Introduction
Liver cancer, also known as hepatic cancer, is a
severe human malignant tumor; it is considered one of the major
causes of cancer-associated mortality worldwide (1). Previous epidemiological evidence
indicates increasing global rates of incidence and mortality of
hepatic cancer, with particularly high incidence rates in East
Asian countries including China (2,3). The
most common liver cancer subtype is hepatocellular carcinoma (HCC),
which accounts for >80% of cases of malignant liver tumors. The
causative factors of HCC include chronic hepatitis B or C virus
infection, hemochromatosis and excessive alcohol consumption
(2,3). The treatment options available
include surgery, radiation therapy and targeted therapy; however,
ideal therapeutic outcomes are primarily witnessed only in patients
with early-stage cancer (3). Over
the previous decades, progress in determining the molecular
mechanisms of cancer progression have provided a foundation for the
development of specific reagents that target cancer cells and also
promising novel treatment strategies including immunotherapy
(4) However, the majority of these
strategies remain unsuitable for clinical application, partially
due to an incomplete understanding of the complex pathological
processes involved in liver cancer progression.
Long noncoding RNAs (lncRNAs), defined as
transcripts >200 nucleotides but not used for translation of
proteins in cells, have previously been revealed to be involved in
various cellular processes and liver cancer tumorigenesis (5). Previously, genome-wide association
studies of large cancer data sets revealed that major
single-nucleotide polymorphisms were associated with noncoding
genes; lncRNAs have been demonstrated to be a novel class of key
noncoding factors in tumor development and progression (6). A large number of studies have
suggested that lncRNAs perform their various biological and
pathological roles by affecting gene transcription, translation and
other molecular processes through interacting with DNA, RNA or
protein components (7–10). This interaction has been indicated
to be dependent on the origin of lncRNAs within the genome and also
their structure (11,12). For example, a number of lncRNAs are
mediated by their ability to form various 3-dimensional structures
and bind different protein components. For example, lncRNA-p21 is
important for the localization of heterogeneous nuclear
ribonucleoprotein complex H to specific promoters, resulting in
global suppression of gene expression (13). At present, a number of lncRNAs have
been demonstrated to be associated with the initiation and
progression of liver cancer including highly upregulated in liver
cancer (HULC) (14), HOX
transcript antisense intergenic RNA (HOTAIR) (15), H19 (16), high expression in HCC (HEIH)
(17), and metastasis-associated
lung adenocarcinoma transcript 1 (MALAT1) (18). An additional study has indicated
that lncRNAs may regulate liver cancer progression through
epigenetic alterations, including DNA methylation and histone
modifications (12). These lncRNAs
have also been considered as biomarkers for early liver cancer
detection and also as potential targets for developing combination
therapies.
The smooth muscle α-actin 2 (ACTA2) gene encodes
smooth muscle-specific α-actin and functions as a key contractile
component of vascular smooth muscle. It has been associated with
the development of various human diseases including aortic
aneurysm, stroke, coronary artery disease, Moyamoya disease and
multi-systemic smooth muscle dysfunction syndrome (19). ACTA2 has previously been indicated
to be expressed as a member of Wnt-induced gene signature in human
HCC and is associated with poor clinical outcomes (20). The lncRNA ACTA2 antisense RNA 1
(ACTA2-AS1) encoded by ACTA2, also known as ZXF1, was identified to
be associated with cell invasion and metastasis in lung
adenocarcinoma (21). ACTA2-AS1
has also been demonstrated to have significantly different
expression patterns in human lung cancer tissues compared with
adjacent normal lung tissues, and suppression of ACTA2-AS1
expression by small interfering RNAs (siRNAs) greatly inhibited
lung cancer cell migration and invasion (21). The expression of ACTA2-AS1 in liver
carcinomas and its role in liver cancer development and progression
remain to be investigated.
The present study addressed the potential role of
ACTA2-AS1:4, one of the transcript variantss of ACTA2-AS1, in liver
cancer development by analyzing its expression in liver cancer
tissues and also multiple liver cancer cell lines. Furthermore,
ACTA2-AS1 was inhibited using specific siRNAs to investigate the
role of ACTA2-AS1 in regulating liver cancer cell proliferation,
invasion and migration. This provided important information for
understanding the function of lncRNAs during hepatic
tumorigenesis.
Materials and methods
HCC biopsies and cultured cell
lines
Biopsies of cancerous tissues and paired healthy
adjacent hepatic tissues were collected from 17 patients with HCC;
these patients were hospitalized in the Department of Hepatobiliary
Surgery, Second Affiliated Hospital of Sun Yat-sen University from
January 2017 to December 2017. The study proceeded following the
receipt of written informed consent from the patients, and tissue
collection and subsequent experiments were approved by the Ethics
Committee of the Sun Yat-sen University and the Fifth Affiliated
Hospital of Guangzhou Medical University. Clinical samples were
independently verified by 2 experienced clinical pathologists.
