Introduction
There is a poor intrinsic capacity for repair in
articular cartilage, and even a minor defect as a result of
mechanical damage may fail to heal, thus degenerating to
osteoarthritis (OA) (1). OA, one
of the most common degenerative joint disorders worldwide, is
characterized by slow destruction and loss of articular cartilage,
mainly affecting the hips and knees (2). In China specifically, the morbidity
rate of symptomatic knee OA is as high as 8.1% (3). Unfortunately, the precise
pathogenesis of OA remains unclear.
C-X-C motif chemokine ligand 12 (CXCL12), also
termed stromal cell-derived factor 1, is a typical bone
marrow-derived chemokine which promotes stem cell homing to injured
areas (4,5) and functions in tissue and/or organ
repair (4,6) via binding to C-X-C chemokine receptor
type 4 (CXCR4), a G protein-coupled receptor. CXCL12 and CXCR4
binding promotes the early osteogenic differentiation of
mesenchymal stem cells by regulating the bone morphogenetic protein
(BMP)-2/mothers against decapentaplegic homolog (Smad)/Runt-related
transcription factor 2 (Runx2)/Osterix axis (7). In addition, CXCL12 from bone
marrow-derived mesenchymal stromal cells has been shown to improve
muscle regeneration (8). On the
other hand, it has been reported that CXCL12 is involved in the
destruction of cartilage in OA (9). The present study investigated whether
CXCL12 was involved in OA development in chondrocytes.
MicroRNAs (miRNAs/miRs), a group of non-coding small
RNAs (~22 nucleotides), are widely expressed in eukaryons and serve
important roles in numerous physiological and pathological
processes. In OA, a number of miRNAs are aberrantly expressed in
tissues, such as miR-33a (3) and
miR-155 (4). However the role of
miR-31 in OA remains to be elucidated, and was therefore
investigated in the present study.
Materials and methods
Clinical specimens
Articular cartilage tissues were collected from 30
OA patients (male, 21; female, 9; age range 52–65 years) undergoing
total knee replacement surgery and 30 non-OA patients (male, 19;
female, 11; age range 54–67 years) with femoral neck fracture
undergoing surgery at the Second People's Hospital of Huai'an
(Huai'an, China) between August 2015 and August 2016. The non-OA
patients had no known history of OA or rheumatoid arthritis. All
patients provided written informed consent and the present study
was approved by the ethics committee of the Second People's
Hospital of Huai'an.
Cell culture
Human chondrocyte cell line CHON-001 was obtained
from American Type Culture Collection (cat no. CRL-2846; Manassas,
VA, USA) and incubated in Dulbecco's modified Eagle's
medium/nutrient mixture F12 (DMEM/F12; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.), 1%
penicillin-streptomycin solution and 25 µg/ml amphotericin B
(Fungizone; Gibco; Thermo Fisher Scientific, Inc.) in an incubator
at 37°C with 5% CO2.
Cell transfection
For cell transfection, 5×104 CHON-001
cells/well were seeded into a 24-well plate. miRNA mimic and
inhibitor were obtained from Shanghai GenePharma Co., Ltd.,
(Shanghai, China). The next day, cells were transfected with 50 nM
miR-31 mimic (forward: 5′AGGCAAGAUGCUGGCAUAGCU3′, reverse:
5′CUAUGCCAGCAUCUUGCCUUU3′), 50 nM miR-31 inhibitor
(5′AGCUAUGCCAGCAUCUUGCCU3′), miR-control (forward:
5′UUCUCCGAACGUGUCACGUTT3′, reverse: 5′ACGUGACACGUUCGGAGAATT3′), 2
µl control plasmid (cat. no. sc-108083; Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA), 2 µl CXCL12-plasmid (cat. no.
sc-422854-ACT; Santa Cruz Biotechnology, Inc.), miR-31 mimic +
control plasmid or miR-31 mimic+CXCL12-plasmid using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.).
After 48 h, the cells were harvested for subsequent
experimentation.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted by TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.). RNA was reverse
transcribed into cDNA using a Reverse Transcription system (Promega
Corporation, Madison, WI, USA). The qPCR reaction mixture was as
follows: SYBR Premix Ex Taq™ II (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA), 2 µl cDNA, 5 µl 2X master mix, 0.5 µl
forward/reverse primer and 2 µl nanopure water. qPCR was performed
on an ABI 7900HT machine (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Amplification conditions were as follows: 95°C
for 5 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1
min. The primer sequences were obtained from Jinsite Science and
Technology (Nanjing) Co., Ltd. (Nanjing, China) and listed in
Table I. Data were calculated by
2−ΔΔCq method (10).
 | Table I.Primer sequences used for quantitative
polymerase chain reaction analysis. |
Table I.
Primer sequences used for quantitative
polymerase chain reaction analysis.
| Gene | Direction | Sequence (5′-3′) |
|---|
| C-X-C motif chemokine
ligand 12 | F |
ATGCCCATGCCGATTCTT |
|
| R |
GCCGGGCTACAATCTGAAGG |
| Type I collagen | F |
CCCTGAGTGGAAGAGTGGAG |
|
| R |
GAGGCGTGAGGTCTTCTGTG |
| Aggrecan | F |
CTAGAGATCAGTGGACTGCCT |
|
| R |
TCTGGAGCTGTGCAGTCTAGTGG |
| microRNA-31 | F |
CTCGGATCCTGTGCATAACTGCCTTCA |
|
| R |
CACAAGCTTGAAGTCAGGGCGAGACAGAC |
| U6 | F |
Universal_RNU6B_Primer |
|
| R | Uni-miR qPCR
Primer |
| GAPDH | F |
CTTTGGTATCGTGGAAGGACTC |
|
| R |
GTAGAGGCAGGGATGATGTTCT |
Western blot analysis
CHON-001 cell lysates were washed with PBS and
homogenized in radioimmunoprecipitation assay buffer (Roche
Diagnostics, Basel, Switzerland). Proteins (30 µg per lane) were
separated by 10% SDS-PAGE and transferred to polyvinylidene
fluoride membranes, which were blocked with 5% milk in Tris
buffered saline with 0.1% Tween-20 for 2 h at room temperature.
