-regulation in human atrial fibrillation.
Heart Rhythm. 6:1802–1809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ehrlich JR: Inward rectifier potassium
currents as a target for atrial fibrillation therapy. J Cardiovasc
Pharmacol. 52:129–135. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Doss JF, Corcoran DL, Jima DD, Telen MJ,
Dave SS and Chi JT: A comprehensive joint analysis of the long and
short RNA transcriptomes of human erythrocytes. BMC Genomics.
16:9522015. View Article : Google Scholar : PubMed/NCBI
|