Introduction
Prostate cancer (PCa) is a common type of malignant
tumour in the urinary system of elderly males, and has become a
major cause of mortality among urological cancers in the USA and
other industrialized countries (1). Therefore, it is necessary to
elucidate the underlying pathophysiological processes of PCa with
the advent of global ageing.
Hepatocyte cell adhesion molecule (HepaCAM) was
first detected in the liver (2),
and it was later identified as a member of the immunoglobulin
superfamily. Members of the immunoglobulin superfamily are
primarily localized at the cell membrane and are composed of three
domains: Extracellular, transmembrane and cytoplasmic (3–5). The
cytoplasmic domain is fundamental to its biological function
(2). Recent studies have indicated
that HepaCAM is present at low levels or is even absent in certain
types of cancer tissues and cell lines (2,3,6). Our
previous studies demonstrated that cell proliferation is
significantly inhibited when HepaCAM is overexpressed in urological
cancer cells (3–5,7).
Furthermore, recent studies have revealed that HepaCAM can promote
apoptosis and inhibit invasion and migration of cancer cells
(7,8). These studies indicate that HepaCAM is
a tumour suppressor candidate that may be downregulated in cancer
development. However, extensive research is required to elucidate
the mechanism of the anti-tumour role of HepaCAM in the
carcinogenesis and progression of PCa, particularly that of the
cytoplasmic domain of HepaCAM.
Androgen receptor (AR), a steroid hormone receptor,
is necessary for the physiological function of the prostate
(9) and serves an important role
in the proliferation, apoptosis, invasion and metastasis of PCa
cells (10,11). After binding to androgens in the
cytoplasm, AR undergoes a series of conformational changes to form
the AR-androgen complex (12),
which translocate from the cytoplasm to the nucleus, and then
activates AR-targeted gene expression (13–15).
Therefore, investigation of the potential activation mechanisms of
AR is crucial in order to identify novel androgen-based treatments
for PCa.
Our previous study demonstrated that HepaCAM
inhibits the nuclear translocation of the androgen-AR complex
(7). Nuclear translocation is a
dynamic equilibrium process in which molecular complexes shuttle
back and forth through the cell nuclear membrane via nuclear pore
complexes (16). Ran proteins
serve a key role in providing energy for this process. Ran is a
guanine triphosphate (GTP)-binding protein that is widely present
in the nucleus and circulates between the cytoplasm and the nucleus
in the form of RanGDP and RanGTP (17,18).
Therefore, it was hypothesized that the anti-tumour effects of
HepaCAM may be partially associated with its influence on the AR
signalling pathway, which involves AR and Ran. The present study
focuses on the biological function of the cytoplasmic domain of
HepaCAM and investigates the association among AR, Ran and the
cytoplasmic domain of HepaCAM in the process of AR nuclear
translocation.
Based on the important role of AR in the progression
of PCa, the present study aimed to ascertain whether HepaCAM
influenced the endonuclear and extranuclear distribution in PCa
tissues and LNCaP cell line, and then to explore the possible
mechanisms by which HepaCAM and its cytoplasmic domain affect
nuclear translocation of AR. The present study may provide further
support for HepaCAM inhibition of PCa.
Materials and methods
Patients and tissues
A total of 46 PCa specimens from patients who had
undergone radical prostatectomy, and 46 benign prostatic
hyperplasia (BPH) specimens from patients who had undergone a
transurethral resection of the prostate between September 2015 and
April 2017 were included. All collected specimens were were fixed,
dehydrated, embedded stained with hematoxylin and eosin (H&E)
or immunohistochemical staining (IHC). None of the patients with
PCa had undergone androgen deprivation therapy (ADT) or neoadjuvant
chemotherapy. All samples were meticulously studied by the same
experienced pathologist without misdiagnosis. The stage and Gleason
grade of the PCa were diagnosed according to UICC guidelines
(19). Informed consent forms were
signed by all patients, and the study was approved by the Research
Ethical Committee of the Chongqing Medical University (Chongqing,
China).
H&E and IHC
The PCa and BPH tissues were fixed with 10% neutral
formalin for 2 days at room temperature, dehydrated in a graded
series of alcohol, embedded in paraffin, and then sectioned with
thickness of 4 µm for histological examination. H&E and IHC
staining was performed according to a standard procedure (20). The primary antibodies were as
follows: Anti-HepaCAM (1:200, ProteinTech Group, Inc., Chicago, IL,
USA; cat. no. 18177-1-AP), anti-AR (1:200, ProteinTech Group, Inc.;
cat. no. 22576-1-AP), anti-Ran (1:200, Abcam, Cambridge, UK; cat.
no. ab157213), horseradish peroxidase-labelled goat anti-rabbit
secondary antibody (1:500; OriGene Technologies, Inc., Rockville,
MD, USA; cat. no. ZF-0316). The semi-quantitative intensity was
defined as follows: 0 (unstained), 1 (yellow), 2 (light brown), and
3 (brown). The immunoreactivity ratio was semi-quantitatively
counted as follows: 0 (0%), 1 (<5%), 2 (5–50%), and 3 (>50%).
The parameters of ratio and intensity were combined to determine
the final scores as follows: Samples with score <3 were
considered negative, and samples with scores >3 were regarded as
positive for statistical convenience.
Cell culture and transfection
The hormone-sensitive PCa LNCaP cell line was
incubated in high-glucose Dulbecco's modified Eagle's medium/F-12
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 100 U/ml penicillin and streptomycin (Thermo
Fisher Scientific, Inc.) at 37°C in a 45% humidified incubator with
5% CO2. An adenovirus containing intact HepaCAM
(ad-HepaCAM) (1×109) was constructed by our group as
previously described (21). An
adenovirus construct with the entire cytoplasmic domain residues
(264–416) of HepaCAM deleted (ad-HepaCAM-mt) (1×109) was
designed and constructed by Shanghai GeneChem Co., Ltd., (Shanghai,
China). The cells were incubated firstly in serum-free medium for
24 h at 37°C, and then, the adenoviruses described above were used
for transfection for ~ 48 h at 37°C for subsequence analysis. Cells
transfected with ad-hepaCAM or ad-hepaCAM-mt were named ad-hepaCAM
group or ad-hepaCAM-mt group respectively; the cells transfected
with vector that served negative control were named vector group,
the cells treated PBS that served blank control were named blank
group.
Ran-targeting siRNA (si-Ran) and a scrambled siRNA
sequence, which served as negative control (NC) were designed and
synthesized by Shanghai GeneChem Co., Ltd. The Ran siRNA sequences
was: 5′-CAGAUUGUUCGGUUUGGCUUGUUUA-3′, the scrambled siRNA sequences
was: 5′-CAGUGUUCGGUUUGGCUUGUAUUUA-3′. When the cell density reached
~30%, si-Ran was transfected into cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for about 48 h at 37°C according to the
manufacturer's protocol. The cells transfected with si-Ran were
named si-Ran group, the cells transfected with ad-hepaCAM and
si-Ran were named ad-hepaCAM + si-Ran group. The cells transfected
with ad-hepaCAM-mt and si-Ran were named ad-hepaCAM-mt + si-Ran
group. To analyze the transfection efficiency, the numbers and
staining intensity of positive cells expressing green fluorescent
protein (GFP) were observed by fluorescence microscopy at different
time points.
