Introduction
Serious burn trauma is destructive damage associated
with a hyper-metabolic and catabolic state. Furthermore, it
compromises the immune system by activating systemic inflammation,
frequently leading to suboptimal innate immunity (1). Damage to the skin barrier and
alterations in immunity elevate the susceptibility of patients with
burns to infections in the burn wound, and may cause systemic
bacterial spread, triggering multi-organ failure, sepsis and
mortality (1). A variety of
patient factors, including depth of burn, extent of injury, immune
status and age, combined with microbial factors, including toxin
production, enzymes, number and type of organisms and mobility, are
determinants of the possibility of aggressive burn wound infection.
Burns with a large surface area are associated with infections in
the burn wound leading to multi-organ failure, sepsis and mortality
(2,3). Infection is a frequent trigger of
complications in patients with serious burns. Burns trigger damage
to the barrier that protects the skin. This provides an optimal
opportunity for microbial colonization and likely spread into the
blood and other organs, which may cause sepsis. Therefore,
infection is the principal factor leading to mortality in patients
with burns who survive the early phase of burn shock (4). Williams et al (5) performed a clinical observational
study and demonstrated that sepsis accounted for 47% of the
mortality in patients with burns at a large pediatric burn center.
The study of Mann et al (6)
reported that patients with burns have a high incidence of sepsis
and associated unfavorable outcomes.
The majority of transcribed RNAs are non-coding in
mammalian cells (7). Numerous RNAs
produce small RNAs, including microRNAs (miRs), a well-known family
of small RNAs, in addition to small nucleolar RNAs, tRNA-derived
stress-induced RNAs, transcription initiation RNAs,
Piwi-interacting RNAs and small interfering RNAs (8). Other transcripts [long non-coding
RNAs (lncRNAs)] consist of >200 nucleotides (9). ncRNAs impact the modulation of genes
at all levels via their interactions with proteins, RNA and DNA,
including protein stability, mRNA translation, mRNA turnover,
pre-mRNA splicing, transcription and chromatin remodeling (10).
A previous study demonstrated that
metastasis-associated lung adenocarcinoma transcript-1 (MALAT1) has
a regulatory role in the generation of inflammatory cytokines in
certain human cancers, including lung, liver, breast, cervical and
bladder cancers (11). Yan et
al (12) first demonstrated
the alteration in MALAT1 expression in the retina in patients with
diabetes. It has been demonstrated that the glucose-mediated
upregulation of the inflammatory mediators tumor necrosis factor
(TNF)-α and interleukin (IL)-6 is modulated by lncRNA MALAT1 via
stimulation of serum amyloid A3 (SAA3); it was identified that
early elevation, followed by a reduction in MALAT1, positively
increases the expression of inflammatory ligands, including SAA3,
finally enhancing the expression of inflammatory cytokines,
including TNF-α and IL-6 (13).
Further duration-dependent studies at different phases of diabetes
using animal models (long-term, short-term and acute) may indicate
whether this lncRNA, MALAT1, is an actual initiator of oxidative
stress and inflammation (13). The
flagellin receptor Toll-like receptor 5 (TLR5) is among the most
frequently-triggered pattern recognition receptors (14). Allelic variations in TLR5 may make
infants prone to sepsis, and high-level production of TLR5 in
septic individuals is positively associated with more serious
disease (14,15). Using computational analysis, TLR5
was identified to be a potential target gene of miR-214 in the
present study, and TLR5 has been reported to be functionally
involved in the pathogenesis of post-burn sepsis; the occurrence of
sepsis is important for the prognosis of patients with burns
(16,17). The present study analyzed the
association between the dysregulation of MALAT1, miR-214 or TLR5
and the risk of post-burn sepsis.
Materials and methods
Mouse model of burn injury and burn
wound infection
The Institutional Animal Care and Use Committee of
Baoji Center Hospital (Baoji, China) approved all procedures
involving animals. A total of 48 female C57BL/6 mice at 8–10 weeks
of age (weight, 21.2±2.8 g) were obtained from the Animal Center of
Baoji Center Hospital and used in the present study. Mice were
housed in the appropriate institutional animal care facility, and
acclimatized for 7 days post-arrival at 25°C with 60% humidity. The
acclimatization was performed under a 12/12 light/dark cycle; mice
were provided food and water ad libitum. A well-established
mouse model with full-thickness cutaneous burn injury was utilized
in the present study. Buprenorphine (0.1 mg/kg) was utilized to
treat the mice subcutaneously for 30 min for analgesia prior to the
burn injury. Isoflurane (2.5%) was used for general anesthesia. The
dorsal surface of the mouse was shaved, and 1 ml normal saline was
used to treat the mice subcutaneously to avoid injury to the
underlying tissues. A 35% total surface area scald burn was caused
by exposing the shaved dorsal area to 97–98°C water for 10 sec
using a molded template with a rectangular opening. A total of 2 ml
Ringer's lactate solution (Kelun Pharmaceutical, Chengdu, China)
was utilized to treat the mice immediately for fluid resuscitation
via the intraperitoneal route, and buprenorphine (0.1 mg/kg) was
used to treat the mice secondarily post-burn procedure.
Burn-injured mice were housed in sterile cages individually, and
allowed food and sterile water. Burn wounds were collected for
analysis under anesthesia. Pseudomonas aeruginosa bacteria
were seeded on the surface of the burn wound. A total of
1×108 colony-forming units/ml (200 µl) Pseudomonas
aeruginosa were incubated on the surface of the burn wound 4
days post-burn to cause wound infection. A hematology analyzer was
used to determine blood cell count of the animal groups.
Cell culture and transfection
THP-1 cells were obtained from the Chinese Cell Bank
of the Chinese Academy of Sciences (Shanghai, China). RPMI medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% heat-inactivated fetal bovine serum (Life
Technologies; Thermo Fisher Scientific, Inc.), 100 mg/ml
streptomycin, 50 U/ml penicillin and 2 mM glutamine (Euroclone
Diagnostica S.r.l., Siziano, Italy) was used to maintain the THP-1
cells, in a humidified incubator with 5% CO2 at 37°C. In
order to perform cell transfections, THP-1 cells were cultured in
24-well plates at a concentration of 2×103 cells per
well, and Attractene Transfection Reagent (Qiagen GmbH, Hilden,
Germany; cat. no. 1051531) was used to transfect 15 pmol/ml
miR-675-5p mimic (Life Technologies; Thermo Fisher Scientific,
Inc.; cat. no. 4464066) or miR-675-5p inhibitor (Life Technologies;
Thermo Fisher Scientific, Inc.; cat. no. 4464084) or scramble
control (miR-NC 5′-CTAAGGTTAAGTCGCCCTCGCT-3′) or 15 pmol/ml MALAT1
shRNA into THP-1 cells. Each reaction was performed in triplicate;
analysis was conducted 48 h following transfection.
THP-1 cell polarization and
lipopolysaccharide (LPS) treatment
A total of 20 ng/ml interferon (IFN)-γ (PeproTech,
Inc., Rocky Hill, NJ, USA) with 10 ng/ml LPS (Sigma-Aldrich, St.
Louis, MO) was used to direct PMA-differentiated THP-1 cells to the
M1 phenotype, and 20 ng/ml IL-4 (PeproTech, Inc.) and 20 ng/ml
IL-13 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used to
direct PMA-differentiated THP-1 cells to the M2 phenotype. THP-1
cells were seeded into 96-well plates at a concentration of
2×103 cells per well, and exposure to polarizing
conditions was performed. Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis was performed to
identify the polarized phenotypes. A total of 1 µg/ml LPS was used
to treat THP-1 cells for 24 h when the cells had grown to 80%
confluence.
RNA isolation and RT-qPCR
TRIzol reagent (RNAprep Pure Cell/Bacteria kit;
Tiangen Biotech Co., Ltd., Beijing, China) was used to isolate the
total RNA from THP-1 cells and peripheral blood mononuclear cells
(PBMCs) from blood samples collected from the experimental mice, in
accordance with the manufacturer's protocol. A PrimeScript™ RT
Reagent kit (Perfect Real-Time; Takara Bio, Inc., Otsu, Japan) was
used to perform reverse transcription to obtain cDNA according to
the manufacturer's protocols, using 2 µg cell-extracted RNA or 300
ng total tissue RNA from the burn wound as the template.