Next, they were incubated with primary antibodies against β-actin
(cat no. 4970; 1:1,000; Cell Signaling Technology, Inc., Danvers,
MA, USA), CXCL12 (cat no. 3530; 1:1,000; Cell Signaling Technology,
Inc.), type I collagen (cat no. ab34710; 1:1,000; Abcam) and
aggrecan (cat no. ab36861; 1:1,000; Abcam) at 4°C overnight.
Membranes were subsequently incubated with horseradish
peroxidase-conjugated anti-rabbit IgG secondary antibody (cat no.
7074; 1:5,000; Cell Signaling Technology, Inc.) for 1 h at room
temperature. Bands were visualized by SuperSignal® West
Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Inc.).
Band density was normalized to β-actin.
MTT assay
Cell viability was assessed by MTT assays at 24, 48
and 72 h after cell transfection. Cells (5×103
cells/well) were seeded in 96-well plates and incubated for 24 h at
37°C. Next, MTT (20 µl; 5 mg/ml) was added to each well and
incubated for another 4 h. Wells were then washed in PBS, dried and
150 µl dimethyl sulfoxide was added. Data were measured by a
microtiter plate reader at 490 nm (Thermo Fisher Scientific,
Inc.).
Cell migration assay
The migratory ability of CHON-001 cells was detected
using 24-well Transwell chambers. Cells (2×104/well)
were plated into the upper chambers and incubated in 200 µl
serum-free DMEM (Gibco; Thermo Fisher Scientific, Inc.). The bottom
chamber contained DMEM (0.5 ml) with 20% FBS (Gibco; Thermo Fisher
Scientific, Inc.). Following a 48 h incubation, cells on the upper
chambers were removed, and the migrated cells on the lower chambers
were fixed with 10% formalin for 30 min at room temperature and
stained with hematoxylin and eosin (H&E) for 15 min at 37°C.
Cell migratory ability was assessed by counting the number of
H&E-stained cells of five fields of view under a light
microscope (Olympus Corporation, Tokyo, Japan).
Bioinformatic analysis
Prediction of miRNA target sites was performed using
microRNA.org (www.microrna.org).
Luciferase reporter assays
A luciferase reporter (Promega Corporation) for the
wild-type (WT) CXCL12 3′untranslated region (UTR) and mutant (MUT)
CXCL12 3′UTR was constructed by site-directed mutagenesis with
Phusion™ High-Fidelity DNA Polymerase (Thermo Fisher Scientific,
Inc.). CHON-001 cells (5×104) were seeded into each well
of a 24-well plate. The next day, cells were co-transfected with 50
nM CXCL12-3′UTR-WT or CXCL12-3′UTR-MUT and miR-31 mimics or control
using Lipofectamine® 3000 (Thermo Fisher Scientific,
Inc.). After 72 h, cells were harvested to determine luciferase
activity with the Dual-Luciferase Reporter Assay system (Beyotime
Institute of Biotechnology, Haimen, China) and a luminometer.
Firefly luciferase activity was normalized to Renilla.
Statistical analysis
Data were analyzed by Student's t-test or one-way
analysis of variance followed by Tukey's test, using SPSS version
21 (IBM Corp., Armonk, NY, USA). Data were expressed as the mean ±
standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
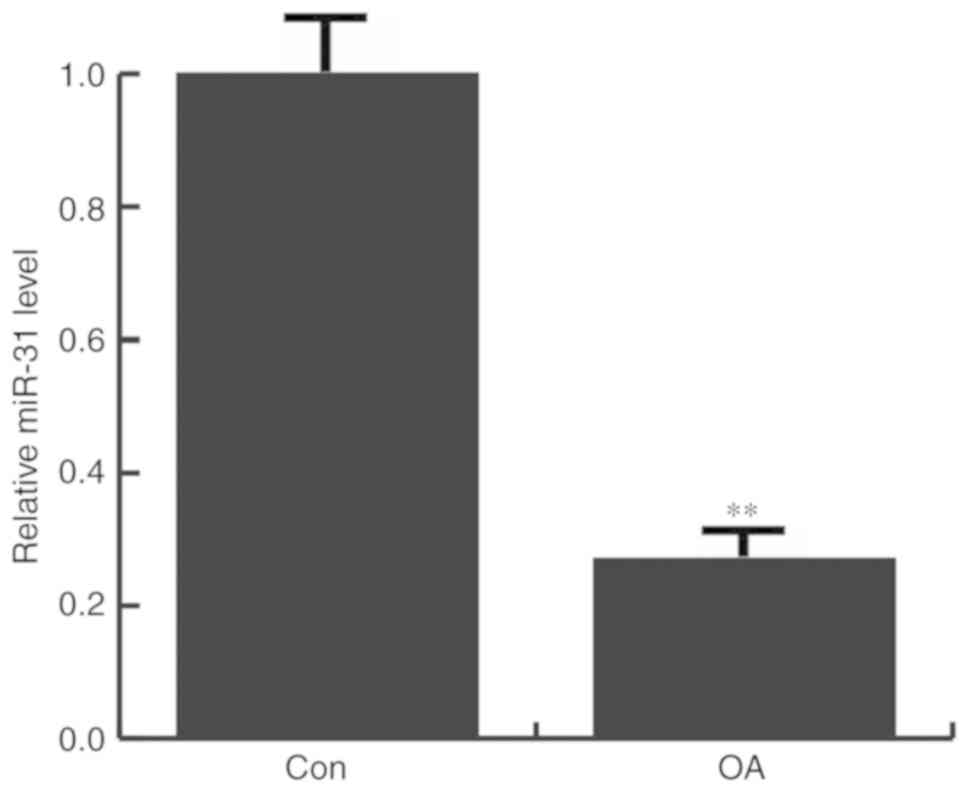
miR-31 is downregulated in OA
patients
The expression of miR-31 in the articular cartilage
tissues of patients with and without OA was determined by RT-qPCR.