MTT assay
Cells (2,000/well) were cultured in 96-wells plates
with 100 µl of culture medium. After cell attachment was complete,
each cell transfection was conducted in five replicate wells At
each time point, 5 mg/ml MTT (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) was added to the treated wells. Subsequently, the treated
cells were maintained for ~3-5 h at 37°C. Subsequently, dimethyl
sulfoxide was added to each well after removing the culture medium.
The 96-well plates were agitated steadily on a rotator platform for
15 min at room temperature. The absorbance at 490 nm was recorded
on an ultraviolet spectrophotometric reader. The experiments were
repeated five times. Wells with treatment-free medium served as the
negative control.
Colony formation assay
Cells in each treatment group were seeded in
six-well plates at a density of 1,000 cells/well with 3 ml culture
medium. After two weeks of incubation, the treated LNCaP cells were
washed with PBS twice and then fixed with 4% paraformaldehyde at
37°C for 20 min. The cell clusters were dyed in 0.1% violet
solution for 20 min at room temperature. The colony numbers were
counted under an inverted microscope (Nikon Corporation, Tokyo,
Japan). The colony forming efficiency (CFE) was calculated with the
following equation: CFE (%)=(the number of colonies/1,000) ×100.
The colony formation assay was repeated three times.
Wound healing and Transwell assay
Approximately 6×104 cells/well were
incubated in six-well plates. PBS, vector, ad-HepaCAM and
ad-HepaCAM-mt were utilized for transfection of LNCaP cells. When
the cells reached ~80% confluency, a straight line was scratched
across the cell layer with a pipette tip. Subsequently, fresh
culture medium was added, and the cultures were maintained for a
further 24 h. The scratch width was measured in micrographs that
were captured at 0, 12 and 24 h. Approximately 4.0×103
cells of each treatment LNCaP culture were inserted into the upper
chamber and high glucose DMEM medium containing 30% FBS was placed
in the lower chamber. The LNCaP cells on the upper membrane were
swept with swabs after being incubated in serum-free medium for 36
h at 37°C. Subsequently, the Transwell membranes were stained with
0.1% crystal violet solution for 15 min at room temperature. The
LNCaP cells that were anchored to the Transwell membranes were
counted using an inverted microscope (Nikon Corporation;
magnification, ×200). The cell number was quantified from five
random fields for each experimental group.
Western blotting
The cells of each treatment group were incubated for
~48 h at 37°C, and then, proteins in the cytoplasm and nucleus were
extracted using nuclear and cytoplasmic extraction reagents,
respectively (Thermo Fisher Scientific, Inc.). Western blotting
assays were performed as previously described (8). Briefly, the protein concentration was
determined by Enhanced Bicinchoninic Protein Assay kit (Beyotime
Institute of Biotechnology, Haimen, China). Protein samples (50
µg), stacked by 5% SDS-PAGE and separated by 10 or 12% SDS-PAGE
were transferred electrophoretically to polyvinylidene fluoride
(PVDF) membranes. The PVDF membranes were blocked with 5% skim milk
for 1 h at 4°C and incubated with a primary antibody at 4°C
overnight. Subsequently, the membrane was incubated with
horseradish peroxidase-labelled goat anti-rabbit secondary antibody
(1:500; OriGene Technologies, Inc., cat. no. ZF-0316) overnight at
4°C. The protein bands were visualized by the enhanced
chemiluminescent kit (Beyotime Institute of Biotechnology; cat. no.
P0018AS) and quantified using Quantity One software 4.6.2 (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The primary antibodies were
diluted as follows: Anti-HepaCAM (1:1,000; ProteinTech, cat. no.
18177-1-AP), anti-GAPDH (1:1,000; OriGene Technologies, Inc., cat.
no. ZF-0316), anti-histone (1:2,000; Abcam, cat. no. ab176842),
anti-AR (1:1,000; ProteinTech, cat. no. 22576-1-AP), anti-c-myc
(1:1000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA, cat. no.
sc-70469), anti-β-actin (1:1,000; Abcam, cat. no. ab8226),
anti-cyclinD1 (1:1,000; Santa Cruz Biotechnology, Inc., cat. no.
sc-70899), anti-E-cadherin (1:2,000; Cell Signaling Technology,
Inc., Danvers, MA, USA, cat. no. 3195), anti-N-cadherin (1:2,000;
Cell Signaling Technology, Inc., cat. no. 4061), and anti-snail
(1:1,000; Cell Signaling Technology, Inc., cat. no. 3895) and
anti-Ran (1:1000; Abcam, cat. no. ab157213). All protein expression
experiments were repeated three times.
Immunofluorescence
Immunofluorescence was conducted as previously
described (22). A total of
1×105 cells/well with predetermined treatment were
seeded on sterile coverslips at 37°C for ~48 h. Cells on the
coverslips were fixed in 4% paraformaldehyde, permeabilized with
0.1% Triton X-100, and blocked with 5% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.). Subsequently, anti-AR (1:500,
ProteinTech, cat. no. 22576-1-AP) or anti-Ran (1:1,000, Abcam, cat.
no. ab157213) was added onto the coverslips and they were incubated
overnight at 4°C. The coverslips were then incubated with the
secondary antibody for 50 min at room temperature (1:500, ZSGB;
OriGene Technologies, Inc., cat. no. ZF-0316) and the cell nuclei
were stained with DAPI (ZSGB; OriGene Technologies, Inc.) for 10
min at room temperature. The immunofluorescent images were obtained
at ×400 magnification using a fluorescence microscope (Eclipse 80i;
Nikon Corporation, Tokyo, Japan).
Prostate specific antigen (PSA)
determination
LNCaP cells (1×105 cells/well) were
seeded in 96-well plate. PSA protein levels in the supernatant of
the LNCaP cells in different treatment groups were determined using
solid sandwich ELISA assays using a PSA ELISA kit, according to the
manufacturer's protocol (Abcam; cat. no. ab113327).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Cells were seeded in 6-well plates at a density of
2×106 cells/well. Total RNA from the LNCaP cells
transfected with si-Ran was extracted using a TRIzol kit (Thermo
Fisher Scientific, Inc.), and reverse transcription reactions were
conducted using the Prime Script RT reagent kit (Takara
Biotechnology Co., Ltd.) according to the manufacturer's protocol.
RT-qPCR was performed with the SYBR PremixEx Taq™ II kit (Takara
Biotechnology Co., Ltd.) with 400 ng total RNA and oligo (dT)
primers in a total 10 µl. Ran knockdown expression was analyzed by
RT-qPCR. The specific primers were as follows: β-actin forward,
5′-GACCTGTACGCCAACACAGT-3′, and reverse,
5′-CTCAGGAGGAGCAATGATCT-3′; Ran forward, 5′-CCAAGGTGGCTACATACTTC-3′
and reverse, 5′-TGGTTGGTGATGGTGGTACT-3′. The conditions consisted
of predenaturing at 95°C for 5 min, followed by 35 cycles of
denaturing at 95°C for 10 sec, annealing at 55°C for 50 sec,
extension at 72°C for 1 min, and a final extension at 72°C for 5
min. The mRNA expression levels were calculated using the
comparative 2−ΔΔCq method (23) and β-actin served as a
calibrator.