LightCycler 480 SYBR Green I Master (Roche, Basel, Switzerland) was
used to perform qPCR analysis to determine the relative expression
of lncMALAT1, miR-214 and TLR5. The qPCR cycling conditions was as
follows: Initial polymerase activation at 95°C for 10 min, followed
by 40 cycles for 15 sec at 95°C and 60 sec at 60°C. The expression
of TLR5 mRNA (F: 5′-CTTCTTGGGCGGCAATAAAC-3′; R:
5′-CCACTAAAGCATAAGTAGGCATC-3′;), lncMALAT1 (F:
5′-GGTAACGATGGTGTCGAGGTC-3′; R: 5′-CCAGCATTACAGTTCTTGAACATG-3′;)
and miR-214 (F: 5′-TTTCCTATGCATATACTTCTTT-3′; R:
5′-CAGTGCGTGTCGTGGAGT-3′;) was calculated relative to the
expression level of the endogenous controls β-actin (F:
5′-ACTCGTCATACTCCTGCT-3′; R: 5′-GAAACTACCTTCAACTCC-3′;) and U6 (F:
5′-CTCGCTTCGGCAGCACA-3′; R: 5′-AACGCTTCACGAATTTGCGT-3′;),
respectively. The value of 2−ΔΔCq (18) was used to describe the relative
gene expression level of TLR5 mRNA and miR-214. Each experiment was
performed at least three times.
Luciferase assay
Target gene prediction was conducted between TRAF3
and miR-214 with online themiRNA database TargetScan release 7.2
(www.targetscan.org). The segment of the
wild-type TLR5 3′ untranslated region (UTR) containing the putative
binding site of miR-214 was synthesized and amplified using PCR
with Applied Biosystems® TaqMan® MicroRNA
Reverse Transcription Kit (Thermo Fisher Scientific, Inc.), and the
PCR products were cloned into the plasmid PGL3-basic (Invitrogen;
Thermo Fisher Scientific, Inc.). A 25 µl reaction system was at
utilized with 5 U Taq Polymerase (Thermo Fisher Scientific, Inc.).
The sequences of the primers used for PCR were as follows: TLR5
forward, 5′-GCTGCAACTGGACCTTTCG-3′ and reverse,
5′-CCCAAACAGTCGAGGATTCAA-3′. The temperature protocol was as
follows: 95°C for 3 min, followed by 30 cycles of 94°C for 40 sec,
56°C for 35 sec and final extension at 72°C for 60 sec. The binding
site of miR-214 located in the TLR5 3′UTR was mutated using a
QuikChange XL mutagenesis kit (Stratagene; Agilent Technologies,
Inc., Santa Clara, CA, USA) to generate a mutant TLR5 3′UTR. The
mutant TLR5 3′UTR was cloned into the plasmid PGL3-basic
(Invitrogen; Thermo Fisher Scientific, Inc.).
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to co-transfect the 2×103
cells per cell THP-1 cells with 0.02 µg control vector containing
Renilla luciferase (Promega Corporation, Madison, WI, USA)
or 0.4 µg firefly luciferase reporter vector containing the
wild-type or mutant TLR5 3′UTR, and miR-214 mimic or miR-NC. After
12 h post-transfection, a dual-luciferase reporter system (Promega
Corporation) was used to detect the luciferase activity of
Renilla and firefly luciferase, according to the
manufacturer's protocol. All tests were performed three times.
Western blot analysis
PBS was used to wash the cells and PBMCs twice, and
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.) was used to lyse the THP-1 cells and PBMCs on ice for 10 min.
A bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Inc.)
was used to measure the concentration of protein, according to the
manufacturer's protocol. Loading buffer was used to resuspend the
protein fractions, which were denatured for 10 min at 100°C.
SDS-PAGE on a 10% gel was used to separate 20 µg/lane proteins,
which were subsequently transferred to polyvinylidene fluoride
membranes (EMD Millipore, Billerica, MA, USA). TBS-Tween 20 buffer
(0.1 % Tween 20) with 5% fat free milk was used to block the
membrane at room temperature for 120 min. The primary rabbit
polyclonal antibody against TLR5 at a dilution of 1:5,000 (cat. no.
DPAB-DC3077, Upstate Biotechnology, Inc., Lake Placid, NY, USA) and
anti-β-actin antibody at a dilution of 1:12,000 (cat. no. 4970S,
Cell Signaling Technology, Inc., Danvers, MA, USA) were utilized to
incubate the membrane at 4°C for 12 h. Subsequently, horseradish
peroxidase-conjugated secondary antibody at a dilution of 1:15,000
(cat. no. AIgYFc-HRP, Upstate Biotechnology, Inc.) was used to
treat the membrane for 2 h at room temperature. Chemiluminescence
(Amersham; GE Healthcare, Chicago, IL, USA) was used to detect the
bands of protein. A total of three independent experiments were
performed.
ELISA analysis
Radioimmunoprecipitation assay buffer (Thermo Fisher
Scientific, Inc.) was used to extract the protein from THP-1 cells
and PBMCs, and the BCA assay (Thermo Fisher Scientific, Inc.) was
used to measure the concentration of protein. Total protein lysates
were isolated from same treatment groups, and these lysates were
also subjected to ELISA assay. Human SAA3 (cat no. CSB-E11836h,
Cusabio Biotech Co., Ltd., Wuhan, China) and ELISA kits were used
to detect the expression levels of TLR5, TNF-α (cat. no. DTA00C,
R&D Systems, Inc., Minneapolis, MN, USA), IL-6 (cat. no. D6050,
R&D Systems, Inc., Minneapolis, MN, USA) and IL-10 (cat. no.
D1000B, R&D Systems, Inc., Minneapolis, MN, USA) based on the
absorbance at 450 nm, which was normalized to the background
absorbance at 568 nm. A total of three independent experiments were
repeated.
Statistical analysis
All data are presented as the mean ± standard error
of the mean. SPSS 19.0 statistical software (IBM Corp., Armonk, NY,
USA) was used to perform the statistical analysis. An unpaired
t-test was used to evaluate the statistical significance of
comparisons between two groups. Two-way analysis of variance and
the Student-Newman-Keuls multiple comparison test were used to
evaluate the statistical significance of comparisons among more
than two groups. P<0.05 was considered to indicate a
statistically significant difference. All experiments were repeated
three times.
Results
Elevation of pro-inflammatory
cytokines caused by burn injury or post-burn infection is
attenuated by treatment with MALAT1
The present study established animal models of burn
injury and post-burn infection, and it was identified that burn
injury caused a significant decrease in the white blood cell count
compared with the sham controls (Fig.
1A) using a hematology analyzer. In addition, post-burn
infection caused a further decrease in white blood cell count
compared with the sham and burn groups, which was eliminated by
treatment with short hairpin (sh)RNA-MALAT1 (shMALAT1) (Fig. 1A). Burn injury caused a significant
increase in TNF-α (Fig. 1B), IL-6
(Fig. 1C) and IL-10 (Fig. 1D), and post-burn infection caused a
further increase in TNF-α (Fig.
1B), IL-6 (Fig. 1C) and IL-10
(Fig. 1D) compared with the burn
groups, which was abrogated by treatment with shMALAT1, indicating
that the burn injury and post-burn sepsis-induced inflammatory
reaction may be attenuated by shMALAT1.