It was found that that miR-31 expression was significantly
decreased in OA patients, compared with the non-OA patients
(Fig. 1).
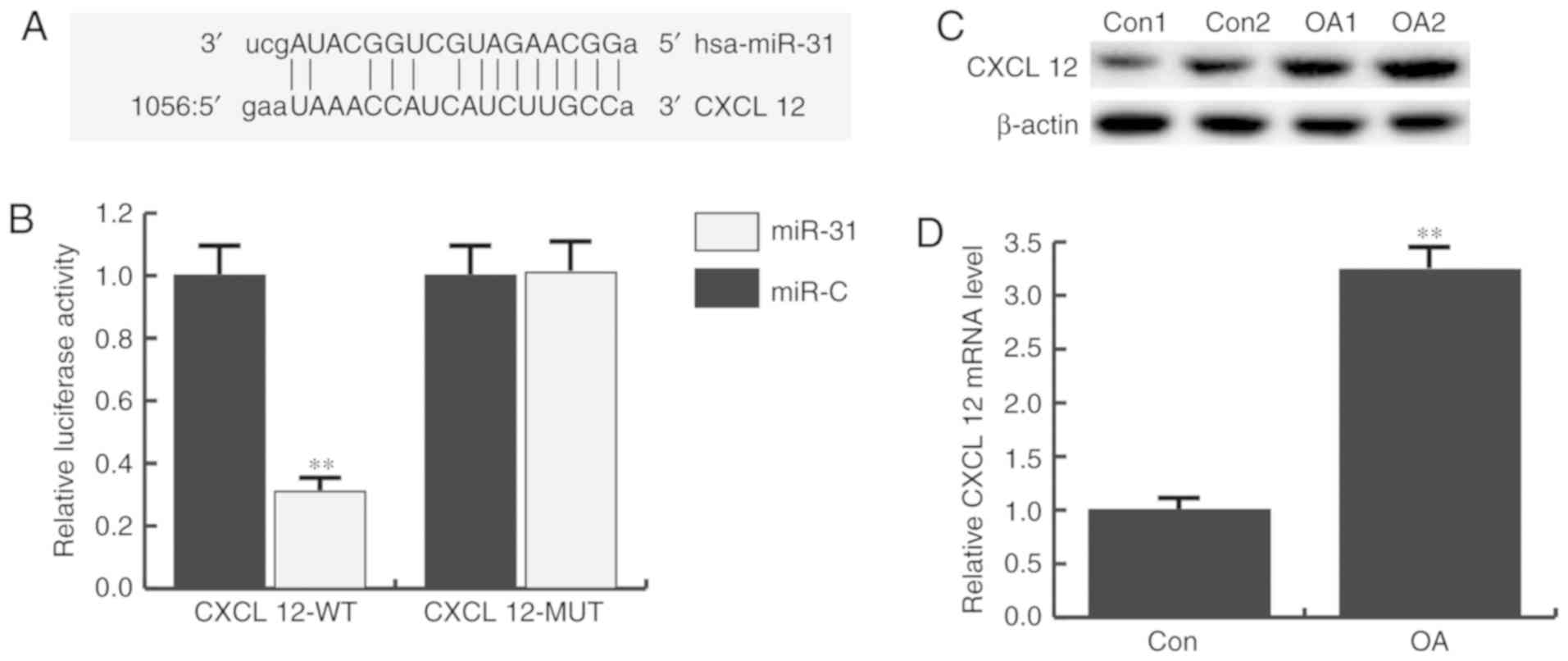
CXCL12 is targeted by miR-31
To investigate the role of miR-31 in OA progression,
bioinformatics software (www.microrna.org) was used to predict the potential
targets of miR-31. A total of 6,829 targets were identified,
including CXCL12 (Fig. 2A). As it
has been previously reported that CXCL12 has critical function in
the development of OA (11,12),
this gene was selected for further analysis.
Next, the correlation between miR-31 and CXCL12 was
verified by dual-luciferase reporter assays, which showed that the
luciferase activity of the CXCL12-WT group significantly declined
following miR-31 mimic transfection compared with the miR-control
group, with no evident alteration in the CXCL12-MUT groups
(Fig. 2B).
In addition, the expression of CXCL12 in the
articular cartilage tissues of patients was determined by RT-qPCR
and western blotting. It was demonstrated that CXCL12 protein
(Fig. 2C) and mRNA (Fig. 2D) expression was significantly
increased in OA patients compared with non-OA patients.