Statistical analysis
Statistical analyses were conducted with SPSS
software, version 20.0 (IBM Corp., Armonk, NY, USA). The data are
presented as the mean ± standard deviation. The data between groups
was compared using a paired t-test or one-way analysis of variance
followed by Tukey's multiple comparisons test where appropriate.
The association between the lower HepaCAM expression and higher
expression of Ran and AR in PCa was analyzed using Cohen's kappa,
and the constituent ratio of positive rate of HepaCAM was analyzed
using χ2 tests. P<0.05 was considered to indicate a
statistically significant difference.
Results
HepaCAM is downregulated or absent in
PCa tissues and negatively associated with upregulation of Ran and
AR
HepaCAM, AR and Ran expression levels were analysed
with IHC in 46 BPH and PCa tissues. In the IHC analysis, 65.22%
(30/46) of the PCa tissues exhibited negative HepaCAM staining, and
34.78% (16/46) exhibited positive staining (Table I). The association between
clinicopathological characteristics and the levels of HepaCAM in
the PCa tissues was analysed, and a statistically significant
difference was only observed between the expression levels of
HepaCAM and the Gleason grade (Table
I; P<0.05). The semi-quantitative staining scores revealed
that the expression levels of HepaCAM were significantly lower in
the PCa tissues compared with the BPH tissues (P<0.01; Fig. 1A and B). Additionally,
significantly higher expression levels of Ran and AR were detected
in the PCa specimens compared with BPH specimens (P<0.01;
Fig. 1C and D). The association
between HepaCAM and AR or Ran was analysed using Cohen's kappa
(Table II). Notably, lower
HepaCAM expression levels were detected in the samples with higher
expression levels of Ran and AR.
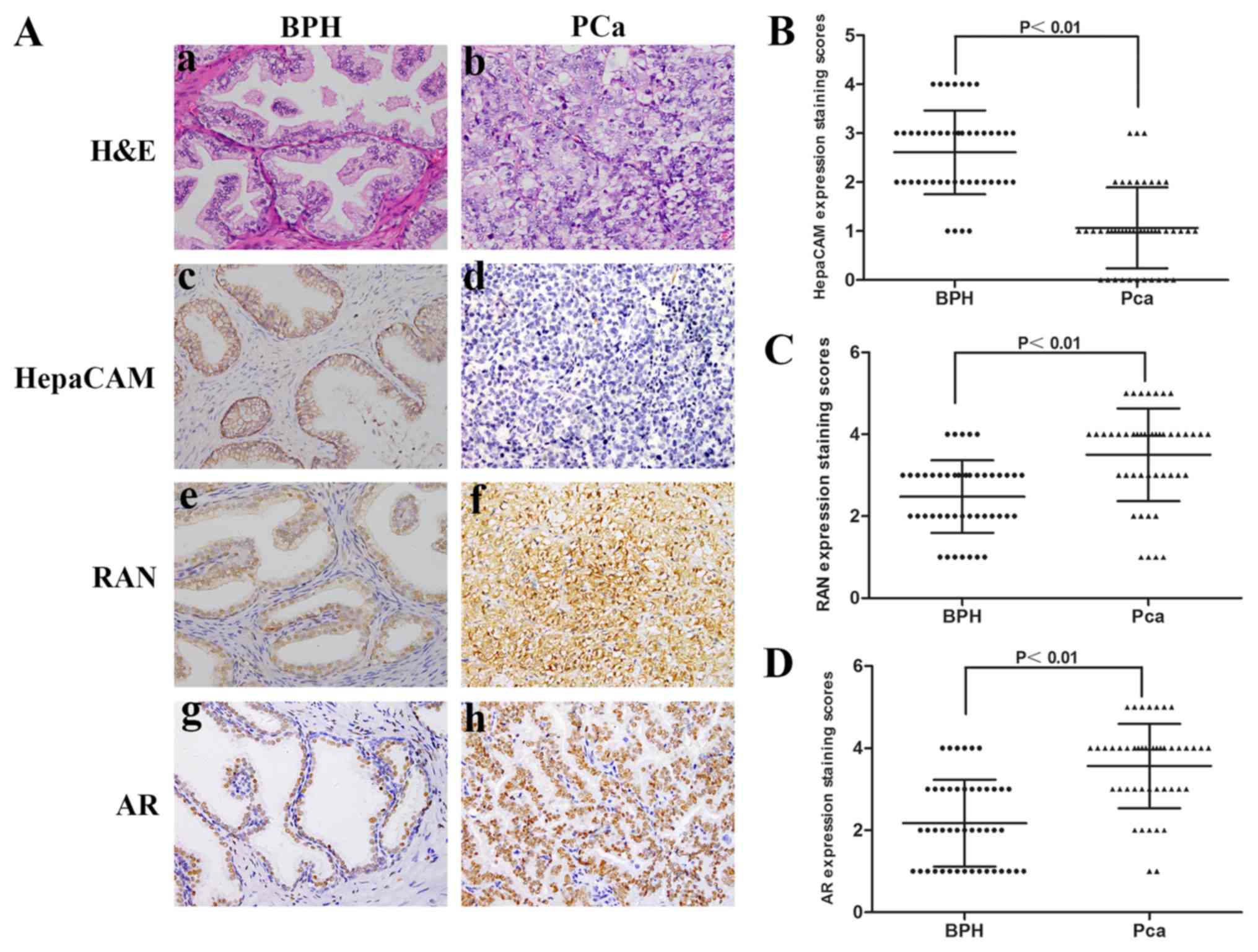 | Figure 1.Low expression of HepaCAM is
associated with high expression of Ran and AR in PCa tissues. (A)
H&E and immunohistochemical staining in PCa. (a, c, e and g)
BPH and (b, d, f and h) PCa. (B) Expression staining scores of
HepaCAM, of (C) Ran and of (D) AR in PCa and BPH. Magnification,
×400. Ad, adenovirus; AR, androgen receptor; BPH, benign prostatic
hyperplasia; HepaCAM, hepatocyte cell adhesion molecule; PCa,
prostate cancer. |
 | Table I.Association between HepaCAM
expression and the clinical characteristics in patients with
prostate cancer. |
Table I.
Association between HepaCAM
expression and the clinical characteristics in patients with
prostate cancer.
|
|
| HepaCAM |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Cases (%) | Negative | Positive | P-value |
|---|
| Total no. | 46 (100) | 30 (65.22) | 16 (34.78) |
|
| Age |
|
|
| 0.125 |
|
<60 | 20 (43.48) | 16 (34.78) | 4 (8.70) |
|
|
≥60 | 26 (56.52) | 14 (30.43) | 12 (26.09) |
|
| Histological
stage |
|
|
| 0.729 |
| Ta-T2 | 34 (73.91) | 22 (47.83) | 12 (26.09) |
|
| T3-T4 | 12 (26.09) | 8 (17.39) | 4 (8.70) |
|
| Gleason grade |
|
|
| 0.024a |
|
<7 | 24 (52.17) | 12 (26.09) | 12 (26.09) |
|
| ≥7 | 22 (47.83) | 18 (39.13) | 4 (8.70) |
|
 | Table II.Association among HepaCAM, Ran and AR
expression in PCa tissues. |
Table II.
