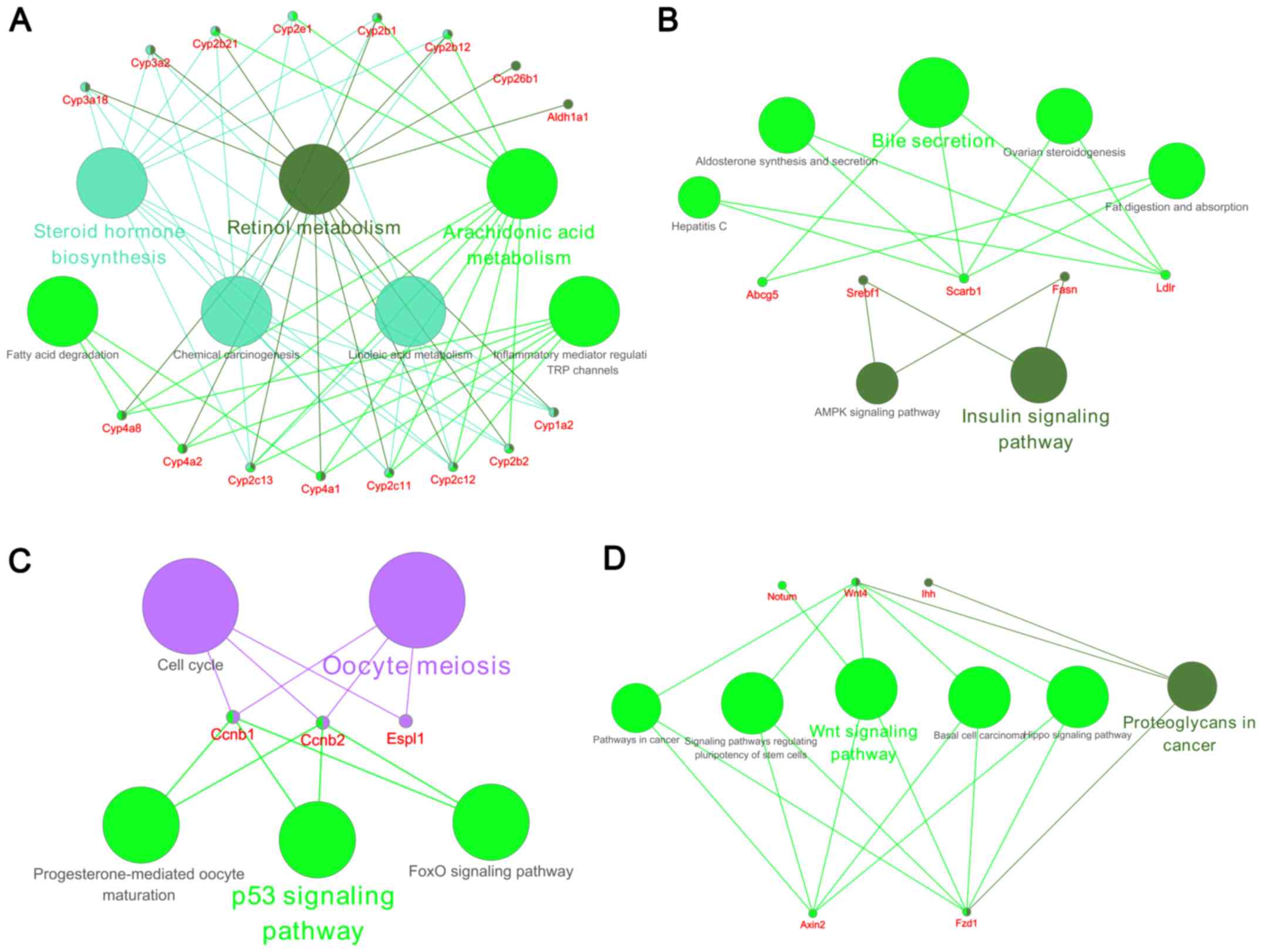
|
1
|
Marrone J, Soria LR, Danielli M, Lehmann
GL, Larocca MC and Marinelli RA: Hepatic gene transfer of human
aquaporin-1 improves bile salt secretory failure in rats with
estrogen-induced cholestasis. Hepatology. 64:535–548. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schreiber AJ and Simon FR:
Estrogen-induced cholestasis: Clues to pathogenesis and treatment.