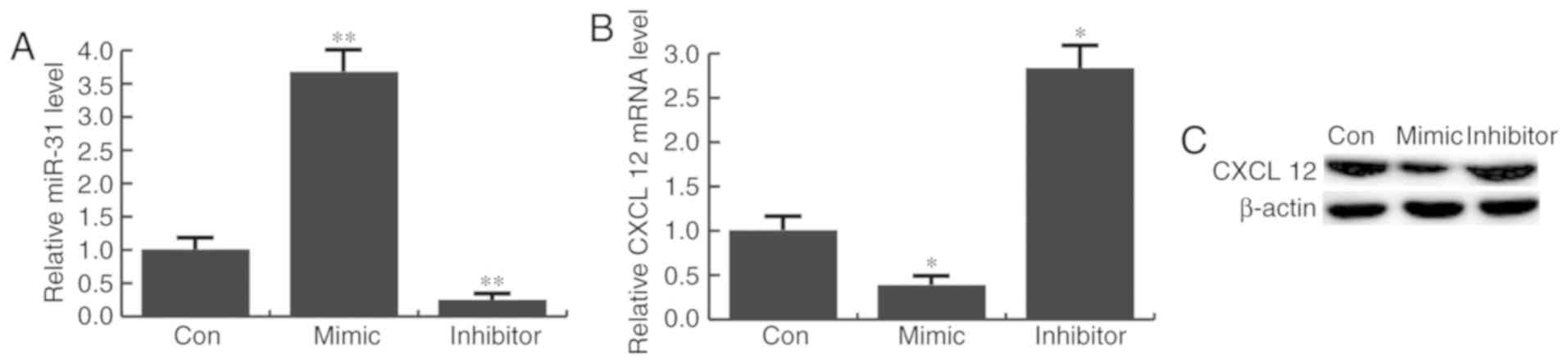
miR-31 inhibits CXCL12 expression in
CHON-001 cells
To further investigated whether miR-31 regulated
CXCL12 expression in CHON-001 cells, they were transfected with
miR-31 mimics, inhibitor or control mimics. Transfection efficiency
was determined 48 h after transfection by RT-qPCR (Fig. 3A). It was revealed that CXCL12
expression was markedly decreased by miR-31 mimic transfection, and
increased by miR-31 inhibitor transfection, compared with the
control group (Fig. 3B and C).
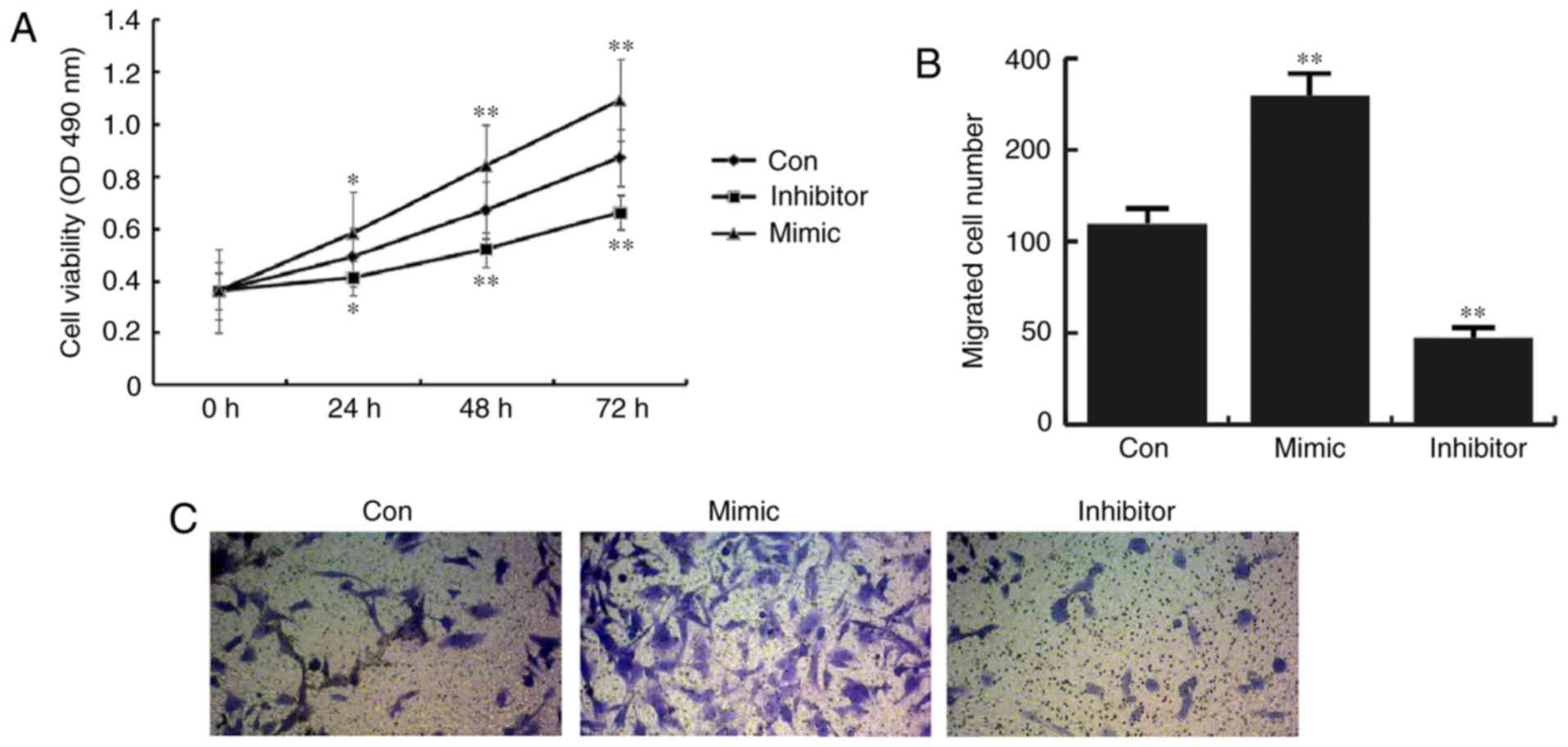
miR-31 increases CHON-001 cell
viability and migration
Compared with the control group, CHON-001 cells
transfected with miR-31 mimics exhibited a significant increase in
cell viability, while miR-31 inhibitor transection results in a
significant decrease in cell viability (Fig. 4A). In addition, it was found that
the number of migrated cells in the miR-31 mimic group was
increased compared with the control group, whereas miR-31 inhibitor
transfection significantly prevented CHON-001 cell migration
(Fig. 4B and C).