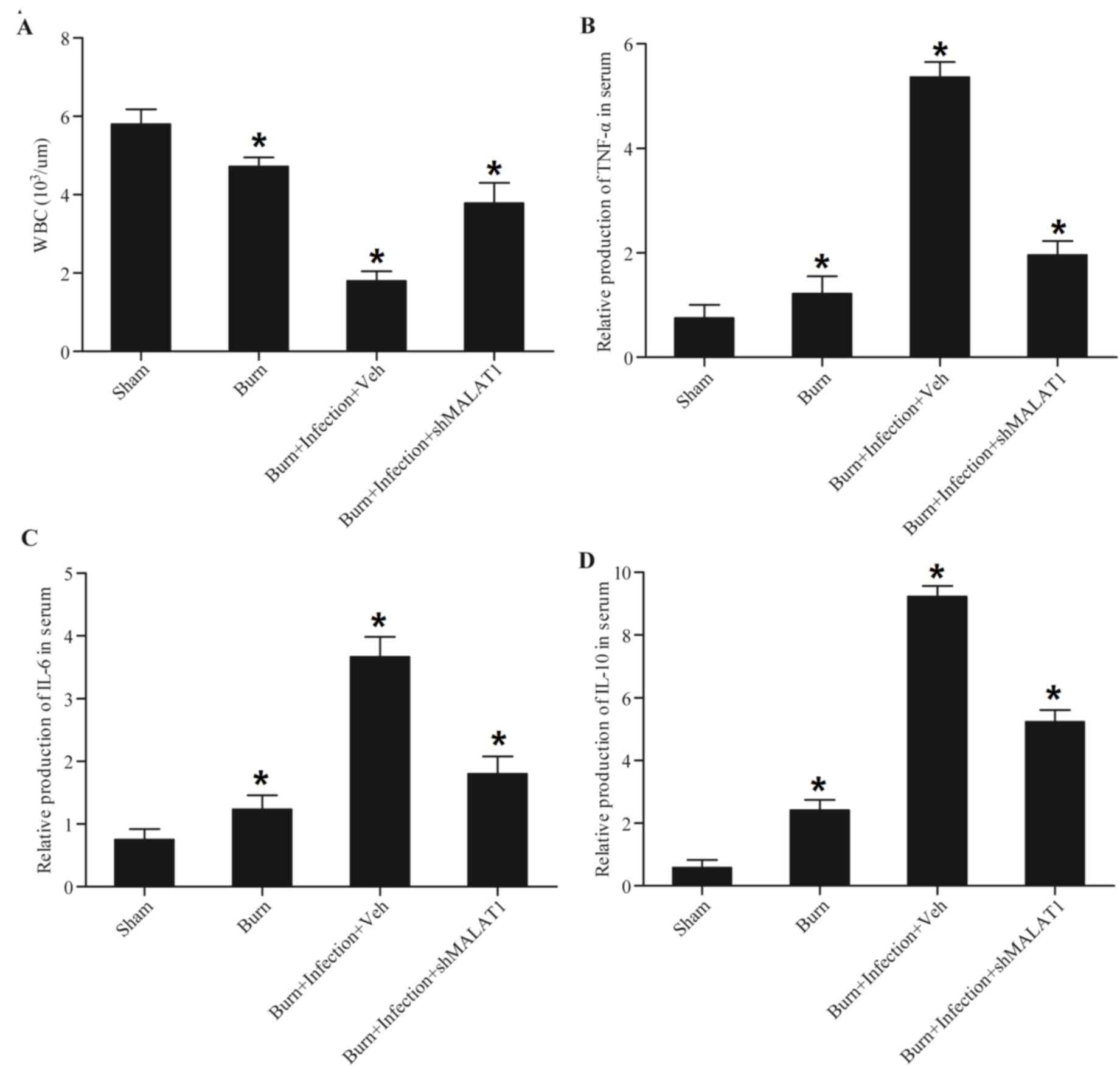 | Figure 1.Elevation of proinflammatory
cytokines caused by burn injury or post-burn infection is
attenuated by treatment with MALAT1. A mouse model of burn injury
and burn wound infection was established, peripheral blood samples
were collected from all mice and monocytes were isolated from the
blood samples. WBC count, TNF-α, IL-6 and IL-10 were detected. Burn
injury induced a decrease in (A) WBC count, and post-burn infection
caused a greater decrease in WBC count, which was abolished
following transfection with shMALAT1. Burn injury induced an
increase in (B) TNF-α, (C) IL-6 and (D) IL-10, and post-burn
infection caused a greater increase in TNF-α, IL-6 and IL-10
expression, which was abolished by transfection with shMALAT1.
*P<0.01 vs. sham. Veh, vehicle; sh, short hairpin; TNF-α, tumor
necrosis factor-α; IL, interleukin; MALAT1, metastasis-associated
lung adenocarcinoma transcript-1; WBC, white blood cell. |
MALAT1, miR-214 and TLR5 are
associated with burn injury and post-burn sepsis
An evident increase in MALAT1 mRNA (Fig. 2A) expression levels was observed in
the burn group compared with the sham control; post-burn infection
caused a further increase in its expression. By contrast, a
significant decrease in miR-214 expression (Fig. 2B) was observed in the burn group
compared with the sham control, and post-burn infection further
enhanced this decrease, which was attenuated by treatment with
shMALAT1. The results for TLR5 expression (Fig. 2C) were comparable to those for
MALAT1. The increase in the expression of TLR5 was attenuated by
treatment with shMALAT1.
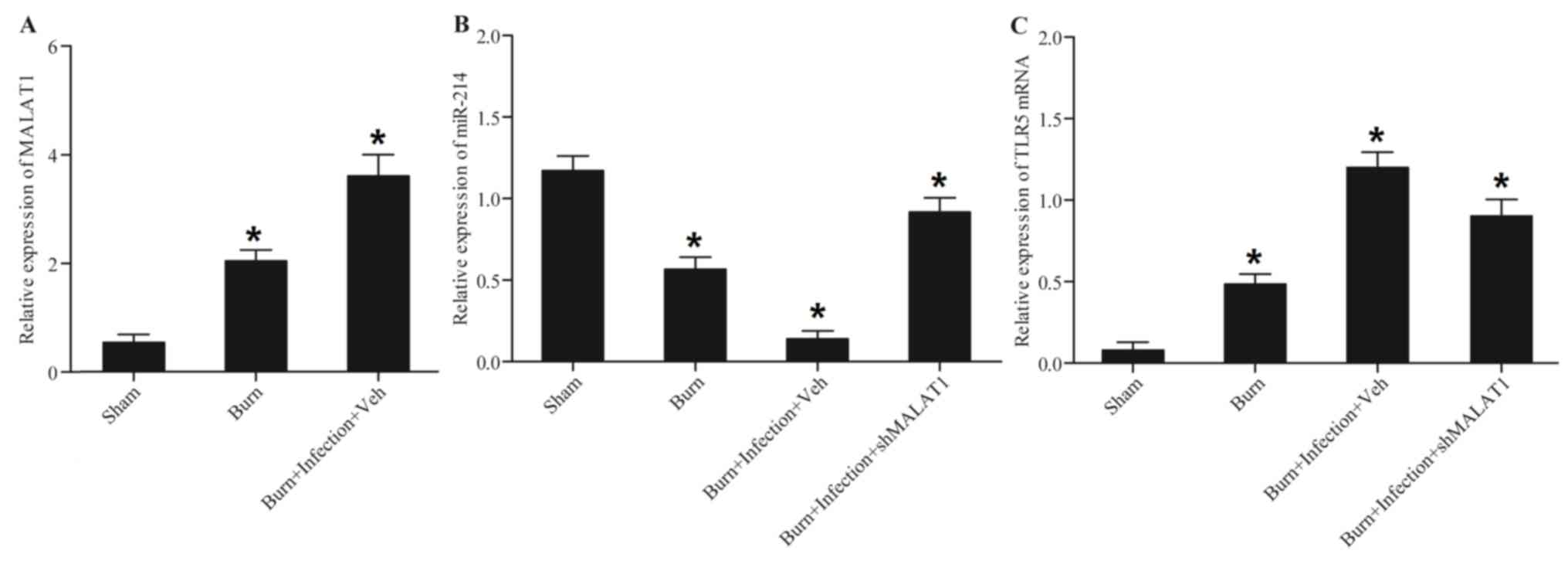 | Figure 2.MALAT1, miR-214 and TLR5 are
associated with burn injury and post-burn sepsis. Peripheral blood
samples were collected from all mice, and monocytes were isolated
from the above blood samples. MALAT1, miR-214 and TLR5 were
detected in the monocytes. (A) The burn and post-burn infection
groups exhibited significantly upregulated MALAT1 expression. (B)
Burn injury inhibited miR-214 expression, and the inhibitory effect
on miR-214 expression was greater in the burn plus infection group;
however, treatment with shMALAT1 restored miR-214 expression. (C)
Burn injury promoted TLR5 expression, and the promoting effect on
TLR5 expression was greater in the burn plus infection group;
however, the effect was abrogated by shMALAT1. *P<0.01 vs. sham.
