Introduction
A biological pacemaker, which may be created via
cell therapy or gene therapy, is able to restore cardiac pacing and
conduction function for patients with bradyarrhythmia (1–3).
Cell-based and gene-based approaches have limitations: Gene
therapies may cause gene mutations and are dependent on the
function of the vectors carrying the genes, while cell therapies
carry the risk of immunosuppression and tumor formation from their
implantation site. Hybrid treatments, which combine gene and cell
therapy to create biological pacemakers, have attracted more
interest in recent years (4–6).
Stem cells are the most common source for deriving pacemaker cells,
since they have great potential for proliferation and pluripotent
differentiation. Adipose tissue-derived stem cells (ADSCs) have
become promising candidate cells for biological pacemakers due to
their accessibility, high harvesting efficiency and myocardial
differentiation potential (7,8).
The sinoatrial node (SAN) is a complex structure,
which is regulated by multiple factors and multiple mechanisms.
Transcription factors serve an important role in the formation and
development of the SAN (9–11). T-box 18 (Tbx18) begins to be
expressed during the formation of the sinus horn myocardium after
embryonic day (E) 9.5, and its expression is maintained. It has
been proposed that Tbx18 is essential for the formation and
differentiation of the mesenchyme of the sinus horn. Between E9.5
and E10.5, insulin gene enhancer binding protein 1 (ISL-1), which
encodes a LIM-domain homeodomain transcription factor, becomes
expressed in the dorsal mesenchyme (12). Although the Tbx18-positive and
ISL-1-positive progenitor cells remain spatially and temporally
separate during heart tube elongation, there exists a small area of
overlapping expression from E8.5 at the right lateral side of the
inflow tract. This Tbx18/ISL-1 co-expressing area will form the SAN
of the heart (13). This raises
the possibility that co-expression of ISL-1 and Tbx18 in ADSCs may
induce reactivation of the embryonic development pathway of the
SAN. The current study aimed to explore whether the combination of
ISL-1 and Tbx18 was sufficient to successfully induce ADSCs to
differentiate into pacemaker cells in order build a new biological
pacemaker in vitro.
Materials and methods
Isolation and culture of ADSCs
All experimental procedures were conducted in
accordance with the Institutional Guidelines for the Care and Use
of Laboratory Animals at Wuhan University (Wuhan, China) and
conformed to the National Institutes of Health (NIH) Guide for the
Care and Use of Laboratory Animals (NIH Publications, no. 8023,
revised 1978; Bethesda, MD, USA). The present study was approved by
the Experimental Animal Committee of Wuhan University (no.
WDRM20171015; Wuhan, China). Adult male Sprague-Dawley (SD) rats
(n=4; age, 3–4 weeks; weight, 40–80 g) were purchased from the
Center for Disease Control and Prevention of Hubei Province (SCX
20150018; Hubei, China). All rats were housed at a temperature of
24°C, relative humidity of 60% and change of air 10 times/h prior
to being sacrificed by cervical dislocation. ADSCs were obtained
using a previously described method with modifications (14). Briefly, SD rat inguinal adipose
tissue was digested in a solution containing 0.1% (w/v) collagenase
type I (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C for
45 min with gentle agitation. Following filtering and
centrifugation at 1,000 × g for 10 min at room temperature, the
supernatant was discarded. The pellet was re-suspended in
Dulbecco's modified Eagle's medium (DMEM)/F12 (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) and 1% penicillin/streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were seeded in 6-well plates at a cell
density of 3×105 cells/ml (Corning, Inc., Corning, NY,
USA) and incubated at 37°C with a 5% CO2 atmosphere. The
medium was changed every 2 days.
Transduction of human ISL-1 and Tbx18
lentiviral vectors
Once ADSCs reached 80–90% confluence, adherent cells
were detached with 0.25% trypsin (Genom, Hangzhou, China;
www.genom.com.cn) solution and passaged. Cells of
passages 3–5 were used for all subsequent experiments. Lentiviruses
overexpressing ISL-1 (Ubi-MCS-ISL-1-3flag-SV40-mCherry; GeneChem,
Inc., Shanghai, China) or Tbx18
(pHBLV-CMV-MCS-Tbx18-3flag-EF1-ZSgreen-puro; Hanbio Biotechnology,
Co., Ltd., Shanghai, China) and 8 µg/ml polybrene were mixed
together and added to the culture medium of cells at different
multiplicity of infection (MOI) values (MOI=0, 20, 50, 80 and 100)
when they reached 30% confluence. The culture medium was replaced
following culturing at 37°C in 5% CO2 for 8–12 h.
Fluorescence microscopy (BX51 systems; Olympus Corporation, Tokyo,
Japan) was used to observe the expression of fluorescent protein
after 48 h. The percentage of fluorescent protein-positive cells
was detected by recording the number of fluorescent cells in at
least five different random fields under fluorescence microscopy. A
total of 7 days post-infection, cells were harvested for evaluation
of hISL-1 and hTBX18 expression by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Isolation and culture of neonatal rat
ventricular cardiomyocytes (NRVMs) and co-culture systems
Primary NRVMs were isolated from 1–3 day-old newborn
SD rats (n=100; weight: 5–15 g), which were purchased from the
Center for Disease Control and Prevention of Hubei Province (SCX
20150018; Hubei, China) and sacrificed by cervical dislocation.
Firstly, neonatal rat heart tissues were digested with 0.125%
trypsin at 37°C for 10 min. Subsequently, the precipitate was
repeatedly digested with a solution containing 0.125% trypsin and
0.08% collagenase II (Biosharp, Wuhan, China) 5–8 times, at 37°C,
for 5 min. Subsequent to being filtered and centrifuged at 1,000 ×
g for 10 min, the supernatant was discarded. The pellets were
re-suspended and seeded in 6-well plates at a cell density of
5×105 cells/ml with fresh DMEM/F-12 supplemented with
15% FBS and 1% penicillin/streptomycin. The harvested NRVMs were
purified via their differential adhesion time to isolate the
cardiomyocytes from the fibroblasts and 0.1 mmol/l
bromodeoxyuridine (Sigma-Aldrich; Merck KGaA) to inhibit the
mitosis of fibroblasts (15). For
co-culture experiments, ADSCs (1×105 cells) and NRVMs
(1×106 cells) were mixed and plated at a ratio of 1:10
onto the 6-well plates (16,17).