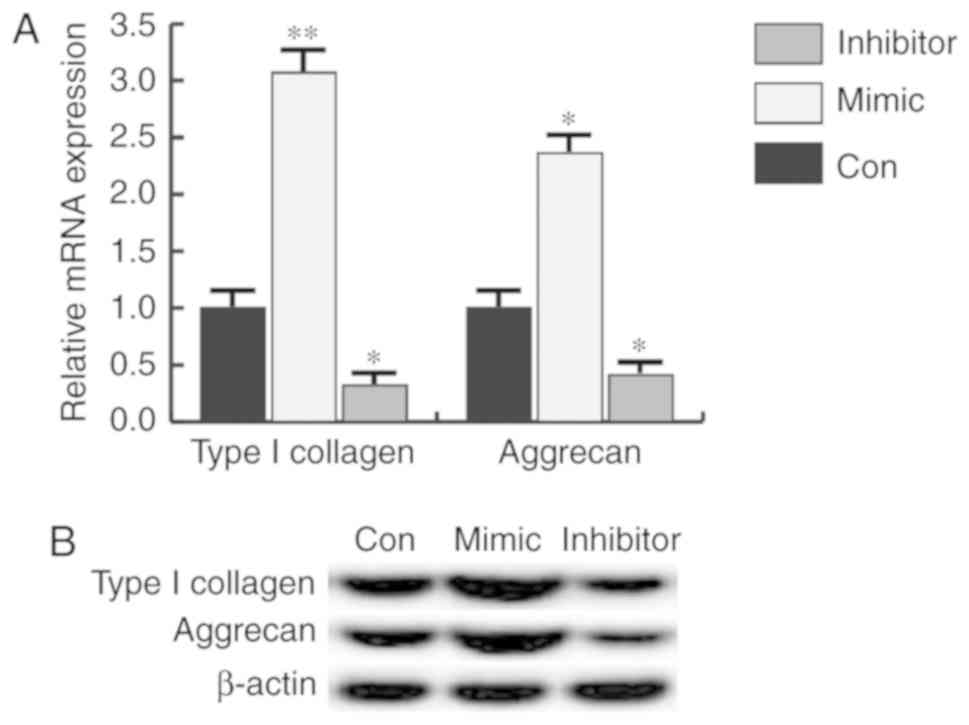
miR-31 promotes the expression of type
I collagen and aggrecan
During the progression of OA, the collagen matrix
undergoes a transitional degradation (13,14).
Therefore, the expression of type I collagen and aggrecan was
detected in the present study. The findings indicated that the
protein and mRNA expression of type I collagen and aggrecan was
increased by miR-31 mimic transfection, and miR-31 inhibitor had
the opposite effect in CHON-001 cells (Fig. 5).
CXCL12 overexpression prevents the
alterations induced by miR-31
To examine whether miR-31 affected CHON-001 cells by
directly targeting CXCL12, control or CXCL12-plasmid was initially
transfected into CHON-001 cells to assess transfection efficiency
by western blotting (Fig. 6A) and
RT-qPCR (Fig. 6B). Following this,
miR-31 mimics + control plasmid or miR-31 mimics+CXCL12-plasmid was
transfected into CHON-001 cells. The protein and mRNA expression of
CXCL12, type I collagen and aggrecan was detected by western blot
and RT-PCR, 48 h after transfection. It was found that the protein
(Fig. 6C) and mRNA expression
(Fig. 6D and E) of type I collagen
and aggrecan was notably enhanced by miR-31 mimics, whereas CXCL12
overexpression eliminated these effects.