miR, microRNA; TLR5, Toll-like receptor 5; sh, short hairpin;
MALAT1, metastasis-associated lung adenocarcinoma transcript-1. |
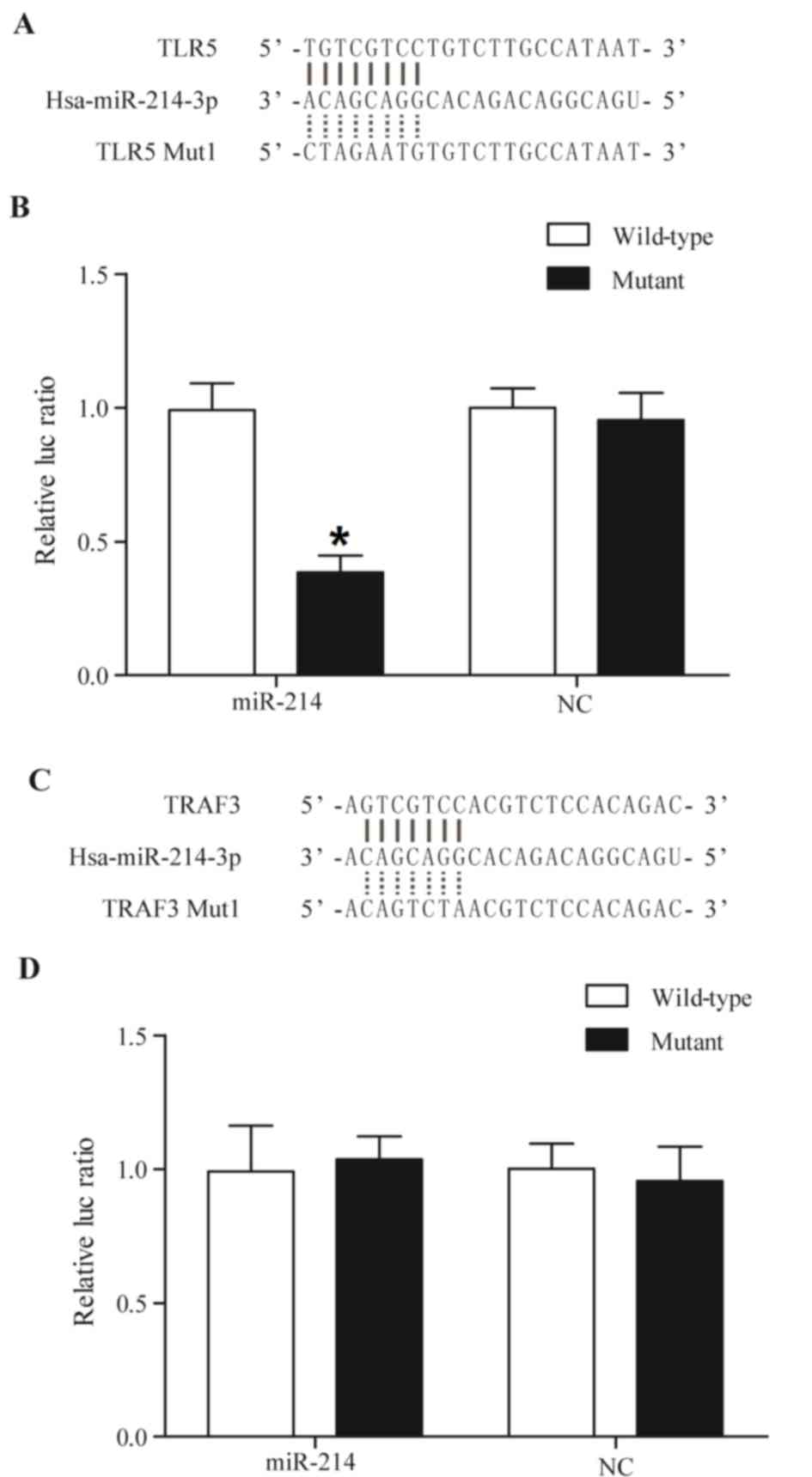
TLR5 is a candidate target gene of
miR-214
TLR5 and TNF receptor-associated factor 3 (TRAF3;
Fig. 3) were predicted to be
target genes of miR-214 using online miRNA databases, including
TargetScan. The putative binding sequences of miR-214 in the
3′-UTRs of TLR5 and TRAF3 are conserved among different species. To
substantiate whether miR214 targets TLR5 and TRAF3 directly,
luciferase reporter vectors were constructed containing a wildtype
or mutant 3′-UTR of TLR5 and TRAF3, in which the sequences
corresponding to the seed sequences were altered. As presented in
Fig. 3B, miR-214 mimics
significantly reduced the luciferase activity of wild-type TLR5
compared with the control, whereas the luciferase activity of
mutant TLR5 (Fig. 3B), wild-type
and mutant TRAF3 (Fig. 3D) in
miR-214-overexpressing cells exhibited no significant difference,
indicating that TLR5 is a virtual target gene of miR-214.
MALAT1 and miR-214 alter the
production of inflammation- associated factors
LPS is considered to be a major toxin in sepsis, and
is the primary cause of the enhanced production of pro-inflammatory
cytokines that is observed in sepsis (19). The expression levels of miR-214,
TLR5, TNF-α, IL-6 and IL-10 were assessed in LPS-treated or
untreated THP-1 cells transfected with MALAT1, anti-miR-214, MALAT1
shRNA and miR-214. As presented in Fig. 4, transfecting with MALAT1 and
anti-miR-214 reduced miR-214 expression (Fig. 4A), while enhancing TLR5 mRNA
(Fig. 4B) and protein (Fig. 4C), TNF-α (Fig. 4D), IL-6 (Fig. 4E) and IL-10 (Fig. 4F) expression compared with the
scramble control.
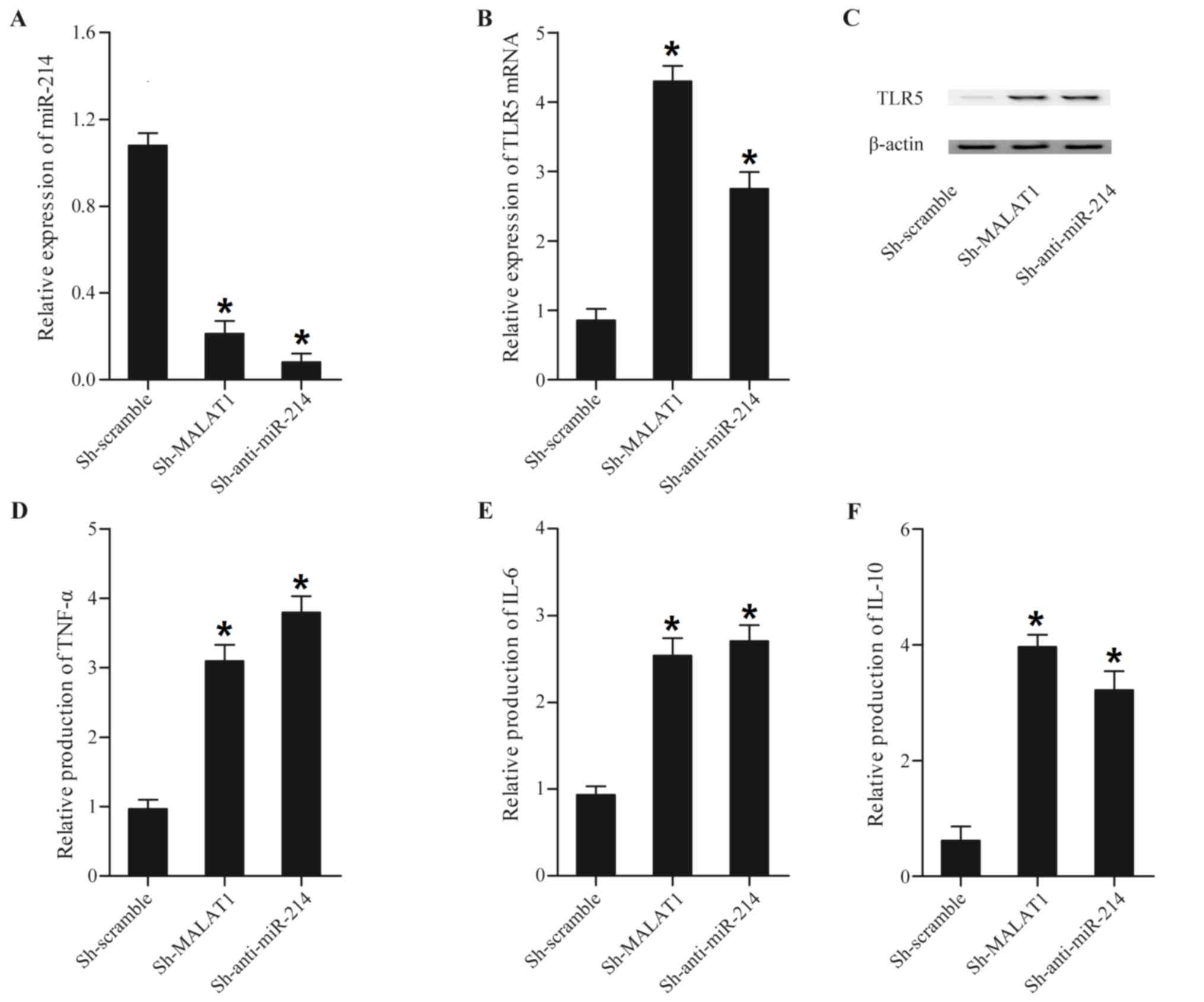 | Figure 4.Expression levels of miR-214, TLR5,
TNF-α, IL-6 and IL-10 in THP-1 cells transfected with Sh-MALAT1 and
Sh-anti-miR-214. Expression levels of miR-214, TLR5, TNF-α, IL-6
and IL-10 in THP-1 cells transfected with Sh-MALAT1 and
Sh-anti-miR-214 were determined using reverse
transcription-quantitative polymerase chain reaction, western blot
and ELISA analyses. (A) Sh-MALAT1 and Sh-anti-miR-214 downregulated
miR-214 expression compared with the scramble control. (B)
Sh-MALAT1 and Sh-anti-miR-214 enhanced TLR5 mRNA expression
compared with the scramble control. (C) TLR5 protein expression was
increased following transfection with constructs containing
Sh-MALAT1 and Sh-anti-miR-214 compared with the scramble control.