NRVMs without any treatment were designated the Blank group and
ADSCs transduced with green fluorescent protein (GFP) the GFP
group. The complete culture medium was replaced every 2 days.
RT-qPCR
Total cellular RNA was extracted from the co-culture
systems after 7 days using TRIzol® (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
Isolated RNA was converted into cDNA using the First Strand cDNA
Synthesis kit (Takara Bio, Inc., Otsu, Japan) in a 15 µl mixture as
follows: 25°C for 5 min, 50°C for 15 min, 85°C for 5 min and 4°C
for 10 min. All primers (Table I)
for PCR amplification were synthesized by Invitrogen (Thermo Fisher
Scientific, Inc.). RT-qPCR was performed with standard
SYBR® Premix Ex Taq™ (Takara Bio, Inc.) on a StepOne™
Real-Time PCR (Thermo Fisher Scientific, Inc.) instrument as
follows: Pre-denaturation at 95°C for 5 min, denaturation at 95°C
for 30 sec, annealing at 60°C for 20 sec, and a final extension at
60°C, a total of 40 cycles. The dissolution curve was from 60–95°C
and the temperature was raised by 1°C per 20 sec. Relative gene
expression was calculated using the 2−ΔΔCq method
(18) following normalization to
GAPDH expression. To ensure accuracy, all results were repeated ≥3
times.
 | Table I.Polymerase chain reaction primers used
in this study. |
Table I.
Polymerase chain reaction primers used
in this study.
| Gene | Primer sequences
(5′-3′) | Product size
(bp) |
|---|
| H-ISL-1 |
| 181 |
|
Forward |
GCGGCAATCAGATTCACGAT |
|
|
Reverse |
GCGCATTTGATCCCGTACAA |
|
| H-Tbx18 |
| 173 |
|
Forward |
TCCAAGGTACTGGGAATGGC |
|
|
Reverse |
TGTGCTGTATCGGTTGAGGG |
|
| R-HCN4 |
| 300 |
|
Forward |
GCATCCACGACTACTACGAAC |
|
|
Reverse |
TCTCCTTGTTGCCCTTAGTG |
|
| R-cTnT |
| 202 |
|
Forward |
GCAGGCTCTTCATGCCCAACT |
|
|
Reverse |
CGCTCTGCCCGACGCTTTT |
|
| R-Tbx3 |
| 168 |
|
Forward |
TTACAGCCCGTATTCCATCCC |
|
|
Reverse |
CGGCTATTCAGTTCCGACCC |
|
| R-Nkx2.5 |
| 248 |
|
Forward |
ACGCCCTTCTCAGTCAAAGA |
|
|
Reverse |
TAAAATGTAGGGGCGGTTGG |
|
| R-Cx43 |
| 242 |
|
Forward |
GGCAAGGTGAAAATGAGGGG |
|
|
Reverse |
AAAGCGAGAGACACCAAGGA |
|
| R-Cx45 |
| 112 |
|
Forward |
TTCTGATAATGTATGGTGTC |
|
|
Reverse |
AGTTCCCTCCTTTTACTGTT |
|
| R-GAPDH |
| 253 |
|
Forward |
ACAGCAACAGGGTGGTGGAC |
|
|
Reverse |
TTTGAGGGTGCAGCGAACTT |
|
Western blotting analysis
Cells were harvested using radioimmunoprecipitation
assay lysis buffer (Aspentech, Houston, America). Protein
concentrations were determined with a bicinchoninic acid protein
kit, according to the manufacturer's instructions. Then, 5% of
concentration gel and 10% of separation gel were chosen and the
protein samples (40 µg protein/lane) were mixed with 5X SDS-PAGE
buffer (Aspentech) in a water bath at 95–100°C for 5 min, prior to
being transferred to a polyvinylidene fluoride membrane. Following
blocking with 5% nonfat milk in tris-buffered saline containing
0.05% Tween-20, the membranes were incubated overnight at 4°C with
primary antibodies against potassium/sodium
hyperpolarization-activated cyclic nucleotide-gated channel 4
(HCN4; 1:1,000; cat. no. ab32675; Abcam, Cambridge, MA, USA),
connexin 43 (Cx43; 1:3,000; cat. no. ab11370; Abcam) and connexin
45 (Cx45; 1:1,000; cat. no. AF5108; Affinity Biosciences,
Cincinnati, OH, USA). The membranes were incubated for 30 min at
room temperature with corresponding secondary antibodies:
Horseradish peroxidase (HRP)-conjugated goat anti-rat (1:10,000,
cat. no. AS1093; Aspen Biological) or HRP-conjugated goat
anti-rabbit (1:10,000, cat. no. AS1107; Aspentech). Visualization
was performed using an enhanced chemiluminescence detection kit
(Aspentech), according to the manufacturer's recommendations. The
level of GAPDH (ab37168, Abcam) was used to normalize the signal
intensities. The image collection and densitometry analyses were
performed with Quantity One analysis software (AlphaEaseFC V;
ProteinSimple, San Jose, CA, USA). Experiments were performed ≥3
times to verify the results.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 15 min
at room temperature. Following three washes with PBS for 5 min,
cells were treated with 0.2% Triton (Sigma-Aldrich; Merck KGaA) for
15 min. The cells were incubated with the primary antibodies
against anti-cardiac troponin T (cTnT; cat. no. ab8295; Abcam) or
HCN4 (cat. no. ab32675; Abcam) overnight at 4°C, followed by
incubation with goat anti-mouse Alexa Fluor® 647 (cat
no. A0473; Beyotime Institute of Biotechnology, Haimen, China) or
goat anti-rat Alexa Fluor® 647 (cat. no. ab150167;
Abcam) secondary antibodies for 50 min at room temperature. DAPI
solution was used to stain the nuclei as above. Fluorescent images
were obtained with a Leica-LCS-SP8-STED confocal laser-scanning
microscopy (Leica Microsystems GmbH, Wetzlar, Germany) and were
redistributed using confocal analysis software (LAS-AF-Lite 2.6.1;
Leica Microsystems GmbH). Images were obtained from three random
visual fields in three different samples in order to assess the
levels of cTnT and HCN4 expression.