Liver cancer cell lines, LM3, MHCC-97L, and MHCC-97L, and the
normal liver cell line, L02, were obtained from the American Type
Culture Collection (Manassas, VA, USA). Liver cancer HepG2 and
Hep3B cell lines were purchased from the Type Culture Collection of
the Cell Bank of Chinese Academy of Sciences (Shanghai, China). For
the analysis of gene expression and cellular function, liver cancer
cells were cultured in Dulbecco's modified Eagle's medium (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing 0.1% fetal
bovine serum (FBS; Thermo Fisher Scientific, Inc.) and antibiotics
at 37°C in a humidified incubator supplied with 5%
CO2.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
For the determination of relative lncRNA levels, an
RT-qPCR assay was performed using specific primers. HCC biopsies
were stored in liquid nitrogen and fully homogenized using a mortar
and pestle to extract RNA. To extract RNA from cultured liver cell
lines and normal hepatic cells, cells were washed 3 times with PBS
and centrifuged at 800 × g for 5 min at 4°C. The PureLink™ RNA Mini
kit (cat. no. 12183018A; Thermo Fisher Scientific, Inc.) was used
to extract RNA from liver cancer tissues and cultured cell lines,
according to the manufacturer's protocol. RNA concentrations in
each sample was determined using a spectrophotometer, and ~1.5 µg
RNA from each sample was used to synthesize cDNA using the
High-Capacity cDNA Reverse Transcription kit (cat. no. 4368813;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. To quantitatively assess lncRNA expression, qPCR was
performed using a SYBR Select Master Mix kit (cat. no. 4472908;
Applied Biosystems; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. To perform RT, the thermocycling
conditions were as follows: 25°C for 10 min, 37°C for 120 min and
85°C for 5 min. The PCR program were set as follows: 95°C for 30
sec, 95°C for 10 sec, 60°C for 30 sec, for 40 cycles. Then
dissolution curve was generated by setting the program as 95°C for
15 sec, 60°C for 60 sec and 95°C for 15 sec. Data were analyzed
using 2−∆∆Cq method (22). GAPDH served as the internal control
for quantitation, and at least 3 independent repeats were performed
for statistical analysis. Sequences of primers used in the present
study were listed in Table I.
 | Table I.Primers used in the present study. |
Table I.
Primers used in the present study.
| Primer name | Sequence (5′-3′) |
|---|
| lnc-MUC20-9-F |
GCCCACCAAGTAAGGAACCC |
| lnc-MUC20-9-R |
AAATCCAGGCCAGACCAATCT |
| ACTA2-AS1:4-F |
GCCCATCAGGCAACTCGTAA |
| ACTA2-AS1:4-R |
TCTTTGCCACCTGTGCAGAC |
| TRPM2-AS:1-F |
CCCGAGGAAGGCTACTGATG |
| TRPM2-AS:1-R |
GGCTGAGTGACGAGAAGCAA |
| H-GAPDH-F |
GAGTCAACGGATTTGGTCGT |
| H-GAPDH-R |
GACAAGCTTCCCGTTCTCAG |
| H-E-Cadherine-F |
GAGAAACAGGATGGCTGAAGG |
| H-E-Cadherine-R |
TGAGGATGGTGTAAGCGATGG |
| H-N-Cad-F |
ATGAAAGACCCATCCACGC |
| H-N-Cad-R |
CCTGCTCACCACCACTAC |
| H-MMP2-F |
TGATGGCATCGCTCAGATCC |
| H-MMP2-R |
GGCCTCGTATACCGCATCAA |
| H-Caspase3-F |
CATGGAAGCGAATCAATGGACT |
| H-Caspase3-R |
CTGTACCAGACCGAGATGTCA |
| H-CyclinD1-F |
GCTGCGAAGTGGAAACCATC |
| H-CyclinD1-R |
CCTCCTTCTGCACACATTTGAA |
Transfection of ACTA2-AS1:4
The expression of ACTA2-AS1:4 was suppressed in LM3
cells, as previously described, with minor modifications (17). These are described as follows. The
ACTA2-AS1:4 small interfering RNA (siRNAs; sense,
5′-GCCUGGUGGUAAAUAUGAATT-3′ and antisense,
5′-UUCAUAUUUACCACCAGGCTT-3′) were synthesized by Shanghai
GenePharma Co., Ltd. (Shanghai, China). LM3 cells, at 80%
confluence, were transfected with the aforementioned siRNAs (200
nM) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Total RNA was
extracted 48 h after transfection, and RT-qPCR was performed using
this RNA. LM3 cells treated with Lipofectamine 2000 but not
transfected with ACTA2-AS1:4 siRNA were used as the negative
control (NC) for the expression inhibition assay.
Cell proliferation and viability
assays
The viability of liver cancer cells was evaluated by
assessing cell proliferation using the colorimetric MTS Cell
Proliferation Assay kit (cat. no. 197010; Abcam, Cambridge, MA,
USA), according to the manufacturer's protocol. Briefly, LM3 cells
and the control L02 cells were cultured in 96-well microtiter
plates to ~80% confluence and then incubated with 20 µg MTS
reagents for 2 h at 37°C in a humidified culture chamber supplied
with 5% CO2. The optical density values were measured at
490 nm using a plate reader and used for evaluation of cell
proliferation and viability. For statistical analysis, at least
three replicates were performed.
Cell cycle progression analysis
Cell cycle progression was evaluated by assessing
the percentage of LM3 cells at G1, S, and G2/M stages following
transfection. Briefly, LM3 cells were washed 3 times with PBS,
fixed with 70% ethanol overnight at 20°C, stained with ~300 µl
propidium iodide (Hangzhou MultiSciences(Lianke)Biotech, Co., Ltd.,
Hangzhou, China) in the dark at 37°C for 30 min, and quantitatively
analyzed using a fluorescence-activated cell sorting (FACS)
cytometer (BD Biosciences, Franklin Lakes, USA). Data were analyzed
using Flowjo software, version 7.6 (FlowJo LLC, Ashland, OR, USA).
At least 3 biological repeats of the cell cycle progression assay
were performed for the statistical quantitation of LM3 cell numbers
at different cell cycle stages.
Cell migration and invasion
assays
The migration and invasion capabilities of liver
cancer cells were assessed using the Transwell culture system. For
analysis of cell migration, transfected LM3 cells cultured at a
density of 1×106/ml in serum-free medium were placed in
the upper chamber, while the lower chamber was filled with culture
medium supplemented with 10% FBS. Following culture at 37°C in a
humidified incubator supplied with 5% CO2, cells that
had migrated to the lower chamber were incubated with 4%
paraformaldehyde for 10 min, stained with 1% crystal violet for 10
min at room temperature, and then counted using a cell counter
under an inverted phase-contrast microscope (magnification, ×200).