(D) The expression of TNF-α in the Sh-MALAT1 and Sh-anti-miR-214
treatment groups was significantly increased compared with the
scramble control. (E) The expression of IL-6 in the Sh-MALAT1 and
Sh-anti-miR-214 treatment groups was upregulated compared with the
scramble control. (F) The expression level of IL-10 in cells was
upregulated subsequent to transfection with Sh-MALAT1 and
Sh-anti-miR-214 compared with the scramble control. *P<0.01 vs.
respective Sh-scramble group. Sh, short hairpin; MALAT1,
metastasis-associated lung adenocarcinoma transcript-1; miR,
microRNA; TLR5, Toll-like receptor 5; TNF-α, tumor necrosis
factor-α; IL, interleukin. |
Cells treated with LPS exhibited a lower level of
miR-214 (Fig. 5A) compared with
that in untreated cells. Furthermore, MALAT1 and anti-miR-214
induced a further decrease in the miR-214 expression level compared
with the scramble control. By contrast, cells treated with LPS
displayed higher levels of TLR5 mRNA (Fig. 5B) and protein (Fig. 5C), TNF-α (Fig. 5D), IL-6 (Fig. 5E) and IL-10 (Fig. 5F) compared with untreated cells.
Furthermore, treatment with MALAT1 and anti-miR-214 induced a
decrease in TLR5 mRNA (Fig. 5B)
and protein (Fig. 5C), TNF-α
(Fig. 5D), IL-6 (Fig. 5E) and IL-10 (Fig. 5F) expression levels compared with
the scramble control. Simultaneously, MALAT1 shRNA and miR-214
significantly promoted the relative expression of miR-214 (Fig. 6A) compared with the scramble
control. MALAT1 shRNA and miR-214 significantly repressed TLR5 mRNA
and protein expression (Fig. 6B),
and transfection of MALAT1 shRNA and miR-214 significantly
downregulated the expression of MALAT1 (Fig. 6C), TNF-α (Fig. 6D), IL-6 (Fig. 6E) and IL-10 (Fig. 6F) compared with the scramble
control.
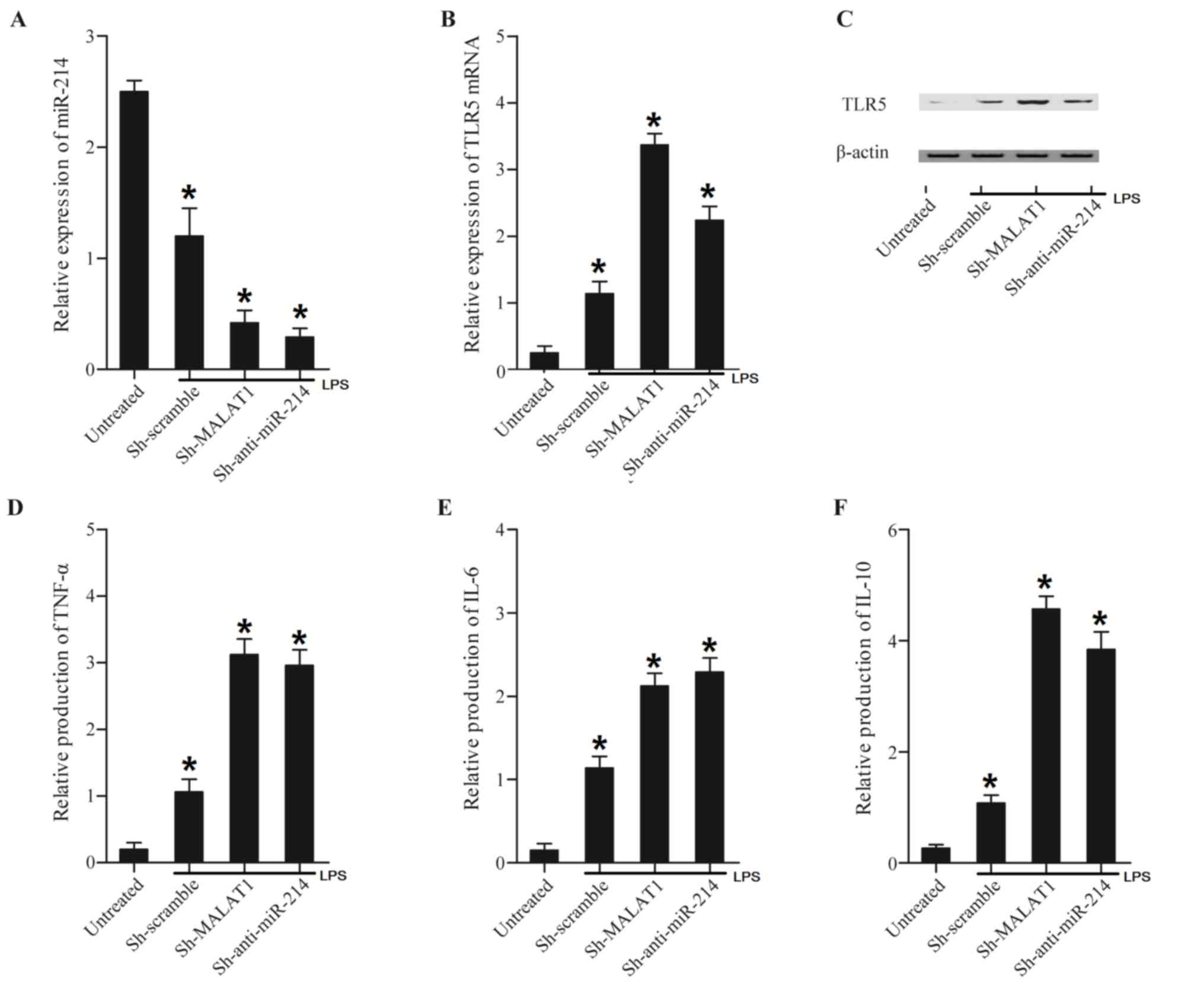 | Figure 5.Treatment of LPS altered expression
levels of miR-214, TLR5, TNF-α, IL-6 and IL-10 in THP-1 cells or
LPS-treated cells transfected with Sh-MALAT1, Sh-anti-miR-214.
Expression levels of miR-214, TLR5, TNF-α, IL-6 and IL-10 in THP-1
cells or LPS-treated cells transfected with Sh-MALAT1,
Sh-anti-miR-214 were determined using reverse
transcription-quantitative polymerase chain reaction, western blot
and ELISA analyses. (A) Treatment with LPS decreased miR-214
expression compared with untreated cells. The miR-214 expression
level in LPS-treated cells transfected with Sh-MALAT1 or
Sh-anti-miR-214 were significantly decreased compared with the
scramble control, and this effect was more marked with
Sh-anti-miR-214. (B) LPS-treated cells displayed a higher level of
TLR5 mRNA compared with untreated cells. TLR5 mRNA expression
levels in LPS-treated cells transfected with Sh-MALAT1 or
Sh-anti-miR-214 were significantly increased compared with the
scramble control, and this effect was more marked with
Sh-anti-miR-214. (C) LPS increased the TLR5 protein expression
level compared with that in untreated cells. TLR5 protein
expression in LPS-treated cells transfected with Sh-MALAT1 and
Sh-anti-miR-214 were further increased. (D) TNF-α expression in
LPS-treated cells was significantly increased compared with
LPS-untreated cells, while TNF-α expression in LPS-treated cells
transfected with Sh-MALAT1 and Sh-anti-miR-214 were further
increased compared with LPS-treated cells. (E) LPS-treated cells
exhibited a higher level of IL-6 compared with LPS-untreated cells,
while IL-6 expression in LPS-treated cells transfected with
Sh-MALAT1 and Sh-anti-miR-214 was further increased compared with
LPS-treated cells. (F) IL-10 expression in LPS-treated cells was