Electrophysiological recordings
To examine the electrophysiological properties of
individual cells derived from ADSCs, whole cell patch-clamp
technique was used to record the funny current (If). The
Axon patch-clamp amplifier 700B (Molecular Devices, LLC, Sunnyvale,
CA, USA), digital 700AD/DA converter and 6.0.4 pClamp (both from
Axon Instruments; Molecular Devices, LLC) were used for recording
and analyzing the data. Following co-culture for 7 days, the cells
were incubated with 180 µmol/l 2-aminoethoxydiphenyl borate
(Sigma-Aldrich; Merck KGaA) for 15 min to block intercellular
electrical conduction. Cells were perfused with a normal Tyrode's
solution containing NaCl 135 mmol/l, KCl 5.4 mmol/l,
CaCl2 1.8 mmol/l, MgCl2 1 mmol/l, glucose 10
mmol/l, Bacl2 1 mmol/l and
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) 10
mmol/l (pH=7.4) with NaOH, while recordings were under way. The
pipette solution contained: KCl 120 mmol/l, Cacl2 1
mmol/l, MgCl2 5 mmol/l, HEPES 10 mmol/l, EGTA 10 mmol/l
(pH=7.35) with KOH. The impedance of the fluid filled electrode was
5 to 7 MΩ. The Clampex version 6.0 software was used to collect
data. The sampling frequency was 10 kHz and the filtering rate was
5 kHz. Holding potential was set at −30 mV, and a set of
hyperpolarizing voltage steps, ranging between −140 mV and −40 mV
for 1.5 sec with 20 mV increments, was applied to elicit
If. CsCl (4 mmol/l) was administered to detect
alterations in the If.
Statistical analysis
All data are expressed as the mean ± standard
deviation. Significant differences among multiple groups were
analyzed by performing one-way analysis of variance followed by
Tukey's multiple comparison tests. Statistical analyses were
performed using SPSS 19.0 software (IBM Corp., Armonk, NY, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Optimum MOI value for transfection and
expression of the human ISL-1 and Tbx18 genes
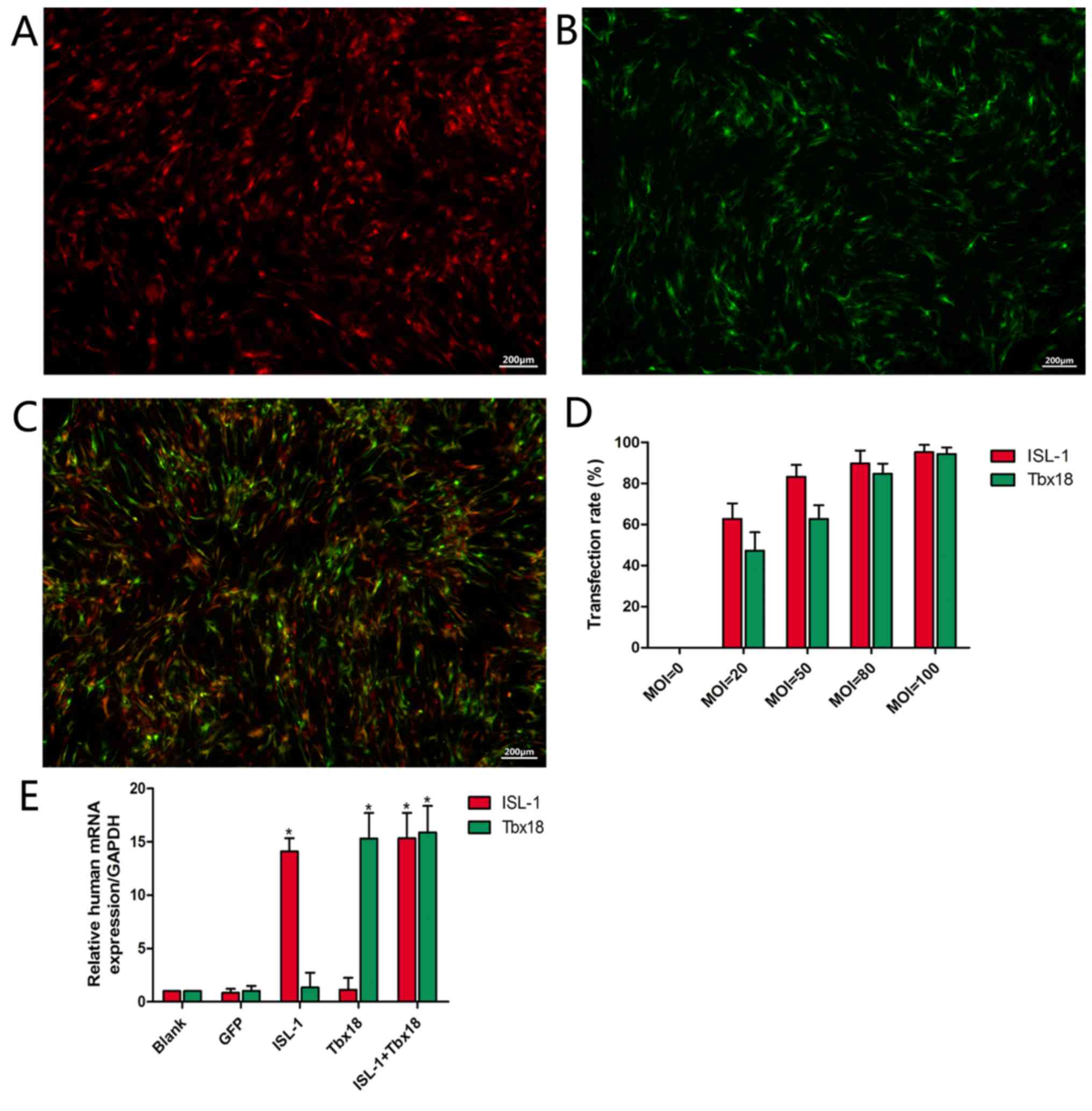
Primary ADSCs became adherent with an irregular
spindle shape and reached 80–90% confluence following culture for
5–7 days. Following passaging, ADSCs were able to reach ~90%
confluence in a homogenous spindle-like fashion within 2–3 days. At
2 days post-transfection, it was observed that infected ADSCs
exhibited red or green fluorescence or both, depending on whether
the cultures were transfected with ISL-1, Tbx18 or both,
respectively (Fig. 1A, B and C,
respectively). The number of fluorescent cells in at least 5
different random fields was observed under fluorescence microscopy.