To detect the invasive capacity of liver cancer cells,
Matrigel® Basement Membrane Matrix (BD Biocoat, BD
Biosciences) was used to coat the Transwell chamber at 37°C for 2
h. The upper chamber was seeded with liver cancer cells, as
aforementioned, and culture medium was placed in the lower chamber.
Similar subsequent experimental steps to fix, stain and count the
invaded cells were performed as aforementioned in the migration
assay protocol. At least 3 biological replicates were performed for
each of the migration and invasion experiments.
Western blot analysis
LM3 cells were washed 3 times with PBS and lysed
using radioimmunoprecipitation (50 mM Tris-base, 150 mM NaCl, 0.1%
SDS, 0.5% sodium seoxycholate, 1% Triton X-100) for total protein
extraction. Following determination of protein concentration via a
Bicinchoninic Acid assay, ~30 µg protein from each group was boiled
for 5 min, separated by 12% SDS-PAGE, and transferred onto a PVDF
membrane. The membrane was then blocked at room temperature for 1 h
with 5% lipid-free milk solution, incubated with primary antibodies
and then washed 3 times with TBS + 0.05% Tween 20 (TBST). The
membrane was then incubated with horseradish peroxide-conjugated
secondary antibodies (1:10,000; cat. no. 2999, Cell Signaling
Technology, Inc., Danvers, MA, USA), washed 3 times with TBST
buffer and finally developed using enhanced chemiluminescent
solution (Amersham; GE Healthcare, Chicago, IL, USA). The following
primary antibodies, purchased from Abcam, were used in this study:
Anti-epithelial cadherin (E-cadherin) antibody (cat. no. ab15148,
diluted 1:1,000); anti-neural cadherin (N-cadherin) antibody (cat.
no. ab18203, diluted 1:1,000); anti-Caspase-3 antibody (cat. no.
ab13847, diluted 1:1,000); anti-Cyclin D1 antibody (cat. no.
ab134175, diluted 1:1,000); anti-MMP2 antibody (cat. no. ab37150,
diluted 1:1,000); anti-GAPDH antibody (cat. no. ab9484, diluted
1:1,000). The relative expression of proteins were quantified using
ImageJ (National Institutes of Health, Bethesda, MD, USA) together
with 64-bit Java 1.8.0_112.
Statistical analysis
Quantitative data in the present study were
statistically analyzed using the SPSS 18.0 software package (SPSS,
Inc., Chicago, IL, USA. A Student's t-test was performed to examine
the difference between two groups. A one-way analysis of variance
followed by Bonferroni method was used to compare the difference
between >2 groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Downregulation of lncRNA expression in
liver cancer
To examine the potential roles of lncRNAs in liver
cancer development and progression, the expression levels of 3
lncRNAs ACTA2-AS1:4, TRPM2-AS1, encoded by the TRPM2 gene, and
MUC20-9 encoded by the mucin 20 gene, were analyzed by qPCR. As
demonstrated in Fig. 1, the
expression level of ACTA2-AS1:4 in cancerous tissues collected from
17 patients with liver cancer was significantly downregulated
compared with the corresponding adjacent non-cancerous tissues
(P<0.05; Fig. 1A). Similarly,
the expression of TRPM2-AS1 was suppressed in liver cancer tissues
(P<0.05; Fig. 1B). The
expression of MUC20-9, however, was increased in liver cancer
tissues (P<0.05; Fig. 1C). The
differentially altered expression of these 3 lncRNAs in liver
cancer tissues suggested that lncRNAs serve distinct roles in liver
cancer initiation and progression. For additional verification, the
expression of ACTA2-AS1:4 in the normal human liver cell line L02
and 5 liver cancer LM3, HepG2, Hep3B, 97H and 97L cell lines were
subsequently analyzed by qPCR; there was a decreased expression of
ACTA2-AS1:4 in all five cancerous cell lines compared with the L02
normal liver control cells (Fig.
1D). The downregulation of ACTA2-AS1:4 in both clinical liver
cancer biopsies and cancer cell lines suggest that ACTA2-AS1:4 is
involved in liver cancer progression.