significantly increased compared with LPS-untreated cells, while
IL-10 expression in LPS-treated cells transfected with Sh-MALAT1
and Sh-anti-miR-214 was further increased compared with LPS-treated
cells. *P<0.01 vs. respective untreated group. LPS,
lipopolysaccharide; miR, microRNA; Sh, short hairpin; MALAT1,
metastasis-associated lung adenocarcinoma transcript-1; TLR5,
Toll-like receptor 5; TNF-α, tumor necrosis factor-α; IL,
interleukin. |
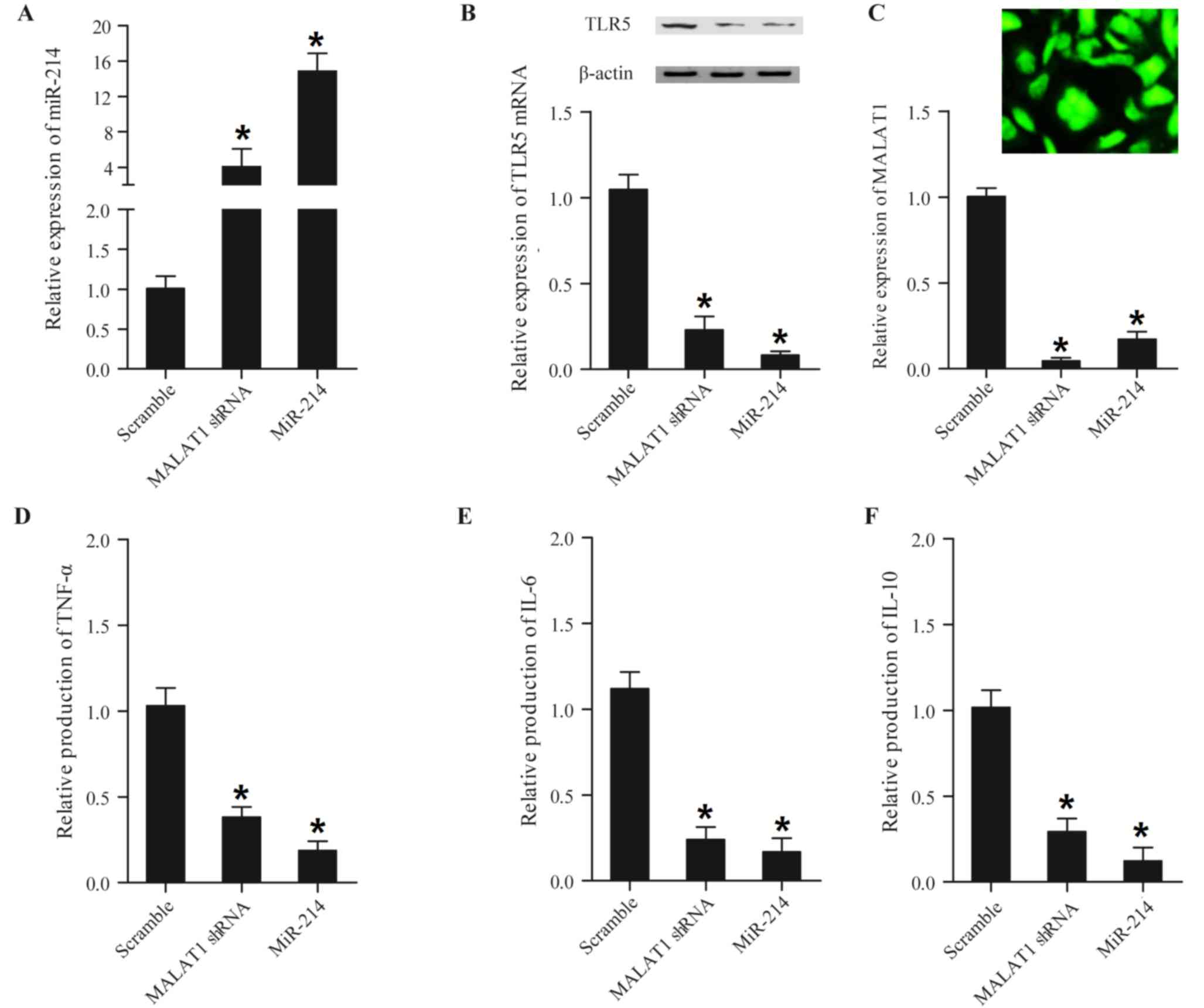 | Figure 6.Expression levels of miR-214, TLR5,
TNF-α, IL-6 and IL-10 in mouse peripheral blood mononuclear cells
transfected with MALAT1 shRNA and miR-214. Expression levels of
miR-214, TLR5, TNF-α, IL-6 and IL-10 in mouse peripheral blood
mononuclear cells transfected with MALAT1 shRNA and miR-214 were
determined using reverse transcription-quantitative polymerase
chain reaction, western blot and ELISA analyses. (A) shRNA and
miR-214 enhanced miR-214 expression compared with the scramble
control, and the effect was strongest with miR-214. (B) TLR5 mRNA
expression (lower panel) and protein expression (upper panel) in
the MALAT1 shRNA or miR-214 group was increased compared with the
scramble control, and the effect was strongest in the miR-214
group. (C) MALAT1 shRNA was successfully transfected into the cells
(upper panel) and MALAT1 shRNA and miR-214 reduced MALAT expression
(lower panel) compared with the scramble control; this effect was
strongest with miR-214 (scale bar: 10 µm). (D) Expression of TNF-α
in the MALAT1 shRNA and miR-214 treatment groups was significantly
decreased compared with the scramble control, and this effect was
strongest in the miR-214 group. (E) Expression of IL-6 in the
miR-214 treatment group was slightly decreased compared with the
MALAT1 shRNA group, and expression in the two treatment groups was
significantly decreased compared with the scramble control. (F)
Expression of IL-10 in the MALAT1 shRNA and miR-214 treatment
groups significantly decreased compared with the scramble control,
and this effect was more marked in the miR-214 group. *P<0.01
vs. respective scramble group. miR, microRNA; Sh, short hairpin;
MALAT1, metastasis-associated lung adenocarcinoma transcript-1;
TLR5, Toll-like receptor 5; TNF-α, tumor necrosis factor-α; IL,
interleukin. |
LPS-treated cells exhibited a lower expression level
of miR-214 (Fig. 7A) and higher
expression levels of TLR5 mRNA (Fig. 7B) and protein (Fig. 7C), TNF-α (Fig. 7D), IL-6 (Fig. 7E) and IL-10 (Fig. 7F) compared with untreated cells.
Furthermore, MALAT1 shRNA and miR-214 induced a further decrease in
miR-214 expression compared with the scramble control, and the
suppression effects of miR-214 on TLR5, TNF-α, IL-6 and IL-10
expression were further enhanced. Finally, MALAT1 shRNA and miR-214
a induced significant decrease in TLR5 mRNA (Fig. 7B) and protein (Fig. 7C), TNF-α (Fig. 7D), IL-6 (Fig. 7E) and IL-10 (Fig. 7F) expression levels compared with
the scramble control, and the inhibitory effects of miR-214 on the
expression levels of the above factors appeared to be increased in
cells treated with LPS compared with those in cells not treated
with LPS. The results collectively suggested that MALAT1 promoted
TLR5, TNF-α, IL-6 and IL-10 expression by inhibiting miR-214.
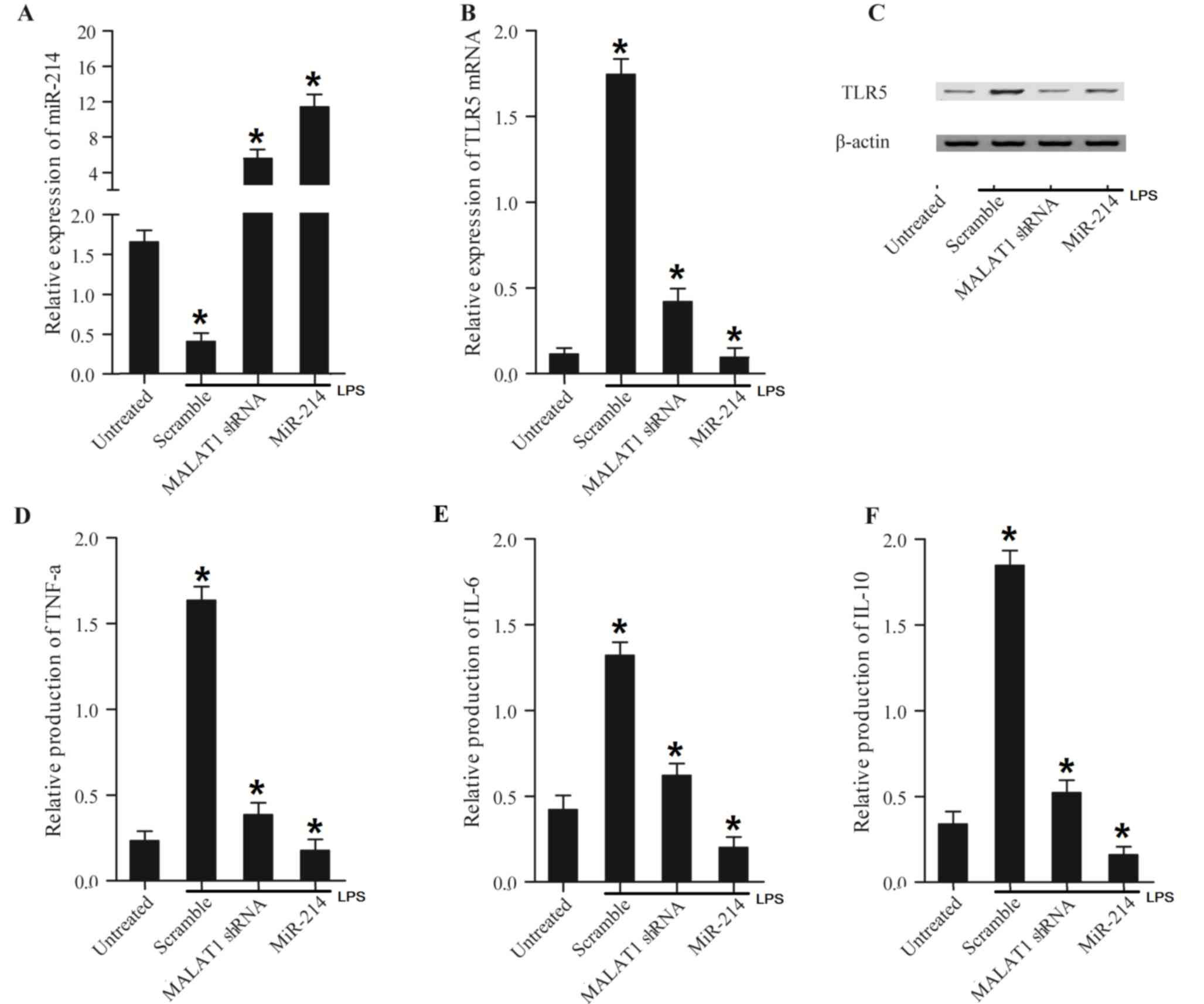 | Figure 7.Treatment of LPS altered expression
levels of miR-214, TLR5, TNF-α, IL-6 and IL-10 in mouse peripheral
blood mononuclear cells transfected with MALAT1 shRNA and miR-214.