Association among HepaCAM, Ran and AR
expression in PCa tissues.
|
| Ran expression |
|
| AR expression |
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| HepaCAM
expression | Positive | Negative | Kappa | P-value | Positive | Negative | Kappa | P-value |
|---|
| Negative | 28 | 2 | −0.442 |
P<0.01a | 29 | 1 | −0.565 |
P<0.01a |
| Positive | 6 | 10 |
|
| 14 | 2 |
|
|
Effect of the constructed adenovirus
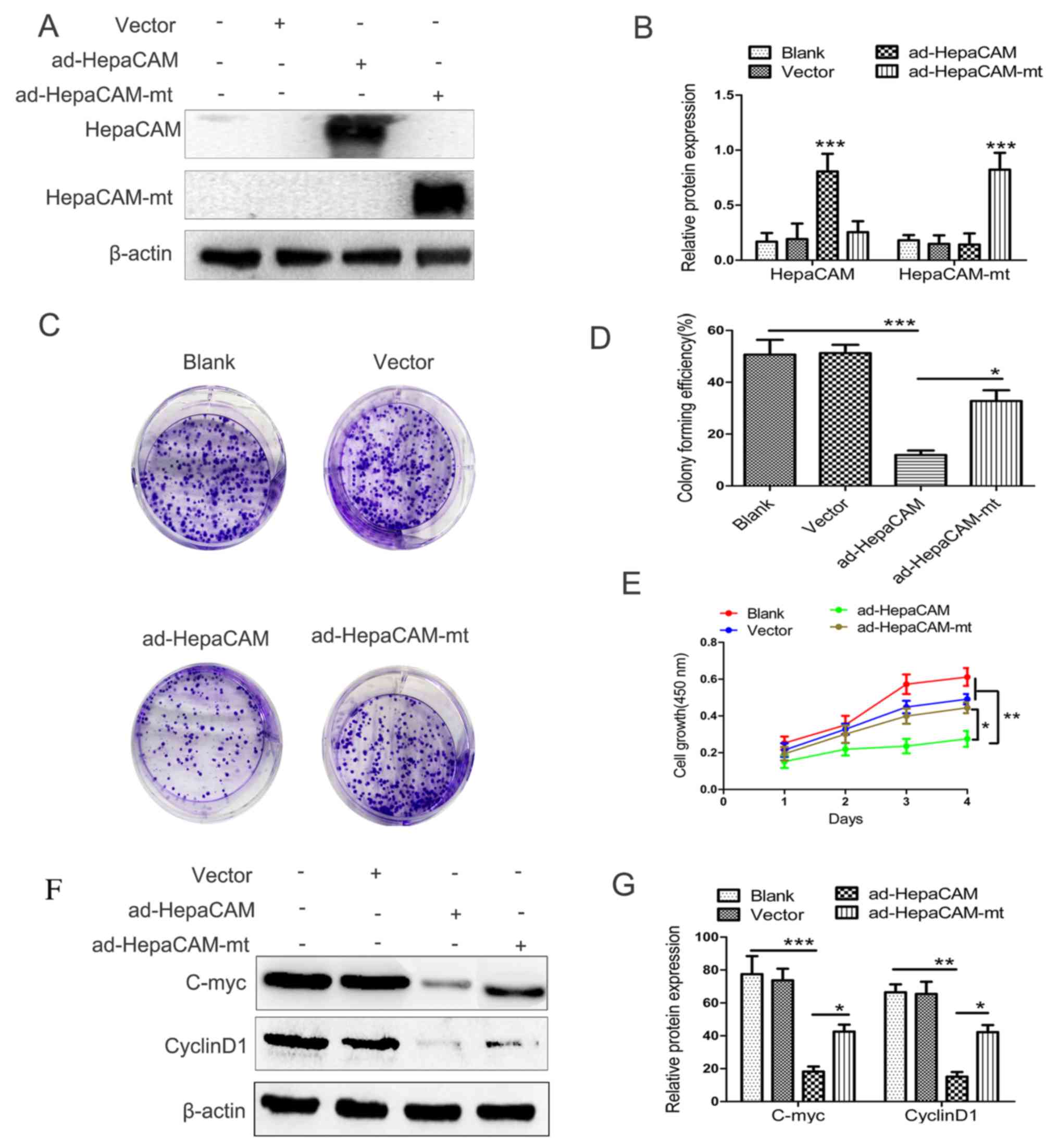
on HepaCAM and HepaCAM-mt protein expression in LNCaP cells
In order to investigate the anti-tumour activity of
HepaCAM and its cytoplasmic domain, ad-HepaCAM and ad-HepaCAM-mt
were used in LNCaP cells. The GFP protein staining under
fluorescence microscope showed high transfection efficiency (data
not shown). Furthermore, western blot analysis was used to
determine HepaCAM and HepaCAM-mt expression in the LNCaP cells in
order to verify transfection efficiency and cell line stability.
The results demonstrated that the strong expression of HepaCAM and
HepaCAM-mt following transfection was established successfully
(P<0.01; Fig. 2A and B).
Effect of HepaCAM and its cytoplasmic
domain on cell proliferation and associated proteins in LNCaP
cells
Colony formation and MTT assays were used to
evaluate whether HepaCAM and its cytoplasmic domain serve an
important role in proliferation of LNCaP cells. The results of the
colony formation assay revealed similar differences in the
viabilities of the cells subjected to the aforementioned treatments
(Fig. 2C and D). The MTT analysis
revealed that the proliferative capacity of LNCaP cells was
suppressed by overexpression of intact HepaCAM. However,
overexpression of HepaCAM-mt did not produce inhibiting effects
equivalent to those of intact HepaCAM. As shown in Fig. 2E, at day 4, the percentage of
viable LNCaP cells in the ad-HepaCAM group was significantly lower
compared with the other three groups (P<0.01).
In addition, the expression levels of
proliferation-associated proteins, including cyclinD1 and c-myc,
were significantly reduced after the cells were transfected with
ad-HepaCAM compared with the other three groups (Fig. 2F and G). These results suggested
that HepaCAM and its cytoplasmic domain serve a key role in the
inhibition of cell proliferation and cell growth in PCa.
Furthermore, c-myc and cyclin D1 may be associated with the tumour
suppression activity of HepaCAM, but this needs to be investigated
further.
Effect of HepaCAM and its cytoplasmic
domain on migration and invasion-associated proteins in LNCaP
cells
The wound healing assays indicated that there was
significant suppression of cellular migration in the HepaCAM group
compared with the other three groups (P<0.01; Fig. 3A and B). The Transwell experiments
also indicated that the migration of LNCaP cells was significantly
lower in the HepaCAM group in comparison with the other three
groups (P<0.01; Fig. 3C and D).
In addition, the E-cadherin expression levels, which are normally
absent from LNCaP cells, were slightly higher in the HepaCAM group
compared with the other three groups. The levels of N-cadherin and
Snail, which are thought to promote migration (24), were significantly attenuated after
transfection with ad-HepaCAM compared with the other three groups
(Fig. 3E and F). These results
indicated that HepaCAM suppresses cell migration via its
cytoplasmic domain in vitro. E-cadherin, N-cadherin and
Snail maybe involved in the suppressive activity of HepaCAM in
LNCaP cells, but this needs to be validated.