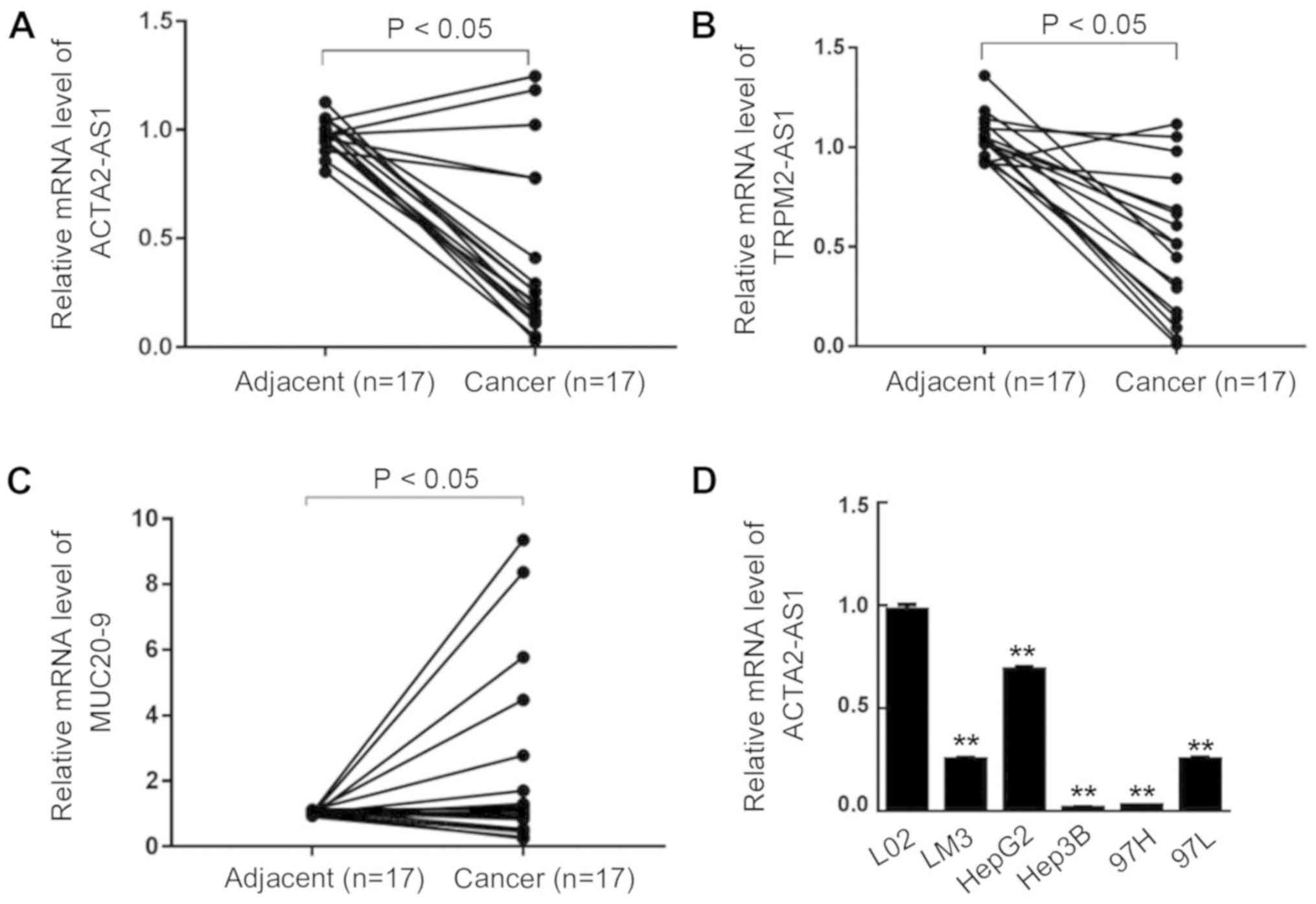 | Figure 1.Long non-coding RNA expression in
hepatocellular carcinoma biopsies and liver cancer cell lines. The
relative expression of (A) ACTA2-AS1:4, (B) TRPM2-AS1 and (C)
MUC20-9 in liver cancer biopsies and adjacent normal hepatic
tissues was determined by RT-qPCR. (D) The relative expression of
ACTA2-AS1:4 in the liver cancer LM3, HepG2, Hep3B, 97H and 97L cell
lines was analyzed by RT-qPCR. The normal liver cell line L02 was
used as a control. **P<0.01 vs. LO2, ACTA2, smooth muscle
α-actin 2; ACTA2-AS1:4, a form of ACTA2-AS1 transcript; TRPM2,
transient receptor potential cation channel subfamily M member 2
reverse; MUC20-9, mucin 20, cell surface associated; RT-qPCR,
reverse transcription quantitative polymerase chain reaction. |
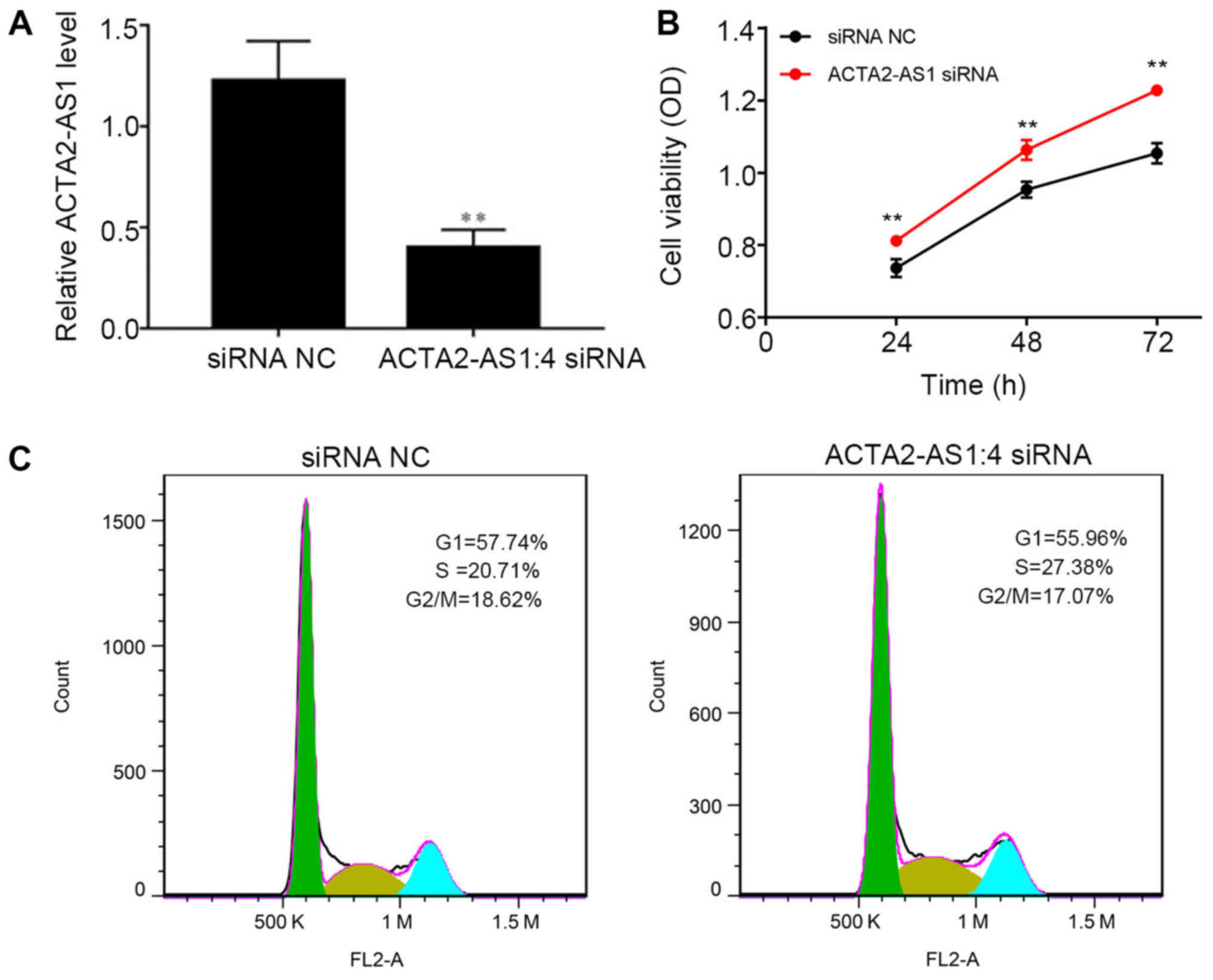
Suppression of ACTA2-AS1:4 promotes
liver cancer cell proliferation
To investigate the potential implication of the
decreased expression of ACTA2-AS1:4 in liver cancer cells compared
with in normal liver cells, the expression of ACTA2-AS1:4 was
knocked down in LM3 cells using siRNA. RT-qPCR analysis confirmed
the suppression of ACTA2-AS1:4 expression in LM3 cells following
transfection (P<0.01; Fig. 2A).