Hepatology. 3:607–613. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stieger B, Fattinger K, Madon J,
Kullak-Ublick GA and Meier PJ: Drug- and estrogen-induced
cholestasis through inhibition of the hepatocellular bile salt
export pump (Bsep) of rat liver. Gastroenterology. 118:422–430.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hillman SC, Stokes-Lampard H and Kilby MD:
Intrahepatic cholestasis of pregnancy. BMJ. 353:i12362016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kondrackiene J and Kupcinskas L:
Intrahepatic cholestasis of pregnancy-current achievements and
unsolved problems. World J Gastroenterol. 14:5781–5788. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Lu L, Victor DW, Xin Y and Xuan
S: Ursodeoxycholic acid and S-adenosylmethionine for the treatment
of intrahepatic cholestasis of pregnancy: A meta-analysis. Hepat
Mon. 16:e385582016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zucchetti AE, Barosso IR, Boaglio AC,
Basiglio CL, Miszczuk G, Larocca MC, Ruiz ML, Davio CA, Roma MG,
Crocenzi FA and Pozzi EJ: G-protein-coupled receptor 30/adenylyl
cyclase/protein kinase A pathway is involved in estradiol
17ss-D-glucuronide-induced cholestasis. Hepatology. 59:1016–1029.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reyes H and Simon FR: Intrahepatic
cholestasis of pregnancy: An estrogen-related disease. Semin Liver
Dis. 13:289–301. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Copple BL, Jaeschke H and Klaassen CD:
Oxidative stress and the pathogenesis of cholestasis. Semin Liver
Dis. 30:195–204. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ozler A, Ucmak D, Evsen MS, Kaplan I,
Elbey B, Arica M and Kaya M: Immune mechanisms and the role of
oxidative stress in intrahepatic cholestasis of pregnancy. Cent Eur
J Immunol. 39:198–202. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sanhal CY, Daglar K, Kara O, Yilmaz ZV,
Turkmen GG, Erel O, Uygur D and Yucel A: An alternative method for
measuring oxidative stress in intrahepatic cholestasis of
pregnancy: Thiol/disulphide homeostasis. J Matern Fetal Neonatal
Med. 31:1477–1482. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Biberoglu E, Kirbas A, Daglar K, Kara O,
Karabulut E, Yakut HI and Danisman N: Role of inflammation in
intrahepatic cholestasis of pregnancy. J Obstet Gynaecol Res.
42:252–257. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kirbas A, Biberoglu E, Ersoy AO, Dikmen
AU, Koca C, Erdinc S, Uygur D, Caglar T and Biberoglu K: The role
of interleukin-17 in intrahepatic cholestasis of pregnancy. J
Matern Fetal Neonatal Med. 29:977–981. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu D, Wu T, Zhang CL, Xu YJ, Chang MJ, Li
XP and Cai HJ: Beneficial effect of Calculus Bovis Sativus on
17α-ethynylestradiol-induced cholestasis in the rat. Life Sci.
113:22–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamamoto Y, Moore R, Hess HA, Guo GL,
Gonzalez FJ, Korach KS, Maronpot RR and Negishi M: Estrogen
receptor alpha mediates 17alpha-ethynylestradiol causing
hepatotoxicity. J Biol Chem. 281:16625–16631. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakagawa R, Muroyama R, Saeki C, Goto K,
Kaise Y, Koike K, Nakano M, Matsubara Y, Takano K, Ito S, et al:
miR-425 regulates inflammatory cytokine production in CD4(+) T
cells via N-Ras upregulation in primary biliary cholangitis. J
Hepatol. 66:1223–1230. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Vohra BP, Zhang Y and Heuckeroth
RO: Transcriptional profiling after bile duct ligation identifies
PAI-1 as a contributor to cholestatic injury in mice. Hepatology.
42:1099–1108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sakamoto T, Morishita A, Nomura T, Tani J,
Miyoshi H, Yoneyama H, Iwama H, Himoto T and Masaki T:
Identification of microRNA profiles associated with refractory
primary biliary cirrhosis. Mol Med Rep. 14:3350–3356. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Carreras FI, Lehmann GL, Ferri D, Tioni
MF, Calamita G and Marinelli RA: Defective hepatocyte aquaporin-8
expression and reduced canalicular membrane water permeability in
estrogen-induced cholestasis. Am J Physiol Gastrointest Liver
Physiol. 292:G905–G912. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Crocenzi FA, Sánchez PE, Pellegrino JM,
Favre CO, Rodríguez GE, Mottino AD, Coleman R and Roma MG:
Beneficial effects of silymarin on estrogen-induced cholestasis in
the rat: A study in vivo and in isolated hepatocyte couplets.
Hepatology. 34:329–339. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak JK and Schmittgen DT: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen J, Zhao KN and Liu GB:
Estrogen-induced cholestasis: Pathogenesis and
therapeuticimplications. Hepatogastroenterology. 60:1289–1296.