The fluorescence intensity was increased at higher MOI values
(Fig. 1D). When the MOI value was
20, comparatively few cells were observed. At a MOI value of 50,
the percentage of fluorescent cells was 83.2±5.8% (ISL-1 group) or
62.7±6.7% (Tbx18 group). When the MOI value was 80, the percentage
of fluorescent cells was 89.7±6.3% (ISL-1 group) or 84.7±4.9%
(Tbx18 group). A small number of fluorescent cells appeared as
roundish cells with vacuoles in the cytoplasm, and gradually began
to float. This phenomenon was more apparent at an MOI of 100. With
the increase in MOI values, the cell toxicity increased gradually.
Thus, the MOI values of 50 (ISL-1) and 80 (Tbx18) were considered
optimal and were adopted in the subsequent experiments. Under
fluorescence microscopy, continuous expression of the viral genes
was observed within 7 days. The mRNA expression of ISL-1 and Tbx18
in the ISL-1+Tbx18 group was significantly higher compared with the
green fluorescent protein (GFP) group at day 7 after transfection
(Fig. 1E). These results indicated
that ISL-1 and Tbx18 were stably overexpressed in ADSCs.
Expression analysis of associated
cardiac genes
To evaluate the role of ISL-1 and Tbx18 in the
differentiation of the SAN, the expression statuses of genes that
are known to be important for the formation and function of the SAN
were investigated. The genes detected in the present study include
HCN4, Cx45, Cx43, homeobox protein Nkx-2.5 (Nkx2.5), Tbx3 and cTnT.
As indicated in Fig. 2A, the mRNA
expression levels of HCN4, Cx45, Tbx3 and cTnT were significantly
increased in the ISL-1, Tbx18 and ISL-1+Tbx18 group when compared
with the GFP group (P<0.05). Conversely, the mRNA levels of Cx43
and Nkx2.5 were significantly downregulated in the ISL-1, Tbx18 and
ISL-1+Tbx18 group when compared with the GFP group (P<0.05). In
particular, the expression of genes in the ISL-1+Tbx18 group was
significantly altered in a manner that promoted the differentiation
of ADSCs into SAN-like cells compared with the GFP group
(P<0.01). Furthermore, the protein levels of HCN4, Cx43 and Cx45
were also examined by western blotting (Fig. 2B and C). The results were
identified to be consistent with those from the RT-qPCR assay.
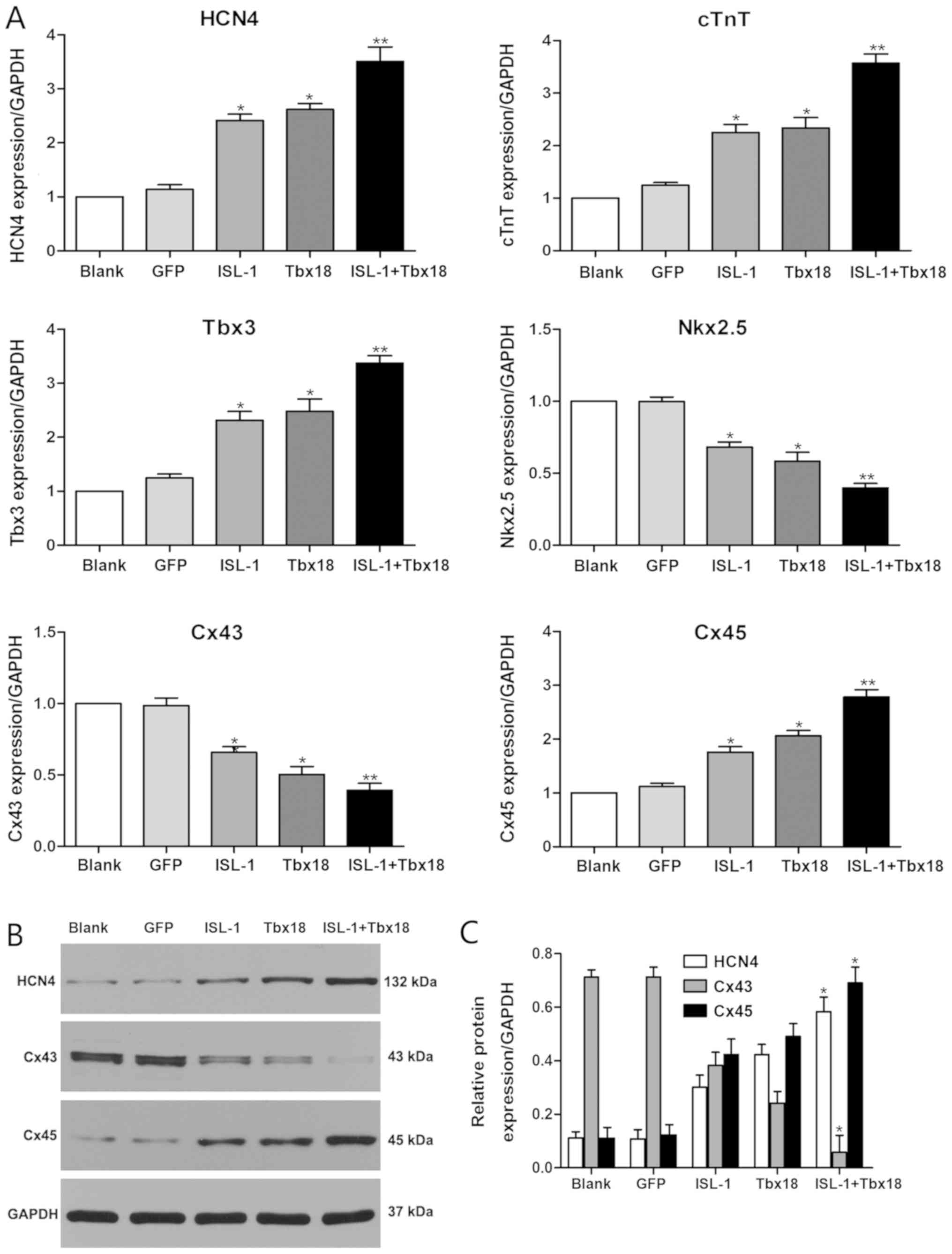 | Figure 2.Expression of associated genes by
RT-qPCR and western blotting after co-culture for 7 days. (A) HCN4,
cTnT, Tbx3, Nkx2.5, Cx43 and Cx45 gene expression was examined
using RT-qPCR. (B) HCN4, Cx43 and Cx45 protein expression examined
using western blotting. (C) Quantitative assessment of HCN4, Cx43
and Cx45 protein levels by integrated optical density analyses.