Levels of miR-214, TLR5, TNF-α, IL-6 and IL-10 in mouse PBMC cells
or LPS-treated cell transfected with MALAT1 shRNA and miR-214 were
determined using reverse transcription-quantitative polymerase
chain reaction, western blot and ELISA analyses. (A) LPS
downregulated miR-214 expression compared with LPS untreated cells.
miR-214 expression levels in LPS-treated cells transfected with
MALAT1 shRNA and miR-214 were increased compared with the scramble
control cells. (B) LPS increased TLR5 mRNA expression compared with
cells not treated with LPS, while the TLR5 mRNA expression level in
LPS-treated cells decreased following transfection with MALAT1
shRNA and miR-214 compared with the scramble group. (C) TLR5
protein expression levels in LPS-treated cells transfected with
MALAT1 shRNA and miR-214 were decreased compared with the scramble
group. (D) LPS-treated cells displayed an increased level of TNF-α
compared with LPS-untreated cells, while TNF-α expression levels in
LPS-treated cells transfected with MALAT1 shRNA and miR-214 were
inhibited compared with the scramble group. TNF-α expression the in
miR-214 group was further decreased compared with the MALAT1 shRNA
group. (E) LPS-treated cells exhibited a higher level of IL-6
compared with LPS-untreated cells, while IL-6 expression in
LPS-treated cells transfected with MALAT1 shRNA and miR-214 was
notably reduced compared with the scramble group. The effect of
miR-214 was stronger compared with that of MALAT1 shRNA. (F) IL-10
expression in LPS-treated cells was significantly increased
compared with LPS-untreated cells, while IL-10 expression in
LPS-treated cells transfected with MALAT1 shRNA and miR-214 was
repressed compared with the scramble group. This effect was more
marked in the miR-214 group compared with the MALAT1 shRNA group.
*P<0.01 vs. respective untreated group. LPS, lipopolysaccharide;
miR, microRNA; Sh, short hairpin; MALAT1, metastasis-associated
lung adenocarcinoma transcript-1; TLR5, Toll-like receptor 5;
TNF-α, tumor necrosis factor-α; IL, interleukin. |
Discussion
The discovery of MALAT1, and other lncRNAs including
p50-associated COX-2 extragenic RNA, Lethe and nuclear factor
(NF)-κB interacting lncRNA, as novel NF-κB modulators, has added
complexity to the understanding of the regulation of this crucial
transcription factor, which is important in inflammation and
immunity (20). It has been
demonstrated that MALAT1 is necessary for the close control of
inflammatory reactions triggered by LPS, revealing the potential
role of MALAT1 in modulating inflammation and innate immunity
(20). Recently, a report
identified that the glucose-mediated increase in the inflammatory
mediators IL-6 and TNF-α is modulated by MALAT1 via stimulation of
SAA3 (13). It has been reported
that MALAT1, which was initially regarded as a prognostic marker
for non-small cell lung cancer, may serve a role in regulating the
expression of inflammatory mediators (20). As a highly conserved long
non-coding RNA, in addition to nuclear-enriched transcript 2,
MALAT1 is deregulated in a range of cancer types. MALAT1 was
identified as a prognostic marker for lung cancer metastasis, and
has also has been associated with a number of other human cancer
types (11). MALAT1 functions as
an oncogene in esophageal squamous cell carcinoma (ESCC), and it
modulates ESCC growth by adjusting the ataxia telangiectasia
mutated serine/threonine kinase-checkpoint kinase 2 pathway
(21). In the present study, it
was identified that burn injury decreased the white blood cell
count, and the suppressive effect of post-burn infection on WBCs
was much stronger, although this was eliminated by shMALAT1. Burn
injury up-regulated MALAT1, TLR5, TNF-α, IL-6 and IL-10 expression
levels, while the suppressive effects of post-burn infection on
TNF-α, IL-6 and IL-10 expression levels were stronger, and were
abrogated by shMALAT1. Furthermore, the present study investigated
whether MALAT1, miR-214 and TLR5 were associated with burn injury
and post-burn sepsis, and revealed that MALAT1 promoted burn
severity by inhibiting miR-214 production and further enhancing
TLR5 expression.
Previous reports have demonstrated the role of
miR-214 in inflammatory responses in vitro. Li et al
(22) demonstrated that miR-214 is
markedly increased in human monocytes and THP-1 cells following
treatment with advanced glycation end products, compromises
phosphatase and tensin homolog (PTEN) expression and postpones the
apoptosis of THP-1 cells. Jindra et al (23) demonstrated that miR-214 is also
markedly elevated in active T cells and is important for enhancing
the proliferation and activation of T cells by affecting PTEN. It
has been indicated that overexpression of miR-214 markedly enhances
IL-6 and TNF-α expression in bone marrow derived macrophages
(24). Gonzales et al
(25) reported that miR-214
expression was substantially increased, which is consistent with
our findings, in an inflammatory macrophage model. Increasing
evidence demonstrates that a large portion of lncRNAs are able to
function as miRNA sponges by sharing common miRNA response
elements, impacting regulation by suppressing available miRNA
activity at the post-transcriptional level. It has been
demonstrated the rs619586A >G single-nucleotide polymorphism is
able to trigger the capture of miR-214 by MALAT1 (15). It has also been revealed that the
miR-214 directly targets MALAT1 with the rs619586G allele and
inhibits the expression of MALAT1, and miR-214 inhibitors reverse
this inhibitory effect; miR-214 inhibitors may release the
inhibitory effect of endogenous miR-214 on MALAT1 with the
rs619586G allele, and hence trigger increased expression of MALAT1
(15). In the present study, it
was demonstrated that miR-214 directly targets the TLR5 3′UTR, and
miR-214 markedly inhibited the luciferase activity of the wild-type
TLR5 3′UTR and had no effect on the luciferase activity of the
mutant TLR5 3′UTR.
TLR5 is present on various cell types including mast
cells, macrophages, monocytes, neutrophils and epithelial cells,
and is the receptor for the bacterial structural protein flagellin
(26). Flagellin signaling through
TLR5 depends on myeloid differentiation primary response protein 88
and interleukin-1 receptor-associated kinase 1 and subsequent
stimulation of the phosphatidylinositol 3-kinase, mitogen activated
protein kinase and NF-κB pathways (27,28).
Similar to other TLR agonists, flagellin has been demonstrated to
trigger dendritic cell maturation and stimulation, thereby
enhancing lymphocyte movement to secondary lymphoid sites (29,30).
Others have demonstrated that spontaneous neutrophil apoptosis is
delayed by flagellin via mediation of induced myeloid leukemia cell
differentiation protein Mcl-1 and suppression of caspase 3
(31). The present study detected
the expression levels of miR-214, TLR5, TNF-α, IL-6 and IL-10 in
cells transfected with Sh-MALAT1, Sh-anti-miR-214 and miR-214 using
RT-qPCR, western blotting and ELISA, and observed that MALAT1 and
anti-miR-214 inhibited miR-214 expression while enhancing TLR5,
TNF-α, IL-6 and IL-10 expression in LPS-untreated or treated cells.
MALAT1 shRNA and miR-214 upregulated miR-214 expression while
inhibiting TLR5, TNF-α, IL-6 and IL-10 expression in LPS-untreated
or treated cells. It was also revealed that LPS caused an evident
decrease in miR-214 expression, while inducing a marked increase on
TLR5, TNF-α, IL-6 and IL-10 levels. Sepsis has been referred to a
systemic inflammation syndrome induced by infection (32). It is currently considered that the
majority of patients who succumb to sepsis have unaddressed immune
inhibition septic foci (33).