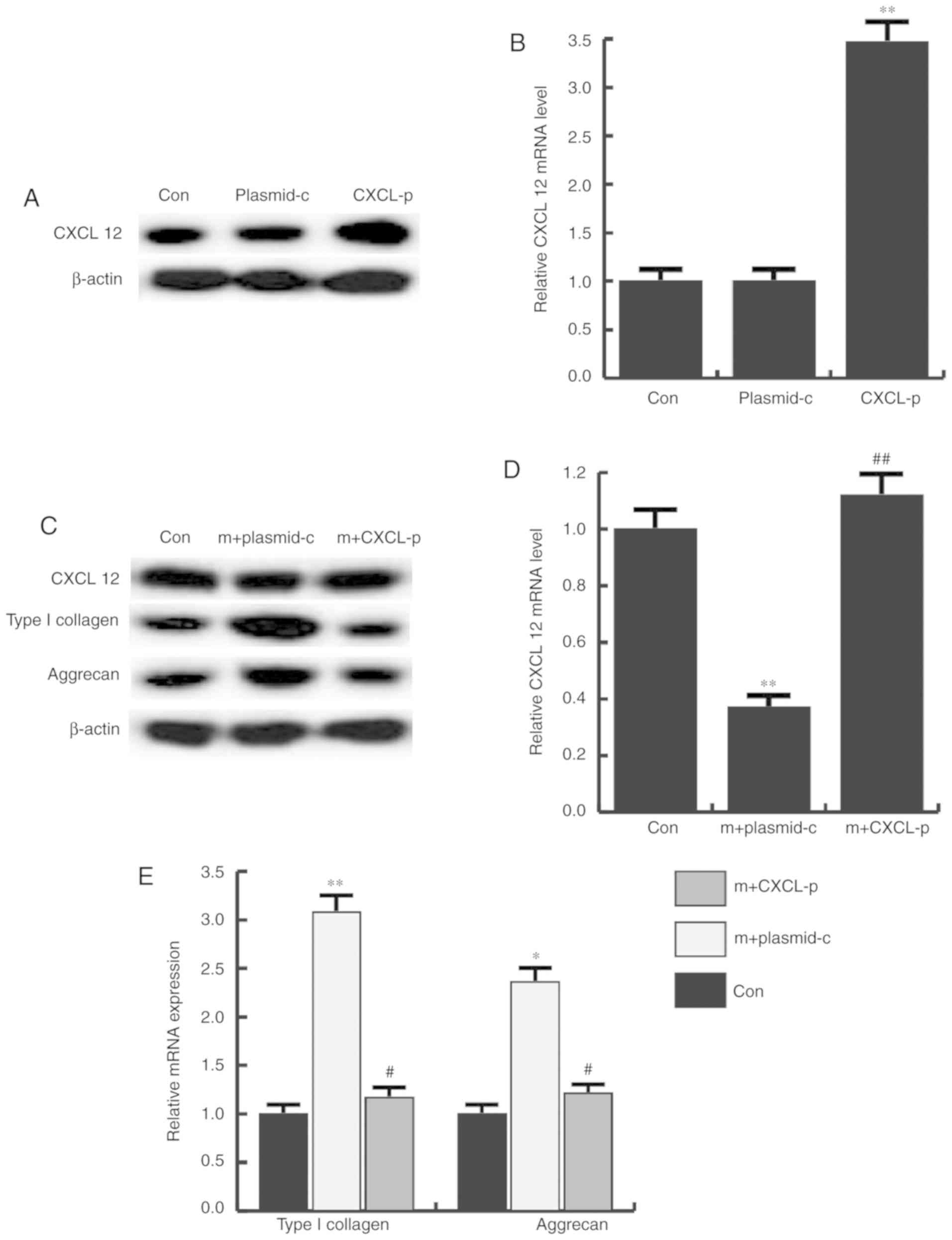 | Figure 6.Effects of CXCL12 plasmid on CXCL12,
type I collagen and aggrecan expression. The protein and mRNA
expression level of CXCL12, type I collagen and aggrecan in each
group was determined 48 h after transfection by western blot
analysis and reverse transcription-quantitative polymerase chain
reaction, respectively. Transfection efficiency of CXCL12 was
confirmed at the (A) protein and (B) mRNA level. (C) CXCL12, type I
collagen and aggrecan protein and (D and E) mRNA expression in
cells transfected with mimics+control plasmid or mimics+CXCL12
plasmid. *P<0.05, **P<0.01 vs. control;
#P<0.05, ##P<0.01 vs. m+plasmid-c Con,
control; plasmid-c, control-plasmid; CXCL-p, CXCL 12-plasmid; m,
microRNA-31 mimics; CXCL12, C-X-C motif chemokine ligand 12. |
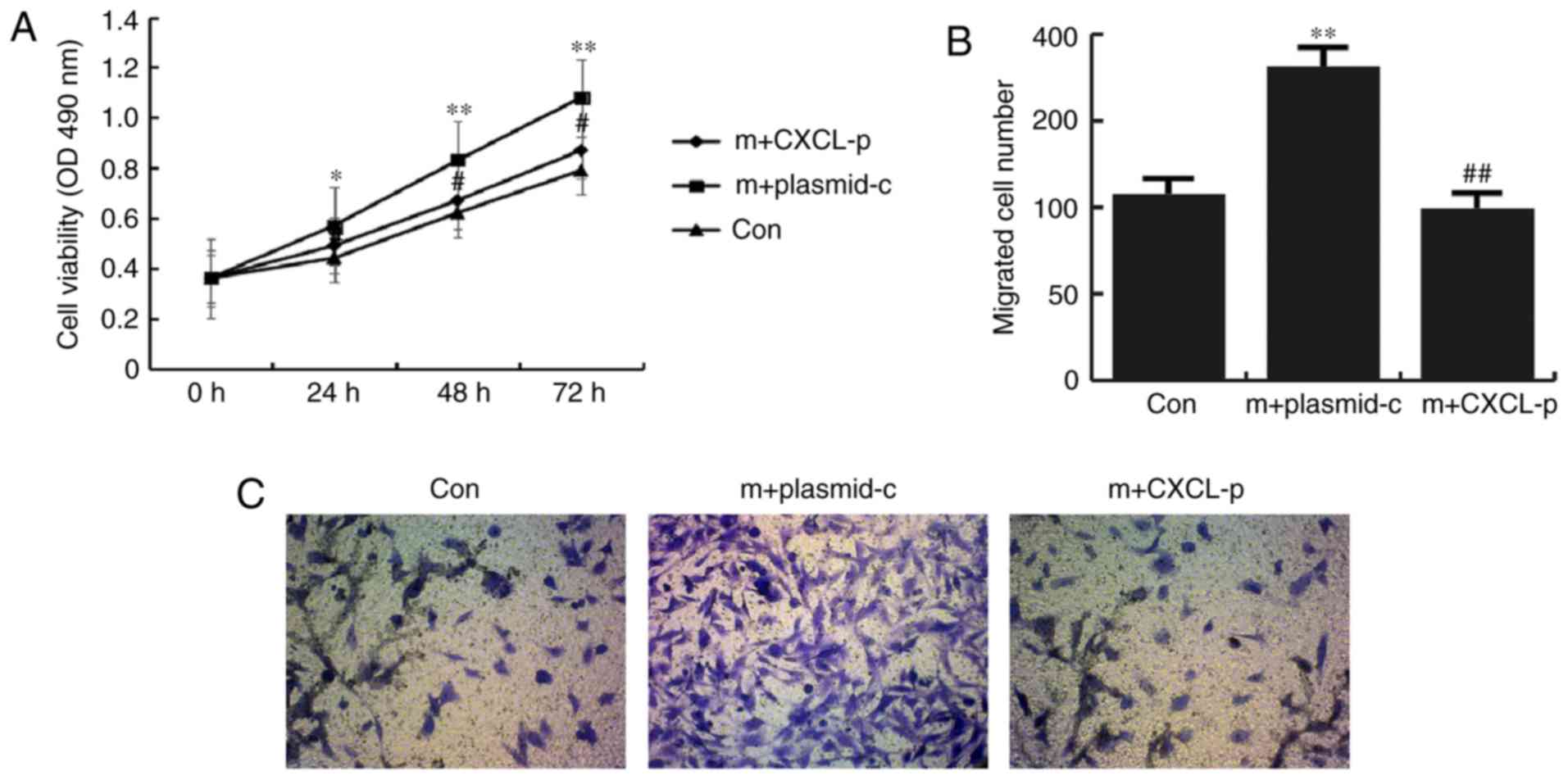
Furthermore, compared with the cells transfected
with miR-31 mimics + control plasmid, cell viability (Fig. 7A) and migration (Fig. 7B and C) significantly decreased in
the miR-31 mimic+CXCL12-plasmid transfected cells, compared with
the mimics + control plasmid transfected cells (Fig. 7). Taken together, these data
indicated that miR-31 promoted CHON-001 cell proliferation and
migration, as well as type I collagen and aggrecan expression by
directly targeting CXCL12.