Similar results were obtained in three independent experiments.
GAPDH was used as the protein control. *P<0.05 and **P<0.01
vs. GFP group. HCN4, hyperpolarization-activated cyclic
nucleotide-gated cation channel 4; cTnT, cardiac troponin T; Tbx3,
T-box 3; Nkx2.5, homeobox protein Nkx-2.5; Cx45, connexin 45; Cx43,
connexin 43; GFP, green fluorescent protein; Tbx18, T-box 18;
ISL-1, insulin gene enhancer binding protein 1; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction |
Combined use of ISL-1+Tbx18
upregulates cardiac marker expression
To evaluate the role of the combination of
ISL-1+Tbx18 in promoting the differentiation of ADSCs into SAN-like
cells, cTnT, a myocardial specific marker, and HCN4, a marker of
SAN function, were detected by immunofluorescence after co-culture
for 7 days. ADSCs were randomly distributed in the culture and had
formed connections with NRVMs. As demonstrated in Fig. 3, the immunofluorescent staining
results revealed that the NRVMs and ADSCs transfected with
ISL-1+Tbx18 were positive for cTnT expression. However, the cTnT
was barely detectable in ADSCs transfected with GFP. Meanwhile,
ADSCs transfected with ISL-1+Tbx18 exhibited abundant positive
staining for HCN4 proteins. However, HCN4 protein expression was
barely detectable in the GFP group. ADSCs transfected with
ISL-1+Tbx18 also exhibited a high percentage of HCN4 and cTnT
compared with cells of the GFP group.
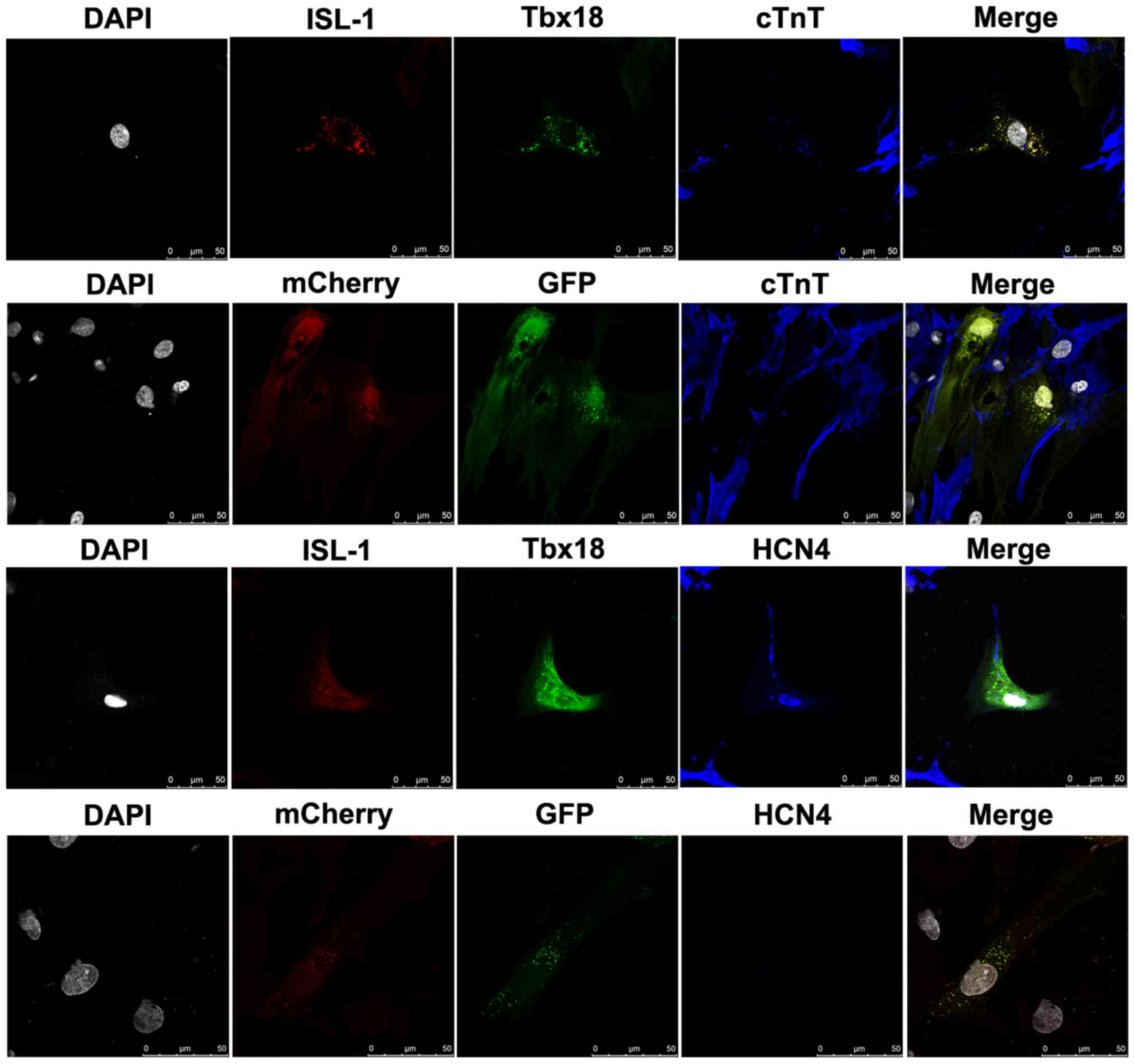 | Figure 3.Cardiac-specific proteins examined by
immunofluorescence staining in differentiated ADSCs after
co-culture for 7 days (magnification, ×400). Grey, nuclei stained
with DAPI; red, ISL-1-ADSCs; green, Tbx18-ADSCs; and blue,
representative positive staining of HCN4 and cTnT. ADSCs, adipose
tissue-derived stem cells; Tbx18, T-box 18; ISL-1, insulin gene
enhancer binding protein 1; HCN4, hyperpolarization-activated
cyclic nucleotide-gated cation channel; cTnT, cardiac troponin T.