These ACTA2-AS1:4-depleted cells exhibited an increase in cell
viability, suggesting that a low level of ACTA2-AS1:4 promotes
liver cell proliferation in vivo (P<0.01; Fig. 2B). Furthermore, the percentage of
transfected LM3 cells at different stages of the cell cycle was
observed. It was indicated that the percentage of
ACTA2-AS1:4-depleted cells in S stage was 27.38%, which was
markedly increased compared with that in the siRNA NC group
(Fig. 2C). The significant
increase in S-stage LM3 cells additionally validates the role of
ACTA2-AS1:4 in regulating liver cancer proliferation. This suggests
that the low expression of ACTA2-AS1:4 in liver cancer cells causes
an increase in cells in S phase, contributing to liver cancer cell
proliferation.
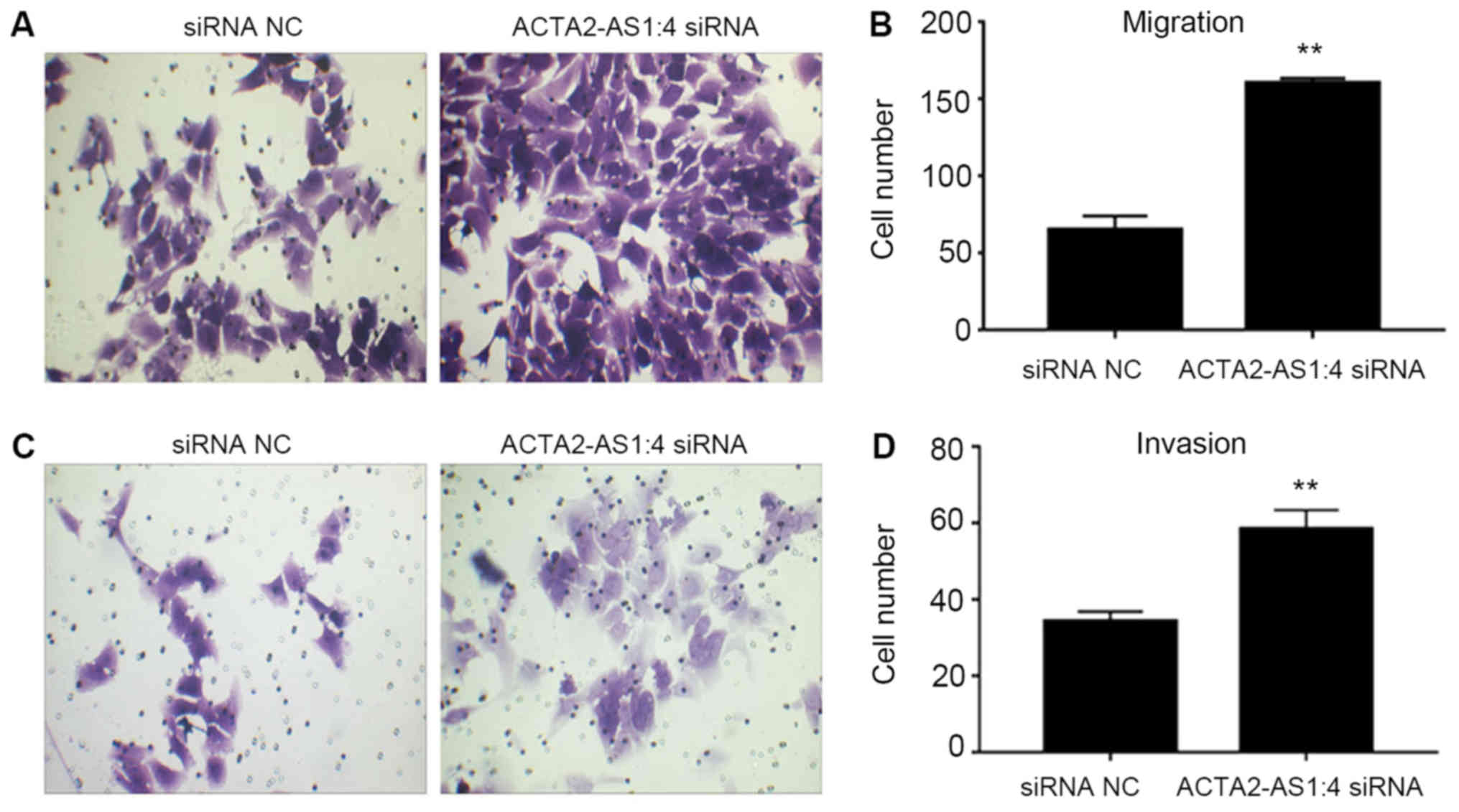
Liver cancer cell migration and
invasion improved by ACTA2-AS1:4 depletion
The migratory and invasive capabilities of LM3 cells
transfected with ACTA2-AS1:4 siRNAs were assessed to provide
additional insight into the cellular properties of liver cancer
cells. The migration of transfected LM3 cells was markedly
increased compared with the NC cells (Fig. 3A and 3B). Similarly, the invasion
of transfected cells was increased compared with the NC (Fig. 3C and 3D). These results suggested
that in addition to the cell proliferation and cell cycle
progression, low expression levels of ACTA2-AS1:4 may also promote
liver cancer cell migration and invasion.