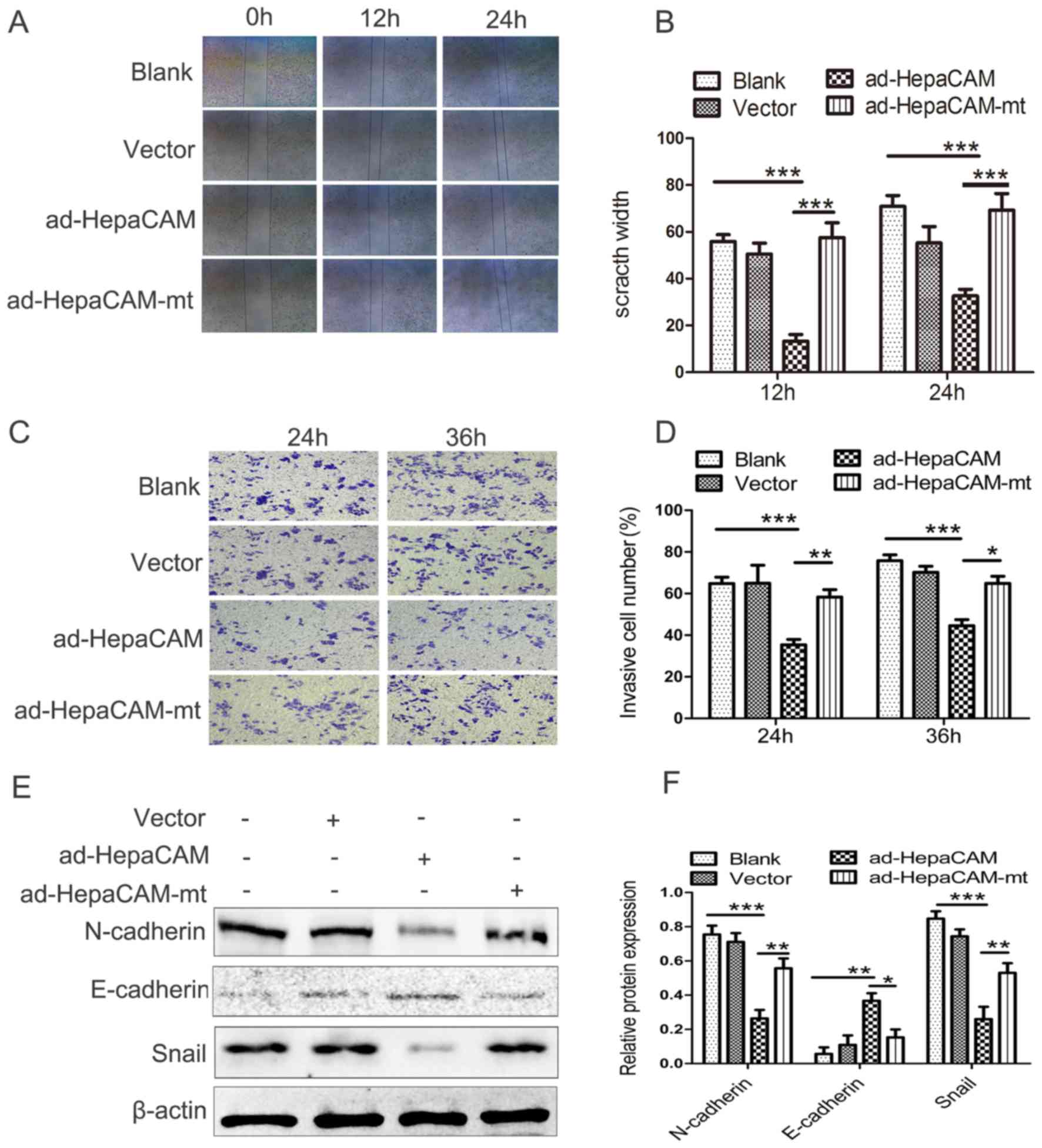 | Figure 3.HepaCAM over-expression inhibits
invasion and migration of LNCaP cells. (A and B) Wound healing
assay of the LNCaP cells with different treatments. (C and D)
Migrated LNCaP cells, as measured by Matrigel migration assay. (E
and F) Expression levels of E-cadherin, N-cadherin, and Snail, as
analysed by western blotting. The data are presented as the mean ±
standard deviation. Magnification, ×200. *P<0.05, **P<0.01,
***P<0.001. Ad, adenovirus; HepaCAM, hepatocyte cell adhesion
molecule; mt, mutant. |
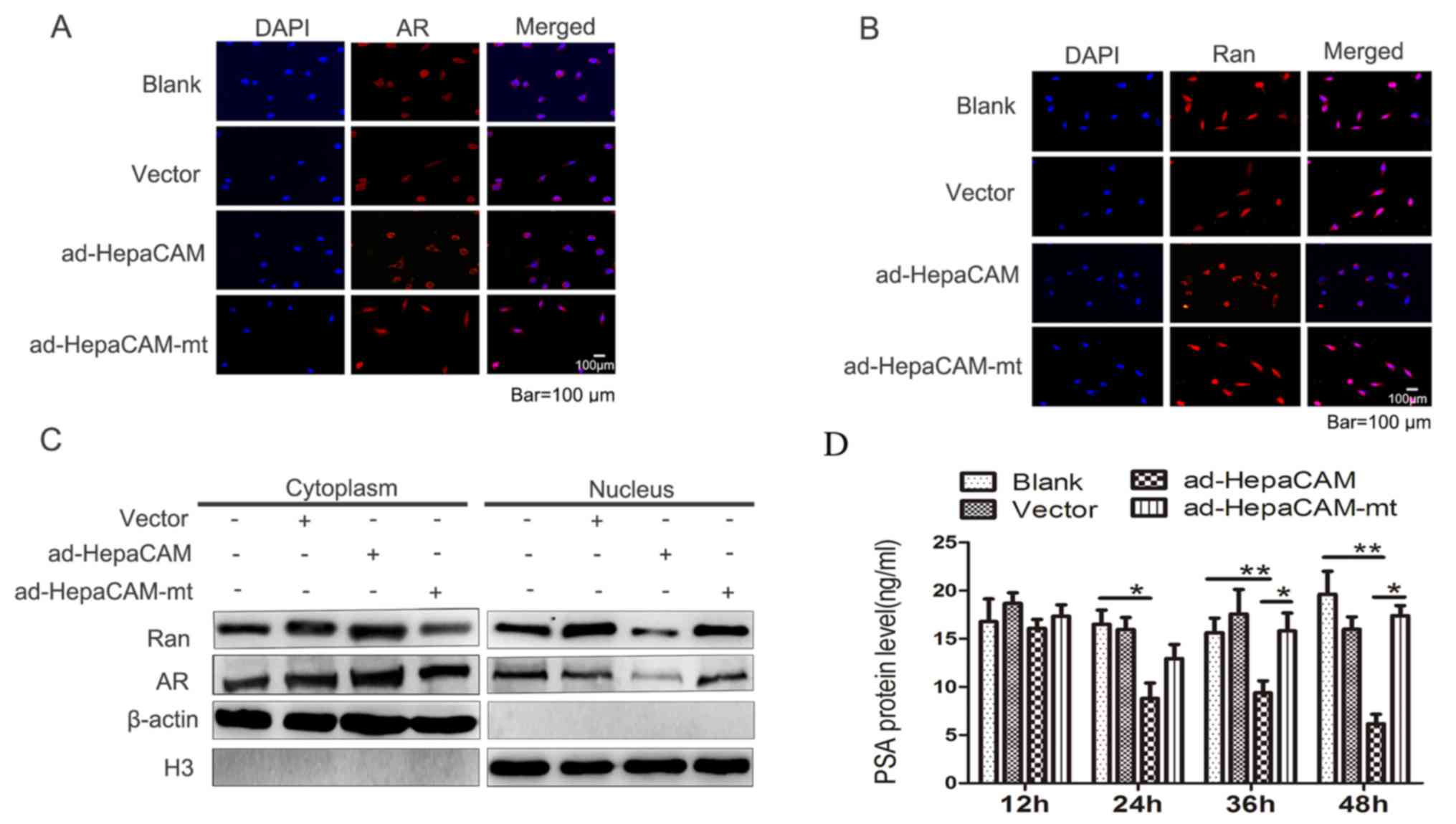
Effect of HepaCAM and its cytoplasmic
domain on nuclear translocation of AR and Ran
Nuclear translocation of AR is one of the most
important steps in AR signalling transduction (25). Ran serves vital roles in modulating
the nuclear translocation of proteins (16). To determine whether HepaCAM and its
cytoplasmic domain had an impact on the nuclear translocation of
Ran and AR, immunofluorescence was employed to assess the
subcellular distribution of AR and Ran between the cytoplasm and
nucleus (Fig. 4A and B,
respectively). The results revealed that AR and Ran expression was
upregulated in the cytoplasm and downregulated in the nucleus after
cells were transfected with ad-HepaCAM (Fig. 4C). No differences were observed