Scale bar, 50 µm. |
Combined use of ISL-1+Tbx18 improves
ADSC automaticity
The intracellular electrical activity of
spindle-shaped fluorescent cells was detected via the patch clamp
technique (Fig. 4A). The
If, a key contributor to spontaneous phase 4
depolarization, was recorded in ISL-1, Tbx18 and ISL-1+Tbx18
groups. If in a single beat had a greater inward current
in the ISL-1+Tbx18 group compared with ISL-1 group and Tbx18 group
(Fig. 4B-D). The If
current is sensitive to CsCl, and was therefore inhibited following
the addition of 4 mmol/l of CsCl to the extracellular fluid
(Fig. 4E), thus proving that ADSCs
successfully generated If. In addition, the inward
current resumed when the CsCl was eluted from the extracellular
fluid. The Hyperpolarization-activated inward current was activated
by the hyperpolarizing steps, ranging from −40 mV to −140 mV, and
had clear dependent characteristics of voltage (Fig. 4F).
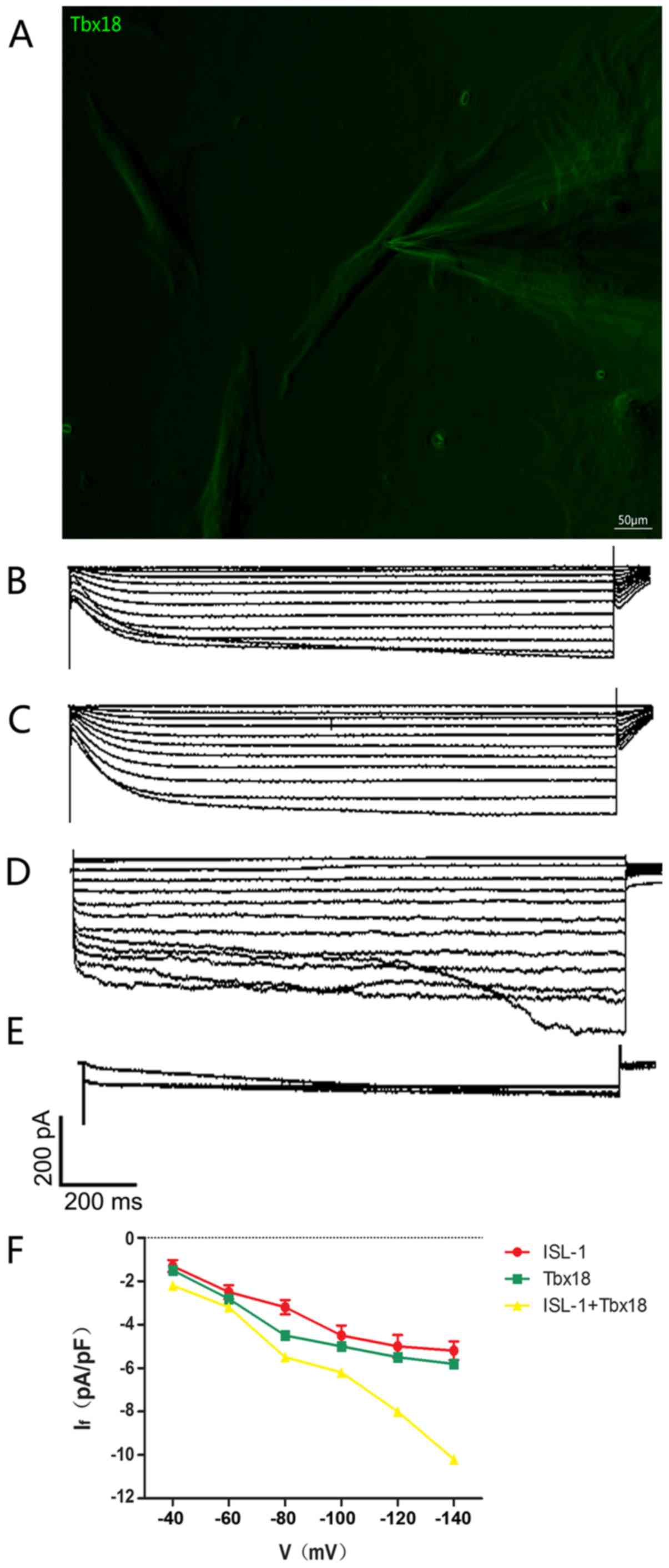 | Figure 4.Single-cell electrophysiology of
ADSCs. (A) Spindle-shaped cells were used for all
electrophysiological recordings (magnification, ×200). (B)
Hyperpolarization-activated inward currents recorded from
ISL-1-ADSCs using the patch clamp technique. (C)
Hyperpolarization-activated inward currents recorded from
Tbx18-ADSCs using the patch clamp technique. (D)
Hyperpolarization-activated inward currents recorded from
ISL-1+Tbx18-ADSCs using the patch clamp technique. (E)
Hyperpolarization-activated inward currents (If) were
blocked by CsCl (4 mmol/l). (F) Current density-voltage association
of ISL-1 (red, n=6), Tbx18 (green, n=6) and ISL-1+Tbx18 (yellow,
n=6) groups. Tbx18, T-box 18; ISL-1, insulin gene enhancer binding
protein 1; ADSCs, adipose tissue-derived stem cells; If,
funny current. Scale bar, 50 µm. |
Discussion
The SAN is a complex structure whose development is
regulated by multiple factors. In recent years, researchers have
aimed to introduce specific genes (19–21)
and transplant pacemaker cells (22–24)
into tissues with a damaged autonomic rhythm or conduction system
in order to repair or replace them. Ideal pacemakers, either
biological or electronic, respond to the complex interactions
between autonomic regulation and physical activity. To the best of
our knowledge, no study on biological pacemaker therapy (gene- or
cell-based) has achieved the target of transforming stem cells or
other cells into pacemaker cells completely. A limiting step in
developing a biological pacemaker is the overall efficiency of
differentiating cells into pacemaker cells. Thus, it is important
to find more effective methods to build biological pacemakers.