Complex mechanisms have been implicated in cell signaling and
bacterial sensing, which are regulated during sepsis (34). Immunity to bacterial infection is
induced when pattern recognition receptors on phagocytes, including
TLRs, identify the pathogen-associated molecular patters produced
by the infecting microorganism (35). TLR5 detects bacterial flagellin. It
is produced in various cell types, including epithelial cells,
dendritic cells, mastocytes and monocytes (35). For instance, in a model of
spontaneous colitis, TLR5-depleted mice exhibit more severe colitis
(36). The production is TLR5 was
elevated among septic patients compared with healthy subjects;
additionally, TLR5 production was reported to be reduced in septic
patients after 7 days at follow-up (14). As for TLR production, TLR5 was
elevated in neutrophils and monocytes from septic patients and TLR4
and TLR2 were reduced in neutrophils in the surviving patients at
follow-up (14).
There are certain limitations to this study: i) The
experiments were only performed in animals and cultured cells, thus
further study on human subject is required to confirm the results;
and ii) the spectrum of inflammatory cytokines has not been
comprehensively studied, and thus certain cytokines, including
NF-κB and its associated components, should be considered in future
studies.
In conclusion, the results of the present study
demonstrated the association between the dysregulation of
MALAT1/miR-214/TLR5 and the risk of post-burn sepsis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FG made substantial contributions to the design of
the present study, analyzed the data and reviewed the manuscript.
RC collected the data and prepared the manuscript; YX made
substantial contributions to the acquisition and analysis of data;
QZ made substantial contributions to the collected of data and
literature, and prepared the manuscript; HG made substantial
contributions to the design of the present study and reviewed the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The Institutional Animal Care and Use Committee of
Baoji Center Hospital (Baoji, China) approved all procedures
involving animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Murray CK: Infections in burns. J Trauma.
62:S732007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pruitt BA Jr, McManus AT, Kim SH and
Goodwin CW: Burn wound infections: Current status. World J Surg.
22:135–145. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Appelgren P, Bjornhagen V, Bragderyd K,
Jonsson CE and Ransjo U: A prospective study of infections in burn
patients. Burns. 28:39–46. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Branski LK, Al-Mousawi A, Rivero H,
Jeschke MG, Sanford AP and Herndon DN: Emerging infections in
burns. Surgical Infections. 10:389–397. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Williams FN, Herndon DN, Hawkins HK, Lee
JO, Cox RA, Kulp GA, Finnerty CC, Chinkes DL and Jeschke MG: The
leading causes of death after burn injury in a single pediatric
burn center. Crit Care. 13:R1832009. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mann EA, Baun MM, Meininger JC and Wade
CE: Comparison of mortality associated with sepsis in the burn,
trauma, and general intensive care unit patient: A systematic
review of the literature. Shock. 37:4–16. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al
RIKEN Genome Exploration Research Group and Genome Science Group
(Genome Network Project Core Group), : The transcriptional
landscape of the mammalian genome. Science. 309:1559–1563. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gutschner T, Hammerle M and Diederichs S:
MALAT1-A paradigm for long noncoding RNA function in cancer. J Mol
Med (Berl). 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan B, Tao ZF, Li XM, Zhang H, Yao J and
Jiang Q: Aberrant expression of long noncoding RNAs in early
diabetic retinopathy. Invest Ophthalmol Vis Sci. 55:941–951. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Puthanveetil P, Chen S, Feng B, Gautam A
and Chakrabarti S: Long non-coding RNA MALAT1 regulates
hyperglycaemia induced inflammatory process in the endothelial
cells. J Cell Mol Med. 19:1418–1425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Silva SC, Baggio-Zappia GL, Brunialti MK,
Assuncao MS, Azevedo LC, Machado FR and Salomao R: Evaluation of
toll-like, chemokine, and integrin receptors on monocytes and
neutrophils from peripheral blood of septic patients and their
correlation with clinical outcomes. Braz J Med Biol Res.
47:384–393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhuo Y, Zeng Q, Zhang P, Li G, Xie Q and
Cheng Y: Functional polymorphism of lncRNA MALAT1 contributes to
pulmonary arterial hypertension susceptibility in Chinese people.
Clin Chem Lab Med. 55:38–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lahiri R, Derwa Y, Bashir Z, Giles E,
Torrance HD, Owen HC, O'Dwyer MJ, O'Brien A, Stagg AJ, Bhattacharya
S, et al: Systemic inflammatory response syndrome after major
abdominal surgery predicted by early upregulation of TLR4 and TLR5.
Ann Surg. 263:1028–1037. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Zhou Q, Wang Y, Liu Z, Dong M, Wang
Y, Li X and Hu D: Negative pressure wound therapy decreases
mortality in a murine model of burn-wound sepsis involving
pseudomonas aeruginosa infection. PLoS One. 9:e904942014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iwagaki A, Porro M and Pollack M:
Influence of synthetic antiendotoxin peptides on lipopolysaccharide
(LPS) recognition and LPS-induced proinflammatory cytokine
responses by cells expressing membrane-bound CD14. Infect Immun.
68:1655–1663. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao G, Su Z, Song D, Mao Y and Mao X: The
long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced
inflammatory response through its interaction with NF-kappaB. FEBS
Lett. 590:2884–2895. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y
and Yang K: Up-regulation of long noncoding RNA MALAT1 contributes
to proliferation and metastasis in esophageal squamous cell
carcinoma. J Exp Clin Cancer Res. 34:72015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li LM, Hou DX, Guo YL, Yang JW, Liu Y,
Zhang CY and Zen K: Role of microRNA-214-targeting phosphatase and
tensin homolog in advanced glycation end product-induced apoptosis
delay in monocytes. J Immunol. 186:2552–2560. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jindra PT, Bagley J, Godwin JG and
Iacomini J: Costimulation- dependent expression of microRNA-214
increases the ability of T cells to proliferate by targeting Pten.
J Immunol. 185:990–997. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao L, Liu YW, Yang T, Gan L, Yang N, Dai
SS and He F: The mutual regulation between miR-214 and A2AR
signaling plays an important role in inflammatory response. Cell
Signal. 27:2026–2034. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gonzales JN, Gorshkov B, Varn MN, Zemskova
MA, Zemskov EA, Sridhar S, Lucas R and Verin AD: Protective effect
of adenosine receptors against lipopolysaccharide-induced acute
lung injury. Am J Physiol Lung Cell Mol Physiol. 306:L497–L507.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Honko AN and Mizel SB: Effects of
flagellin on innate and adaptive immunity. Immunol Res. 33:83–101.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moors MA, Li L and Mizel SB: Activation of
interleukin-1 receptor-associated kinase by gram-negative
flagellin. Infect Immun. 69:4424–4429. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gohda J, Matsumura T and Inoue J: Cutting
edge: TNFR- associated factor (TRAF) 6 is essential for
MyD88-dependent pathway but not toll/IL-1 receptor
domain-containing adaptor-inducing IFN-beta (TRIF)-dependent
pathway in TLR signaling. J Immunol. 173:2913–2917. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Means TK, Hayashi F, Smith KD, Aderem A
and Luster AD: The Toll-like receptor 5 stimulus bacterial
flagellin induces maturation and chemokine production in human
dendritic cells. J Immunol. 170:5165–5175. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mizel SB and Bates JT: Flagellin as an
adjuvant: Cellular mechanisms and potential. J Immunol.
185:5677–5682. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Salamone GV, Petracca Y, Fuxman Bass JI,
Rumbo M, Nahmod KA, Gabelloni ML, Vermeulen ME, Matteo MJ, Geffner
JR and Trevani AS: Flagellin delays spontaneous human neutrophil
apoptosis. Lab Invest. 90:1049–1059. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bone RC, Sibbald WJ and Sprung CL: The
ACCP-SCCM consensus conference on sepsis and organ failure. Chest.
101:1481–1483. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Boomer JS, To K, Chang KC, Takasu O,
Osborne DF, Walton AH, Bricker TL, Jarman SD II, Kreisel D,
Krupnick AS, et al: Immunosuppression in patients who die of sepsis
and multiple organ failure. JAMA. 306:2594–2605. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Salomao R, Brunialti MK, Rapozo MM,
Baggio-Zappia GL, Galanos C and Freudenberg M: Bacterial sensing,
cell signaling, and modulation of the immune response during
sepsis. Shock. 38:227–242. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Janeway CA Jr and Medzhitov R: Innate
immune recognition. Annu Rev Immunol. 20:197–216. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vijay-Kumar M, Sanders CJ, Taylor RT,
Kumar A, Aitken JD, Sitaraman SV, Neish AS, Uematsu S, Akira S,
Williams IR and Gewirtz AT: Deletion of TLR5 results in spontaneous
colitis in mice. J Clin Invest. 117:3909–3921. 2007.PubMed/NCBI
|