between the HepaCAM-mt group and the blank or vector groups. This
suggested that expression of intact HepaCAM altered the
distribution of AR and Ran between the cytoplasm and the nucleus
and demonstrated that there was a synergic effect between AR and
Ran.
Effect of HepaCAM and its cytoplasmic
domain on inhibition of PSA in LNCaP cells
The levels of PSA partially reflect the activation
of AR targeting genes (26). The
analysis of PSA levels revealed that they were significantly lower
in the HepaCAM group compared with the other three groups at 24–48
h. However, following transfection with HepaCAM, no significant
difference was identified in the PSA levels between the HepaCAM-mt
group and the blank or vector groups at 24–48 h (Fig. 4D). These data suggested that the
function of AR may be inhibited by HepaCAM, which maybe via its
cytoplasmic domain.
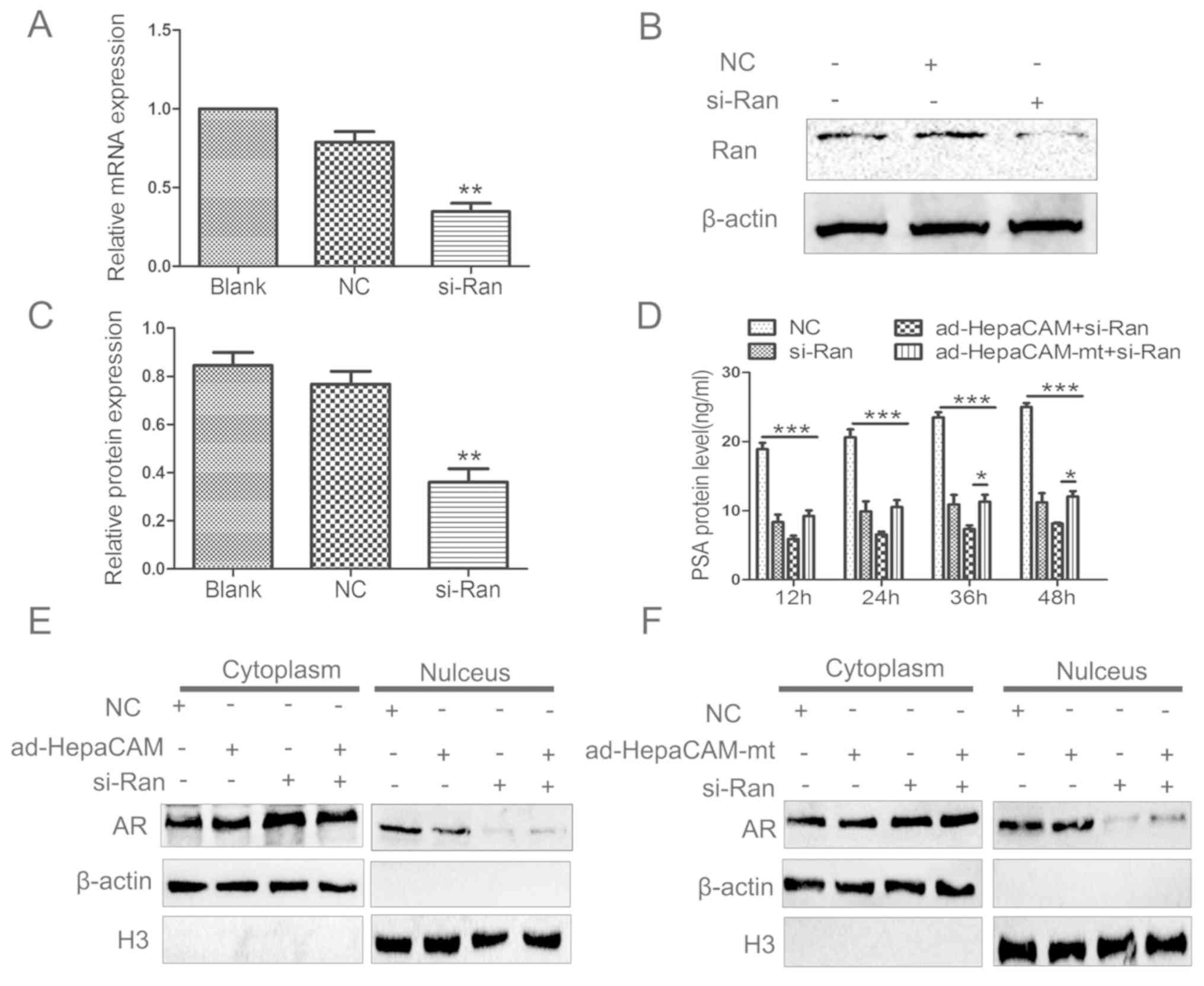
HepaCAM and its cytoplasmic domain
affect the nuclear translocation of AR through Ran protein
si-Ran was employed to determine whether Ran serves
a role in the nuclear translocation of AR. The results revealed
that there was significant downregulation in the total expression
of Ran mRNA and protein levels in the si-Ran group compared with
the NC group (Fig. 5A-C). This
indicates that an active si-Ran was successfully constructed and
transfected into the LNCaP cells.