There is a small area of Tbx18/ISL-1 co-expression at the right
lateral side of the inflow tract. This area exhibits expression of
the SAN-specific gene Tbx3, but does not express Nkx2.5 (12,13).
This suggests that the co-expression of ISL-1 and Tbx18 coincides
with the formation of the SAN in terms of temporal and spatial
expression. These important characteristics formed the basis of the
current study.
In the present study, ADSCs were successfully
transformed into spontaneously beating cells that exhibited
behavior similar to that of pacemaker cells. The combination of
ISL-1 and Tbx18 resulted in a higher differentiation efficiency
compared with transfections of a single transcription factor. The
following lines of evidence support this result: i) Transduced
cells exhibited the distinctive morphology of pacemaker-like cells;
ii) characteristic mRNA alterations, including upregulation of
HCN4, Tbx3, Cx45 and cTnT, and suppression of Cx43 and Nkx2.5; iii)
characteristic protein modifications, including upregulation of
HCN4 and Cx45, and suppression of Cx43; iv) the location of HCN4
and cTnT expression as identified by immunofluorescence; v) the
If recorded in ISL-1-and Tbx18-transduced cells. The
present study not only confirmed the ability of combined ISL-1 and
Tbx18 to convert ADSCs into pacemaker-like cells, but also
investigated the physiological relevance of this conversion. To the
best of our knowledge, this was the first study to combine ISL-1
and Tbx18 transfection in stem cells. The results indicated that
co-expression of ISL-1 and Tbx18 may significantly improve the
number and quality of ADSCs differentiating into pacemaker-like
cells, and may allow the construction of a new and stable
biological pacemaker in vitro.
Nevertheless, the current study has a number of
limitations. In this experiment, lentiviruses were selected as
vectors, and these are known to integrate into the genome of the
host. Moreover, screenings of stable cell lines or long-term
observations were not performed. Due to the low efficiency of
lentivirus transfection, no further animal experiments were
performed. Compared with the degree of overexpression of exogenous
genes, the mRNA expression levels of associated genes were not high
in this experiment., which presumably was due to differences in
species.
In previous years, researchers attempted to
reprogram adult mammalian fibroblasts into embryonic-like cells via
exposure to genetic transcription factors (25–27).
Induced pluripotent stem cells (iPSCs) have great potential for
differentiation into cells of ventricular, atrial and nodal cell
lineages, and may avoid the limitations of immunosuppression and
tumor formation (28). These
characteristics give iPSCs the capacity for superior integration
into host cardiac tissue in comparison to transplantation with
adult tissue stem cells. The aim of the current study was to use
iPSCs to create a biological pacemaker. There are numerous factors
involved in the embryonic development of the SAN, and further
research is required to determine whether there is a more effective
method than the one described here. It also remains to be
investigated whether the combination of ISL-1 and Tbx18 creates
stable biological pacemaker activity in vivo, in addition to
examining whether better combinations are available. This may help
assessing the safety and validity of this method prior to its
application in patients with SAN dysfunction.
Acknowledgements
The authors are grateful to Wuhan University School
of Basic Medical Science and the Medical Research Center for
Structural Biology for their assistance in conducting the
experiments on the fluorescent images.
Funding
This study was supported by the Fundamental Research
Funds for the Central Universities of China (grant no.
2042015kf0229).
Availability of data and materials
All data generated and analyzed during this study
are included in this published article.
Authors' contributions
JZ and CH conceived and designed the study. JZ
performed the experiments and wrote the paper. CH revised the
manuscript and gave final approval of the version to be published.
All authors read and approved the manuscript.
Ethics approval and consent to
participate
Experimental procedures were conducted in accordance
with the Institutional Guidelines for the Care and Use of
Laboratory Animals at Wuhan University (Wuhan, China) and conformed
to the NIH Guide for the Care and Use of Laboratory Animals (NIH
Publications, no. 8023, revised 1978; Bethesda, MD, USA). The
present study was approved by the Experimental Animal Committee of
Wuhan University (no. WDRM20171015; Wuhan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cho HC and Marbán E: Biological therapies
for cardiac arrhythmias: Can genes and cells replace drugs and
devices? Circ Res. 106:674–685. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boink GJ, Christoffels VM, Robinson RB and
Tan HL: The past, present, and future of pacemaker therapies.
Trends Cardiovasc Med. 25:661–673. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cingolani E, Goldhaber JI and Marbán E:
Next-generation pacemakers: From small devices to biological
pacemakers. Nat Rev Cardiol. 15:139–150. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ionta V, Liang W, Kim EH, Rafie R,
Giacomello A, Marbán E and Cho HC: SHOX2 overexpression favors
differentiation of embryonic stem cells into cardiac pacemaker
cells, improving biological pacing ability. Stem Cell Reports.