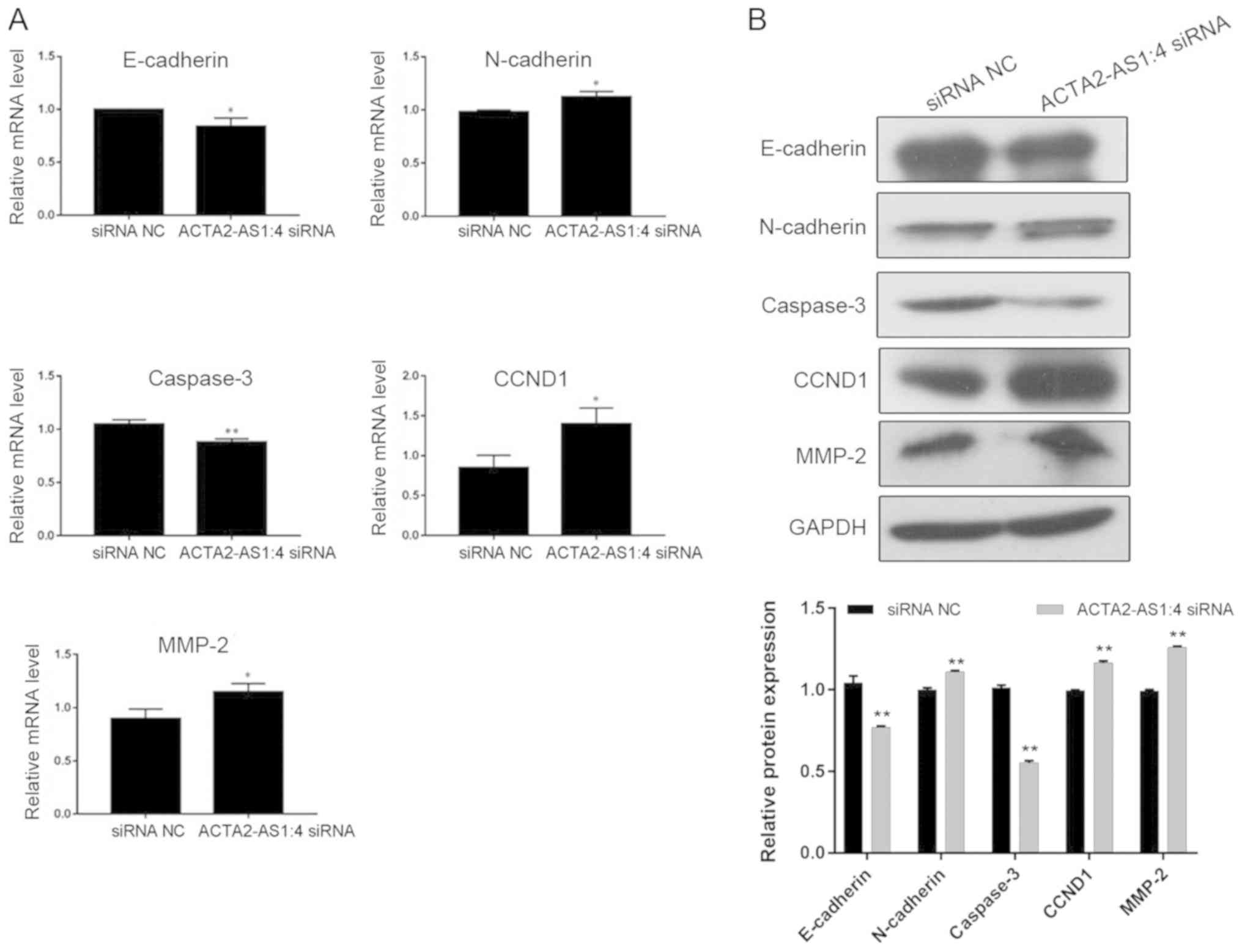
Molecular alterations in transfected
liver cancer cells
To explore the molecular mechanisms mediating the
alteration of multiple cellular properties induced by the
suppression of ACTA2-AS1:4, a number of well-known markers
associated with cell proliferation, apoptosis and migration were
additionally analyzed in the present study. RT-qPCR assays
indicated that the expression of E-cadherin was significantly
decreased alongside the depletion of ACTA2-AS1:4 in LM3 cells,
while N-cadherin expression increased (Fig. 4A). Additionally, expression of
caspase 3, a crucial factor in cell apoptosis, significantly
decreased (Fig. 4A) and cyclin D1
(CCND1) and matrix metalloproteinase 2 (MMP2) expression markedly
increased following transfection of LM3 cells. The alteration of
these gene expression levels was confirmed by the protein abundance
of N-cadherin, CCND1 and MMP2 in transfected cells compared with
non-transfected control cells (Fig.
4B). Taken together, the statistically significant changes in
the expression levels of these key markers suggested that changes
in ACTA2-AS1:4 expression serve as regulators of liver cancer
progression through multiple downstream molecular pathways.