PSA ELISAs were also used to determine the activity
of AR. The PSA expression levels were suppressed in the si-Ran,
ad-HepaCAM+si-Ran, and ad-HepaCAM-mt+si-Ran groups at 12–48 h
compared with the NC group. There was a significant reduction among
the si-Ran, ad-HepaCAM+si-Ran and ad-HepaCAM-mt+si-Ran groups at
12–48 h. Additionally, a statistically significant difference was
identified between the ad-HepaCAM+si-Ran and ad-HepaCAM-mt+si-Ran
groups at 36 or 48 h (Fig. 5D).
This suggested that HepaCAM may inhibited PSA via Ran first. With
time, the inhibition efficiency became higher, suggesting that
HepaCAM may inhibit PSA in some other way.
As shown in Fig.
5E, when HepaCAM was overexpressed, AR was mainly in the
cytoplasm rather than in the nucleus of the LNCaP cells. However,
when the cells were treated with ad-HepaCAM-mt, there was a similar
distribution of AR in the cytoplasm and the nucleus (Fig. 5F). These data indicate that the
cytoplasmic domain regulates AR distribution between the cytoplasm
and nucleus. To further investigate whether Ran has an involvement
in AR localization, si-Ran was used to transfect the LNCaP cells.
Western blotting demonstrated that the levels of AR were higher in
the cytoplasm and lower in the nucleus of the si-Ran group. When
both ad-HepaCAM and si-Ran were applied, the AR distribution in the
cytoplasm and nucleus was similar to that of the si-Ran group
(Fig. 5E). The same results were
observed when the si-Ran group was compared with the
ad-HepaCAM-mt+si-Ran group (Fig.
5F). Therefore, Ran seems to be essential for AR nuclear
distribution, and this may be inhibited by HepaCAM through its
cytoplasmic domain.
Discussion
Our previous studies indicated that HepaCAM serves a
significant role in various pathophysiological processes in urinary
tract cancers, including tumour cell proliferation, apoptosis,
migration, and invasion (7,23,27).
In the present study, there was a reduction or absence of HepaCAM
expression in the majority of the PCa samples. This abnormal
expression of HepaCAM was negatively associated with the Gleason
grade, which is associated with high-risk types of PCa that have a
poorer prognosis. Therefore, it is reasonable to presume that
HepaCAM may be an inhibiting factor in the progression of PCa.
However, the mechanism of the anti-tumour activity of HepaCAM in
PCa remains to be elucidated.
HepaCAM is a transmembrane protein with a
phosphorylated cytoplasmic domain (28). In the present study, the function
of the cytoplasmic domain of HepaCAM in the inhibition of PCa was
investigated. Ad-HepaCAM and ad-HepaCAM-mt were constructed and
transfected into PCa cells. The results revealed that
overexpression of intact HepaCAM effectively inhibited the growth,
proliferation, migration and invasion of cultured LNCaP cells.
Furthermore overexpression of intact HepaCAM also downregulated the
expression of c-myc and cyclin D1, which are upregulated in the
proliferation of LNCaP cells. This downregulation of c-myc and
cyclin D1 was also been demonstrated in the colorectal cancer cells
(29). In the present study,
overexpression of intact HepaCAM was also revealed to downregulate
the expression of N-cadherin and Snail proteins, which are
upregulated in the migration and invasion of LNCaP cells. This
phenomenon has been demonstrated in castration-resistant PCa in a
precious study (30). However,
overexpression of HepaCAM-mt in the present study did not induce an
equivalent anti-tumour effect compared to intact HepaCAM. These
data suggest that the anti-tumour activity of HepaCAM in PCa may
act through its phosphorylated cytoplasmic domain.
The prostate is a canonical AR-dependent organ, the
AR signalling pathway serves a crucial role in the growth and
progression of PCa (31). When
androgen binds to the AR in the cytoplasm, the complex is
translocated from the cytoplasm to the nucleus, where it targets
and promotes downstream gene expression, producing a series of
physiological responses (32). In
our previous study (7,20), HepaCAM expression significantly
reduced the levels of PSA, which is strongly associated with the
activation of AR. Further immunofluorescence experiments indicated
that overexpression of HepaCAM disrupts the endonuclear and
extranuclear distribution of AR in LNCaP cells. The accumulation of
cytoplasmic AR and reduction of nuclear AR indicated that HepaCAM
may inhibit AR nuclear translocation which may inhibit AR
signalling pathway. But this needs to be validated. Furthermore,
overexpression of HepaCAM-mt did not disturb AR translocation and
had no influence on AR distribution, which was similar to the
control groups. Therefore, the present study suggested that
HepaCAM-mediated anti-neoplastic effects were partly caused by the
suppression of AR nuclear translocation, which may be accomplished
through its phosphorylated cytoplasmic domain.
Energy is consumed when the AR traverses the nuclear
pore complex to activate AR signalling (33). This energy is provided by the
concentration difference of Ran across the nuclear membrane
(34). In the present study
overexpression of HepaCAM affected Ran distribution in LNCaP cells.
The levels of Ran were higher in the cytoplasm and lower in the
nucleus compared with the control cells. By contrast,
overexpression of HepaCAM-mt did not result in an altered
distribution of Ran, which was similar to the control groups. The
differences in Ran distribution were consistent with those of AR
distribution in the cytoplasm and nucleus, suggesting that Ran is
closely associated with suppression of AR nuclear translocation and
subsequent deactivation of the AR signalling pathway. To further
investigate the role of Ran in the nuclear translocation of AR,
si-Ran was used to silence the expression of Ran. The results
revealed that AR was mainly localized in the cytoplasm rather than
the nucleus when combined with ad-HepaCAM or ad-HepaCAM-mt. Based
on these results, it could be proposed that HepaCAM affects Ran
through its cytoplasmic domain and consequently affects AR nuclear
translocation, but this needs to be investigated further.
The present study demonstrated that HepaCAM is
associated with a reduction in cell proliferation and migration in
a PCa cell line. Nuclear translocation of AR and the AR signalling
pathway serve a crucial role in the carcinogenesis and tumour
development of PCa (10,11). HepaCAM may affect Ran through its
cytoplasmic domain and consequently affect AR nuclear
translocation. Further studies are required in order to elucidate
whether HepaCAM affects the activity of Ran and AR and to determine
the binding force between HepaCAM and Ran and AR. Targeting
molecules of the HepaCAM/Ran/AR axis may be a potential ADT
strategy for PCa.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81072086), and the
Scientific and Technological Research Program of Chongqing
Municipal Education Committee (grant no. KJ110305).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
QD, XW and CL conceived the experiments. QD, ZQ, ZD
and WS conducted the experiments and analyzed the data in the
researches of the clinical samples. QD, LL and NL conducted the
experiments and analyzed the data. QD and XW wrote the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Informed consent forms were signed by all patients,
and the study was approved by the Research Ethical Committees of
Chongqing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chung Moh M, Hoon Lee L and Shen S:
Cloning and characterization of hepaCAM, a novel Ig-like cell
adhesion molecule suppressed in human hepatocellular carcinoma. J
Hepatol. 42:833–841. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moh MC, Zhang T, Lee LH and Shen S:
Expression of hepaCAM is downregulated in cancers and induces
senescence-like growth arrest via a p53/p21-dependent pathway in
human breast cancer cells. Carcinogenesis. 29:2298–2305. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu B, He Y, Wu X, Luo C, Liu A and Zhang
J: Exploration of the correlations between interferon-γ in patient
serum and HEPACAM in bladder transitional cell carcinoma, and the
interferon-γ mechanism inhibiting BIU-87 proliferation. J Urol.