4:129–142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saito Y, Nakamura K, Yoshida M, Sugiyama
H, Ohe T, Kurokawa J, Furukawa T, Takano M, Nagase S, Morita H, et
al: Enhancement of spontaneous activity by HCN4 overexpression in
mouse embryonic stem cell-derived cardiomyocytes-a possible
biological pacemaker. PLoS One. 10:e01381932015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang M, Zhang GG, Wang T, Wang X, Tang YH,
Huang H, Barajas-Martinez H, Hu D and Huang CX: TBX18 gene induces
adipose-derived stem cells to differentiate into pacemaker-like
cells in the myocardial microenvironment. Int J Mol Med.
38:1403–1410. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dmitrieva RI, Minullina IR, Bilibina AA,
Tarasova OV, Anisimov SV and Zaritskey AY: Bone marrow- and
subcutaneous adipose tissue-derived mesenchymal stem cells:
Differences and similarities. Cell Cycle. 11:377–383. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Joo HJ, Kim JH and Hong SJ: Adipose
tissue-derived stem cells for myocardial regeneration. Korean Circ
J. 47:151–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wiese C, Grieskamp T, Airik R, Mommersteeg
MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF,
Kispert A and Christoffels VM: Formation of the sinus node head and
differentiation of sinus node myocardium are independently
regulated by Tbx18 and Tbx3. Circ Res. 104:388–397. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Colombo S, de Sena-Tomás C, George V,
Werdich AA, Kapur S, MacRae CA and Targoff KL: Nkx genes establish
second heart field cardiomyocyte progenitors at the arterial pole
and pattern the venous pole through Isl1 repression. Development.
145:2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Blaschke RJ, Hahurij ND, Kuijper S, Just
S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J,
Hardt SE, et al: Targeted mutation reveals essential functions of
the homeodomain transcription factor Shox2 in sinoatrial and
pacemaking development. Circulation. 115:1830–1838. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Christoffels VM, Mommersteeg MT, Trowe MO,
Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler
K, Harvey RP, Moorman AF, et al: Formation of the venous pole of
the heart from an Nkx2-5-negative precursor population requires
Tbx18. Circ Res. 98:1555–1563. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mommersteeg MT, Domínguez JN, Wiese C,
Norden J, de Gier-de Vries C, Burch JB, Kispert A, Brown NA,
Moorman AF and Christoffels VM: The sinus venosus progenitors
separate and diversify from the first and second heart fields early
in development. Cardiovasc Res. 87:92–101. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lopez MJ and Spencer ND: In vitro adult
rat adipose tissue-derived stromal cell isolation and
differentiation. Methods Mol Biol. 702:37–46. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lokuta A, Kirby MS, Gaa ST, Lederer WJ and
Rogers TB: On establishing primary cultures of neonatal rat
ventricular myocytes for analysis over long periods. J Cardiovasc
Electrophysiol. 5:50–62. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu Y, Liu T, Song K, Ning R, Ma X and Cui
Z: ADSCs differentiated into cardiomyocytes in cardiac
microenvironment. Mol Cell Biochem. 324:117–129. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Choi YS, Dusting GJ, Stubbs S,
Arunothayaraj S, Han XL, Collas P, Morrison WA and Dilley RJ:
Differentiation of human adipose-derived stem cells into beating
cardiomyocytes. J Cell Mol Med. 14:878–889. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miake J, Marbán E and Nuss HB: Biological
pacemaker created by gene transfer. Nature. 419:132–133. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bucchi A, Plotnikov AN, Shlapakova I,
Danilo P Jr, Kryukova Y, Qu J, Lu Z, Liu H, Pan Z, Potapova I, et
al: Wild-type and mutant HCN channels in a tandem
biological-electronic cardiac pacemaker. Circulation. 114:992–999.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Boink GJ, Nearing BD, Shlapakova IN, Duan
L, Kryukova Y, Bobkov Y, Tan HL, Cohen IS, Danilo P Jr, Robinson
RB, et al: Ca(2+)-stimulated adenylyl cyclase AC1 generates
efficient biological pacing as single gene therapy and in
combination with HCN2. Circulation. 126:528–536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ruhparwar A, Tebbenjohanns J, Niehaus M,
Mengel M, Irtel T, Kofidis T, Pichlmaier AM and Haverich A:
Transplanted fetal cardiomyocytes as cardiac pacemaker. Eur J
Cardiothorac Surg. 21:853–857. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kehat I, Khimovich L, Caspi O, Gepstein A,
Shofti R, Arbel G, Huber I, Satin J, Itskovitz-Eldor J and Gepstein
L: Electromechanical integration of cardiomyocytes derived from
human embryonic stem cells. Nat Biotechnol. 22:1282–1289. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xue T, Cho HC, Akar FG, Tsang SY, Jones
SP, Marbán E, Tomaselli GF and Li RA: Functional integration of
electrically active cardiac derivatives from genetically engineered
human embryonic stem cells with quiescent recipient ventricular
cardiomyocytes: Insights into the development of cell-based
pacemakers. Circulation. 111:11–20. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu H, Zhu F, Yong J, Zhang P, Hou P, Li
H, Jiang W, Cai J, Liu M, Cui K, et al: Generation of induced
pluripotent stem cells from adult rhesus monkey fibroblasts. Cell
Stem Cell. 3:587–590. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T,
Hao E, Hayek A, Deng H and Ding S: Generation of rat and human
induced pluripotent stem cells by combining genetic reprogramming
and chemical inhibitors. Cell Stem Cell. 4:16–19. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang J, Wilson GF, Soerens AG, Koonce CH,
Yu J, Palecek SP, Thomson JA and Kamp TJ: Functional cardiomyocytes
derived from human induced pluripotent stem cells. Circ Res.
104:e30–e41. 2009. View Article : Google Scholar : PubMed/NCBI
|