Discussion
Noncoding sequences account for >90% of the whole
genome, according to a bioinformatics analysis following completion
of the human genome project (23)
At present, a number of functional noncoding sequences have been
proved to be involved in various physiological and pathological
processes, which include microRNAs and lncRNAs. Increasing evidence
demonstrates that lncRNAs are key factors during tumor initiation
and progression (12). According
to previous data, the effects of lncRNAs have been implicated in
tumorigenesis and malignant transformation processes; these include
cell proliferation, differentiation, apoptosis, migration and
invasion, which are closely associated with cancer metastasis
(24). A recent study described
the characterization of a novel lncRNA that promotes breast cancer
brain metastases (BCBMs) through activation of the c-Jun N-terminal
kinase/signal transducer and activator of transcript 3 pathway,
providing a potential target in BCBMs that are resistant to
conventional therapies (25) The
identification of MALAT1, HEIH, HULC and HOTAIR as important
regulators of HCC pathology has identified a novel avenue of study
for understanding liver cancer development (26,27).
However, an estimation based on the ENCODE project has suggested
that as many as >28,000 lncRNAs may be encoded by the human
genome (28), suggesting that the
results from the present study pertaining to the roles of lncRNAs
in liver cancer pathology remain limited.
To investigate the role of lncRNAs in liver cancer,
the expression levels of 3 lncRNAs, ACTA2-AS1:4, TRPM2-AS1 and
MUC20-9, known to be implicated in the development of other human
cancer types (21), but not
hepatic carcinoma, were initially examined. It was observed that
ACTA2-AS1:4 and TRPM2-AS1 were upregulated and MUC20-9 was
downregulated in liver cancer tissues and cultured liver cancer
cell lines compared with normal liver tissues and normal cultured
cells. This indicated that lncRNAs may serve a role in liver cancer
pathology and that each lncRNA in liver cancer cells may serve
their own role in cancer progression and/or initiation. Knockdown
of noncoding RNA using specific siRNAs has not been widely used to
study the in vivo function of lncRNAs (12). To validate the cellular effects of
ACTA2-AS1:4 during tumor development and malignant transformation,
ACTA2-AS1:4 expression was additionally inhibited via
siRNA-mediated knockdown in liver cancer cells in the present
study. As expected, the downregulation of ACTA2-AS1:4 markedly
promoted liver cancer cell proliferation, altered cell cycle
progression and resulted in an increase in cellular migration and
invasion, compared with non-transfected controls, suggesting that
lncRNAs are critical in various cellular processes associated with
liver cancer initiation, development and metastasis. Finally, the
pleiotropic cellular functions of ACTA2-AS1:4 in liver cancer cells
were additionally verified by the changes in the expression of
multiple key regulatory proteins involved in different signaling
pathways. In the present study, significant changes in ACTA2-AS1:4
expression and cellular alterations induced by ACTA2-AS1:4
knockdown provided direct evidence of the role of ACTA2-AS1:4 in
promoting complex cellular processes involved in liver cancer
development. These data are similar to other cancer-associated
lncRNAs described previously (12). The RT-qPCR assays performed in the
present study indicated that the expression of E-cadherin was
significantly downregulated alongside the decrease of ACTA2-AS1:4
expression in liver cancer LM3 cells, while the N-cadherin
expression exhibited marked increased; therefore, we hypothesized
that ACTA2-AS1:4 expression may affect the epithelial-mesenchymal
transition of the liver cancer cells. These results also suggested
that ACTA2-AS1:4 may be explored as a novel biomarker for early
liver cancer detection and development of novel targeted therapies
for patients with liver cancer.
Notably, the present study is limited due to unclear
molecular mechanisms underlying the pro-cancer function of
ACTA2-AS1:4 in liver cancer cells, which warrants additional
investigation. The interaction of lncRNAs with functional proteins
serves as one of the major molecular mechanisms by which lncRNAs
perform their physiological and pathological roles (12). For example, the lncRNA breast
cancer anti-estrogen resistance 4 was identified to regulate breast
cancer metastasis through chemokine-induced interaction with two
transcription factors, resulting in p300-dependent histone
acetylation and activation of the Hedgehog/zinc finger protein GLI2
transcriptional program, which finally promotes cancer cell
migration and invasion (29).
Therefore, it is reasonable to hypothesize that future screening of
proteins interacting with ACTA2-AS1:4 during liver cancer
progression may provide more information about underlying molecular
processes. Potentially, and more importantly, future studies of
epigenetic markers may also provide novel perspectives on the
molecular mechanisms involved in the role of ACTA2-AS1:4 during
liver cancer progression, considering the association of lncRNAs
with epigenetic regulation described previously (12).
Furthermore, how ACTA2-AS1:4 expression was
downregulated in liver cancer cells is an additional key topic for
future study, as comprehensive understanding of the regulation of
cancer-associated lncRNAs expression may also provide novel targets
for early cancer diagnosis and treatment. A previous study revealed
that the lncRNA maternally expressed gene 3 may be regulated by CpG
hypomethylation at its promoter region and also histone-H3 lysine-4
trimethylation in response to menin, (30), suggesting that epigenetic
regulation may be a key method of modulating lncRNAs expression
during cancer development and progression. Combined with the
regulation of an epigenetic-associated pathway by lncRNAs, as
aforementioned, it is possible that alterations in ACTA2-AS1:4
expression may affect the induction of pathways, such as the
epigenetic-associated pathway, which in turn may regulate its own
transduction. We propose that such a feedback loop may be an
essential component of the complicated networks involved in liver
cancer pathology.