188:1346–1353. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xun C, Luo C, Wu X, Zhang Q, Yan L and
Shen S: Expression of hepaCAM and its effect on proliferation of
tumor cells in renal cell carcinoma. Urology. 75:828–834. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang QL, Luo CL, Wu XH, Wang CY, Xu X,
Zhang YY, Liu Q and Shen SL: HepaCAM induces G1 phase arrest and
promotes c-Myc degradation in human renal cell carcinoma. J Cell
Biochem. 112:2910–2919. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song X, Wang Y, Du H, Fan Y, Yang X, Wang
X, Wu X and Luo C: Overexpression of HepaCAM inhibits cell
viability and motility through suppressing nucleus translocation of
androgen receptor and ERK signaling in prostate cancer. Prostate.
74:1023–1033. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang X, Chen E, Tang M, Yang X, Wang Y,
Quan Z, Wu X and Luo C: The SMAD2/3 pathway is involved in
hepaCAM-induced apoptosis by inhibiting the nuclear translocation
of SMAD2/3 in bladder cancer cells. Tumour Biol. 37:10731–10743.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wen S, Chang HC, Tian J, Shang Z, Niu Y
and Chang C: Stromal androgen receptor roles in the development of
normal prostate, benign prostate hyperplasia, and prostate cancer.
Am J Pathol. 185:293–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Scher HI and Sawyers CL: Biology of
progressive, castration-resistant prostate cancer: Directed
therapies targeting the androgen-receptor signaling axis. J Clin
Oncol. 23:8253–8261. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Taplin ME and Balk SP: Androgen receptor:
A key molecule in the progression of prostate cancer to hormone
independence. J Cell Biochem. 91:483–890. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen Y, Clegg NJ and Scher HI:
Anti-androgens and androgen-depleting therapies in prostate cancer:
New agents for an established target. Lancet Oncol. 10:981–991.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Attard G, Cooper CS and de Bono JS:
Steroid hormone receptors in prostate cancer: A hard habit to
break? Cancer Cell. 16:458–462. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dehm SM and Tindall DJ: Androgen receptor
structural and functional elements: Role and regulation in prostate
cancer. Mol Endocrinol. 21:2855–2863. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Knudsen KE and Scher HI: Starving the
addiction: New opportunities for durable suppression of AR
signaling in prostate cancer. Clin Cancer Res. 15:4792–4798. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Adam SA and Gerace L: Cytosolic proteins
that specifically bind nuclear location signals are receptors for
nuclear import. Cell. 66:837–847. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rexach M and Blobel G: Protein import into
nuclei: Association and dissociation reactions involving transport
substrate, transport factors, and nucleoporins. Cell. 83:683–692.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Weis K, Dingwall C and Lamond AI:
Characterization of the nuclear protein import mechanism using Ran
mutants with altered nucleotide binding specificities. EMBO J.
15:7120–7128. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sanda MG, Cadeddu JA, Kirkby E, Chen RC,
Crispino T, Fontanarosa J, Freedland SJ, Greene K, Klotz LH,
Makarov DV, et al: Clinically localized prostate cancer:
AUA/ASTRO/SUO Guideline. Part I: Risk stratification, shared
decision making, and care options. J Urol. 199:683–690. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Wu X, Ou L, Yang X, Wang X, Tang
M, Chen E and Luo C: PLCepsilon knockdown inhibits prostate cancer
cell proliferation via suppression of Notch signalling and nuclear
translocation of the androgen receptor. Cancer Lett. 362:61–69.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Q, Luo C, Wu X, Du H, Song X and Fan
Y: hepaCAM and p-mTOR closely correlate in bladder transitional
cell carcinoma and hepaCAM expression inhibits proliferation via an
AMPK/mTOR dependent pathway in human bladder cancer cells. J Urol.
190:1912–1918. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang L, Hou Y, Sun Y, Zhao L, Tang X, Hu
P, Yang J, Zeng Z, Yang G, Cui X and Liu M: c-Ski activates
cancer-associated fibroblasts to regulate breast cancer cell
invasion. Mol Oncol. 7:1116–1128. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Becker KF, Rosivatz E, Blechschmidt K,
Kremmer E, Sarbia M and Höfler H: Analysis of the E-cadherin
repressor Snail in primary human cancers. Cells Tissues Organs.
185:204–212. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bennett NC, Gardiner RA, Hooper JD,
Johnson DW and Gobe GC: Molecular cell biology of androgen receptor
signalling. Int J Biochem Cell Biol. 42:813–827. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taplin ME and Balk SP: Androgen receptor:
A key molecule in the progression of prostate cancer to hormone
independence. J Cell Biochem. 91:483–490. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Quan Z, He Y, Luo C, Xia Y, Zhao Y, Liu N
and Wu X: Interleukin 6 induces cell proliferation of clear cell
renal cell carcinoma by suppressing hepaCAM via the STAT3-dependent
up-regulation of DNMT1 or DNMT3b. Cell Signal. 32:48–58. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moh MC, Zhang C, Luo C, Lee LH and Shen S:
Structural and functional analyses of a novel ig-like cell adhesion
molecule, hepaCAM, in the human breast carcinoma MCF7 cells. J Biol
Chem. 280:27366–27374. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Geng HT, Cao RJ, Cheng L and Liu CY:
Overexpression of hepatocyte cell adhesion molecule (hepaCAM)
inhibits the proliferation, migration, and invasion in colorectal
cancer cells. Oncol Res. 25:1039–1046. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du Z, Li L, Sun W, Wang X, Zhang Y, Chen
Z, Yuan M, Quan Z, Liu N, Hao Y, et al: HepaCAM inhibits the
malignant behavior of castration-resistant prostate cancer cells by
downregulating Notch signaling and PF-3084014 (a γ-secretase
inhibitor) partly reverses the resistance of refractory prostate
cancer to docetaxel and enzalutamide in vitro. Int J Oncol.
53:99–112. 2018.PubMed/NCBI
|
|
31
|
Bluemn EG and Nelson PS: The
androgen/androgen receptor axis in prostate cancer. Curr Opin
Oncol. 24:251–257. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ardiani A, Gameiro SR, Kwilas AR, Donahue
RN and Hodge JW: Androgen deprivation therapy sensitizes prostate
cancer cells to T-cell killing through androgen receptor dependent
modulation of the apoptotic pathway. Oncotarget. 5:9335–9348. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shank LC, Kelley JB, Gioeli D, Yang CS,
Spencer A, Allison LA and Paschal BM: Activation of the
DNA-dependent protein kinase stimulates nuclear export of the
androgen receptor in vitro. J Biol Chem. 283:10568–10580. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Matchett KB, McFarlane S, Hamilton SE,
Eltuhamy YS, Davidson MA, Murray JT, Faheem AM and El-Tanani M: Ran
GTPase in nuclear envelope formation and cancer metastasis. Adv Exp
Med Biol. 773:323–351. 2014. View Article : Google Scholar : PubMed/NCBI
|