Discussion
In the current study, it was demonstrated that
miR-31 was downregulated in patients with OA, compared with
controls. miR-31 promoted human chondrocyte CHON-001 cell viability
and migration, as well as and the expression of type I collagen and
aggrecan, which provided evidence for miR-31 as a potential target
in cartilage defection therapy.
OA is characterized by slow destruction and loss of
articular cartilage, which predominantly affects the hips and knees
(2). Unfortunately, the precise
pathogenesis of OA remains unclear. miRNAs have received increasing
attention for their contribution to signaling network disruption in
OA (15). Many specific miRNAs
have been identified to be critical during OA pathogenesis
(16). For example, miR-384-5p
downregulation alleviates OA through inhibiting cartilage cell
apoptosis via the NF-κB signaling pathway by targeting SOX9
(17). MiR-34a has been identified
to facilitate the development of OA by enhancing chondrocyte
apoptosis and senescence (18).
MiR-449a upregulation was found to promote chondrocyte
extracellular matrix degradation in OA (19). It has been reported that
downregulation of miR-31 enhances the osteogenic differentiation of
human mesenchymal stem cells by targeting SATB2 (20). miR-31-5p expression is reduced in
cartilage-derived mesenchymal stem cells from OA degraded cartilage
compared to normal cartilage, and may regulate osteogenic
differentiation (5). During bone
mesenchymal stem cell osteogenic differentiation, miR-31 expression
is markedly decreased, and inhibition of miR-31 enhances osteogenic
differentiation (21).
Furthermore, miR-31 has been reported to enhance cell proliferation
during differentiation by targeting CCAAT enhancer binding protein
α (C/EBP-α) (22). However, to the
best of our knowledge, the role of miR-31 in the development of OA
remained largely unclear. In the present study, it was found that
miR-31 was significantly downregulated in OA patients. In addition,
the 3′UTR of CXCL12 was targeted by miR-31, which was predicted via
microRNA.org and verified with dual-luciferase
reporter assays. Additionally, it was indicated that miR-31
negatively regulated CXCL12 expression in human chondrocyte cells.
CXCL12 has been revealed to take part in tissues and organ repair
(4,6). Inhibition of the CXCL12/CXCR4
signaling axis prevents aggrecanase expression and reduces
cartilage degeneration (23,24).
Furthermore, CXCL12 expression in the plasma and synovial fluid of
OA patients may serve as an effective biomarker for the severity of
OA (25).
CHON-001 cells were subsequently transfected with
miR-31 mimics, miR-31 inhibitor or mimic control, and cell
viability was detected by MTT assays 24, 48 and 72 h
post-transfection. CHON-001 cell migration assays were performed
and the expression of type I collagen and aggrecan was also
determined. The results suggested that miR-31 enhanced the cell
viability and migration of CHON-001 cells, and the expression of
type I collagen and aggrecan was increased. Furthermore, to explore
whether miR-31 affected CHON-001 cells by directly targeting
CXCL12, cells were transfected with miR-31 mimics + control plasmid
or miR-31 mimics+CXCL12-plasmid, and cell viability, migration, and
type I collagen and aggrecan expression were subsequently
determined. The findings revealed that the effects of miR-31 on
CHON-001 cell viability and migration, as well as type I collagen
and aggrecan expression were eliminated by CXCL12
overexpression.
Taken together, the results indicated that miR-31
was down-regulated in OA, and miR-31 promoted the growth and
migration of chondrocytes. The data suggested a crucial role of
miR-31 in OA, suggesting that miR-31 may serve as a potential
target for OA treatment. However, whether there is a correlation
between the expression of miR-31 and its target gene CXCL12 in OA
patients, as well as the clinical significance of miR-21 has not
been elucidated. Furthermore, the specific mechanisms by which
miR-31 functions in OA development remain unclear. Therefore, the
authors of the present study will perform more in-depth study on
the role of miR-31 in OA in the near future.
Acknowledgements
The authors thank the Life Science College of
Huaiyin Normal University for providing technical guidance in the
experiments, and the reviewers for their invaluable help in
improving the manuscript.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YD and SL contributed to the concept and design of
the study; YD, SL, XX and MD accessed and analyzed the data; QZ and
XZ contributed to data analysis and prepared the paper. All authors
collaborated to interpret the results and develop the
manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent and
the present study was approved by the ethics committee of the
Second People's Hospital of Huai'an.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Redman SN, Oldfield SF and Archer CW:
Current strategies for articular cartilage repair. Eur Cell Mater.
9:23–32. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tsezou A: Osteoarthritis year in review
2014: Genetics and genomics. Osteoarthritis Cartilage.