Taken together, the present study describes the
identification of a novel liver cancer-associated lncRNA
ACTA2-AS1:4:4, which is markedly downregulated in liver cancer
tissues and cell lines. The siRNA-mediated inhibition of
ACTA2-AS1:4:4 promoted liver cancer cell proliferation, cell cycle
progression, migration and invasion, supported by changes in the
expression of key components of tumor-associated signaling
pathways. These data provide a basis for the application of lncRNAs
in the early detection of liver cancer, and potentially the
development of novel treatment strategies.
Acknowledgements
Not applicable.
Funding
Not funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contribution
R-JZ performed the measurements, analyzed and
interpreted data. H-ZL made substantial contributions to conception
and design, and were involved in drafting, revising the manuscript
and interpreting all data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study proceeded following the receipt of written
informed consent from the patients, and tissue collection and
subsequent experiments were approved by the Ethics Committee of the
Sun Yat-sen University and the Fifth Affiliated Hospital of
Guangzhou Medical University.
Patient approval for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sia D, Villanueva A, Friedman SL and
Llovet JM: Liver cancer cell of origin, molecular class, and
effects on patient prognosis. Gastroenterology. 152:745–761. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Castelli G, Pelosi E and Testa U: Liver
cancer: Molecular characterization, clonal evolution and cancer
stem cells. Cancers (Basel). 9:E1272017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ghouri YA, Mian I and Rowe JH: Review of
hepatocellular carcinoma: Epidemiology, etiology, and
carcinogenesis. J Carcinog. 16:12017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Roth GS and Decaens T: Liver
immunotolerance and hepatocellular carcinoma: Patho-physiological
mechanisms and therapeutic perspectives. Eur J Cancer. 87:101–112.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin C and Yang L: Long noncoding RNA in
cancer: Wiring signaling circuitry. Trends Cell Biol. 28:287–301.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cloutier SC, Wang S, Ma WK, Al Husini N,
Dhoondia Z, Ansari A, Pascuzzi PE and Tran EJ: Regulated formation
of lncRNA-DNA hybrids enables faster transcriptional induction and
environmental adaptation. Mol Cell. 62:1482016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yoon JH, Abdelmohsen K and Gorospe M:
Post-transcriptional gene regulation by long noncoding RNA. J Mol
Biol. 425:3723–3730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Szcześniak MW and Makałowska I: lncRNA-RNA
interactions across the human transcriptome. PLoS One.
11:e1503532016. View Article : Google Scholar
|
|
10
|
Li JH, Liu S, Zheng LL, Wu J, Sun WJ, Wang
ZL, Zhou H, Qu LH and Yang JH: Discovery of protein-lncRNA
interactions by integrating large-scale CLIP-seq and RNA-Seq
datasets. Front Bioeng Biotechnol. 2:882015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zampetaki A, Albrecht A and Steinhofel K:
Long Non-coding RNA structure and function: Is there a link? Front
Physiol. 9:12012018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mehra M and Chauhan R: Long noncoding RNAs
as a key player in hepatocellular carcinoma. Biomarkers Cancer.
9:1179299X177373012017. View Article : Google Scholar
|
|
13
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B Virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li H, An J, Wu M, Zheng Q, Gui X, Li T, Pu
H and Lu D: LncRNA HOTAIR promotes human liver cancer stem cell
malignant growth through downregulation of SETD2. Oncotarget.
6:27847–27864. 2015.PubMed/NCBI
|
|
16
|
Conigliaro A, Costa V, Lo Dico A, Saieva
L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M,
et al: CD90+ liver cancer cells modulate endothelial cell phenotype
through the release of exosomes containing H19 lncRNA. Mol Cancer.
14:1552015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu M, Lin Z, Li X, Xin X, An J, Zheng Q,
Yang Y and Lu D: HULC cooperates with MALAT1 to aggravate liver
cancer stem cells growth through telomere repeat-binding factor 2.
Sci Rep. 6:360452016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cooper K and Brown S: ACTA2 mutation and
postpartum hemorrhage: A case report. BMC Med Genet. 18:1432017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Désert R, Mebarki S, Desille M, Sicard M,
Lavergne E, Renaud S, Bergeat D, Sulpice L, Perret C, Turlin B, et
al: ‘Fibrous nests’ in human hepatocellular carcinoma express a
Wnt-induced gene signature associated with poor clinical outcome.
Int J Biochem Cell Biol. 81:195–207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang L, Zhou XF, Pan GF and Zhao JP:
Enhanced expression of long non-coding RNA ZXF1 promoted the
invasion and metastasis in lung adenocarcinoma. Biomed
Pharmacother. 68:401–407. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Edwards YJ, Walter K, Mc-Ewen G, Vavouri
T, Kelly KA, Abnizova I, Woolfe A, Goode DK, Goodson M, North P, et
al: Characterisation of conserved non-coding sequences in
vertebrate genomes using bioinformatics, statistics and functional
studies. Comp Biochem Physiol Part D Genomics Proteomics. 1:46–58.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang S, Liang K, Hu Q, Li P, Song J, Yang
Y, Yao J, Mangala LS, Li C, Yang W, et al: JAK2-binding long
noncoding RNA promotes breast cancer brain metastasis. J Clin
Invest. 127:4498–4515. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large Intervening Non-Coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: Novel insights into hepatocelluar
carcinoma. Cancer Lett. 344:20–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tragante V, Moore JH and Asselbergs FW:
The ENCODE project and perspectives on pathways. Genet Epidemiol.
38:275–280. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xing Z, Lin A, Li C, Liang K, Wang S, Liu
Y, Park PK, Qin L, Wei Y, Hawke DH, et al: lncRNA directs
cooperative epigenetic regulation downstream of chemokine signals.
Cell. 159:1110–1125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Modali SD, Parekh VI, Kebebew E and
Agarwal SK: Epigenetic regulation of the lncRNA MEG3 and its target
c-MET in pancreatic neuroendocrine tumors. Mol Endocrinol.
29:224–237. 2015. View Article : Google Scholar : PubMed/NCBI
|