22:2017–2024. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tang X, Wang S, Zhang Y, Niu J, Tao K,
Zhang Y and Lin J: The prevalence of symptomatic knee
osteoarthritis in China: Results from China health and retirement
longitudinal study. Arthritis Rheumatol. 68:648–653. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dowthwaite GP, Bishop JC, Redman SN, Khan
IM, Rooney P, Evans DJ, Haughton L, Bayram Z, Boyer S, Thomson B,
et al: The surface of articular cartilage contains a progenitor
cell population. J Cell Sci. 117:889–897. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xian CJ and Foster BK: Repair of injured
articular and growth plate cartilage using mesenchymal stem cells
and chondrogenic gene therapy. Curr Stem Cell Res Ther. 1:213–229.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ji YH, Ji JL, Sun FY, Zeng YY, He XH, Zhao
JX, Yu Y, Yu SH and Wu W: Quantitative proteomics analysis of
chondrogenic differentiation of C3H10T1/2 mesenchymal stem cells by
iTRAQ labeling coupled with on-line two-dimensional LC/MS/MS. Mol
Cell Proteomics. 9:550–564. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fox JM, Chamberlain G, Ashton BA and
Middleton J: Recent advances into the understanding of mesenchymal
stem cell trafficking. Br J Haematol. 137:491–502. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Marquez-Curtis LA and Janowska-Wieczorek
A: Enhancing the migration ability of mesenchymal stromal cells by
targeting the SDF-1/CXCR4 axis. Biomed Res Int 2013.
5610982013.
|
|
9
|
Li Q, Guo Y, Chen F, Liu J and Jin P:
Stromal cell derived factor-1 promotes human adipose tissue-derived
stem cell survival and chronic wound healing. Exp Ther Med.
12:45–50. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei F, Moore DC, Wei L, Li Y, Zhang G, Wei
X, Lee JK and Chen Q: Attenuation of osteoarthritis via blockade of
the SDF-1/CXCR4 signaling pathway. Arthritis Res Ther. 14:R1772012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lisignoli G, Toneguzzi S, Piacentini A,
Cristino S, Grassi F, Cavallo C and Facchini A: CXCL12 (SDF-1) and
CXCL13 (BCA-1) chemokines significantly induce proliferation and
collagen type I expression in osteoblasts from osteoarthritis
patients. J Cell Physiol. 206:78–85. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maroudas AI: Balance between swelling
pressure and collagen tension in normal and degenerate cartilage.
Nature. 260:808–809. 1976. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chou MC, Tsai PH, Huang GS, Lee HS, Lee
CH, Lin MH, Lin CY and Chung HW: Correlation between the MR T2
value at 4.7 T and relative water content in articular cartilage in
experimental osteoarthritis induced by ACL transection.
Osteoarthritis Cartilage. 17:441–447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iliopoulos D, Malizos KN, Oikonomou P and
Tsezou A: Integrative microRNA and proteomic approsteoarthritisches
identify novel osteoarthritis genes and their collaborative
metabolic and inflammatory networks. PLoS One. 3:e37402008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sumiyoshi K, Kubota S, Ohgawara T, Kawata
K, Nishida T, Shimo T, Yamashiro T and Takigawa M: Identification
of miR-1 as a micro RNA that supports late-stage differentiation of
growth cartilage cells. Biochem Biophys Res Commun. 402:286–290.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang W, Cheng P, Hu W, Yin W, Guo F, Chen
A and Huang H: Inhibition of microRNA-384-5p alleviates
osteoarthritis through its effects on inhibiting apoptosis of
cartilage cells via the NF-κB signaling pathway by targeting SOX9.
Cancer Gene Ther. 25:326–338. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang W, Hsu P, Zhong B, Guo S and Zhang
C, Wang Y, Luo C, Zhan Y and Zhang C: MiR-34a enhances chondrocyte
apoptosis, senescence and facilitates development of osteoarthritis
by targeting DLL1 and regulating PI3K/AKT pathway. Cell Physiol
Biochem. 48:1304–1316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu J, Zou M, Ping A, Deng Z and Cai L:
MicroRNA-449a upregulation promotes chondrocyte extracellular
matrix degradation in osteoarthritis. Biomed Pharmacother.
105:940–946. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie Q, Wang Z, Bi X, Zhou H, Wang Y, Gu P
and Fan X: Effects of miR-31 on the osteogenesis of human
mesenchymal stem cells. Biochem Biophys Res Commun. 446:98–104.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deng Y, Wu S, Zhou H, Bi X, Wang Y, Hu Y,
Gu P and Fan X: Effects of a miR-31, Runx2, and Satb2 regulatory
loop on the osteogenic differentiation of bone mesenchymal stem
cells. Stem Cells Dev. 22:2278–2286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Y, Wang Y, He X, Zhang S, Wang K, Wu H
and Chen L: LncRNA TINCR/miR-31-5p/C/EBP-α feedback loop modulates
the adipogenic differentiation process in human adipose
tissue-derived mesenchymal stem cells. Stem Cell Res. 32:35–42.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu W, Shi J, Zhang J, Lv Z, Guo F, Huang
H, Zhu W and Chen A: CXCL12/CXCR4 axis regulates aggrecanase
activation and cartilage degradation in a post-traumatic
osteoarthritis rat model. Int J Mol Sci. 17:E15222016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dong Y, Liu H, Zhang X, Xu F, Qin L, Cheng
P, Huang H, Guo F, Yang Q and Chen A: Inhibition of CXCL12/CXCR4
signalling in subchondral bone attenuates post-traumatic
osteoarthritis. Int J Mol Sci. 17:E9432016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He W, Wang M, Wang Y, Wang Q and Luo B:
Plasma and synovial fluid CXCL12 levels are correlated with disease
severity in patients with knee osteoarthritis. J Arthroplasty.
31:373–377. 2016. View Article : Google Scholar : PubMed/NCBI
|