2013.PubMed/NCBI
|
|
24
|
Glantz A, Marschall HU and Mattsson LA:
Intrahepatic cholestasis of pregnancy: Relationships between bile
acid levels and fetal complication rates. Hepatology. 40:467–474.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lammert F, Marschall HU, Glantz A and
Matern S: Intrahepatic cholestasis of pregnancy: Molecular
pathogenesis, diagnosis and management. J Hepatol. 33:1012–1021.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Y, Yang X, Wang Z, Li M, Ning Y, Chen
S, Yin L and Li X: Estrogen sulfotransferase (SULT1E1) regulates
inflammatory response and lipid metabolism of human endothelial
cells via PPARgamma. Mol Cell Endocrinol. 369:140–149. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu X, Xue R, Yang C, Gu J, Chen S and
Zhang S: Cholestasis-induced bile acid elevates estrogen level via
farnesoid X receptor-mediated suppression of the estrogen
sulfotransferase SULT1E1. J Biol Chem. 293:12759–12769. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yalcin EB, More V, Neira KL, Lu ZJ,
Cherrington NJ, Slitt AL and King RS: Downregulation of
sulfotransferase expression and activity in diseased human livers.
Drug Metab Dispos. 41:1642–1650. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li L and Falany CN: Elevated hepatic
SULT1E1 activity in mouse models of cystic fibrosis alters the
regulation of estrogen responsive proteins. J Cyst Fibros. 6:23–30.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deo AK and Bandiera SM: Identification of
human hepatic cytochrome p450 enzymes involved in the
biotransformation of cholic and chenodeoxycholic acid. Drug Metab
Dispos. 36:1983–1991. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Saini SP, Sonoda J, Xu L, Toma D, Uppal H,
Mu Y, Ren S, Moore DD, Evans RM and Xie W: A novel constitutive
androstane receptor-mediated and CYP3A-independent pathway of bile
acid detoxification. Mol Pharmacol. 65:292–300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li T and Chiang JY: Rifampicin induction
of CYP3A4 requires pregnane X receptor cross talk with hepatocyte
nuclear factor 4alpha and coactivators, and suppression of small
heterodimer partner gene expression. Drug Metab Dispos. 34:756–764.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Honda A, Ikegami T, Nakamuta M, Miyazaki
T, Iwamoto J, Hirayama T, Saito Y, Takikawa H, Imawari M and
Matsuzaki Y: Anticholestatic effects of bezafibrate in patients
with primary biliary cirrhosis treated with ursodeoxycholic acid.
Hepatology. 57:1931–1941. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Back P: Therapeutic use of phenobarbital
in intrahepatic cholestasis. Inductions in bile acid metabolism.
Pharmacol Ther. 33:153–155. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao X, Sheng L, Wang L, Hong J, Yu X,
Sang X, Sun Q, Ze Y and Hong F: Mechanisms of nanosized titanium
dioxide-induced testicular oxidative stress and apoptosis in male
mice. Part Fibre Toxicol. 11:472014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Räisänen SR, Lehenkari P, Tasanen M,
Rahkila P, Harkonen PL and Väänänen HK: Carbonic anhydrase III
protects cells from hydrogen peroxide-induced apoptosis. FASEB J.
13:513–522. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Miethke AG, Zhang W, Simmons J, Taylor AE,
Shi T, Shanmukhappa SK, Karns R, White S, Jegga AG, Lages CS, et
al: Pharmacological inhibition of apical sodium-dependent bile acid
transporter changes bile composition and blocks progression of
sclerosing cholangitis in multidrug resistance 2 knockout mice.
Hepatology. 63:512–523. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Parkkila S, Halsted CH, Villanueva JA,
Väänänen HK and Niemelä O: Expression of testosterone-dependent
enzyme, carbonic anhydrase III, and oxidative stress in
experimental alcoholic liver disease. Dig Dis Sci. 44:2205–2213.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zimmerman UJ, Wang P, Zhang X, Bogdanovich
S and Forster R: Anti-oxidative response of carbonic anhydrase III
in skeletal muscle. IUBMB Life. 56:343–347. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cabiscol E and Levine RL: Carbonic
anhydrase III. Oxidative modification in vivo and loss of
phosphatase activity during aging. J Biol Chem. 270:14742–14747.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hong F and Yang S: Ischemic
preconditioning decreased leukotriene C4 formation by depressing
leukotriene C4 synthase expression and activity during hepatic I/R
injury in rats. J Surg Res. 178:1015–1021. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Martínez-Clemente M, Ferré N,
González-Périz A, López-Parra M, Horrillo R, Titos E,
Morán-Salvador E, Miquel R, Arroyo V, Funk CD and Clària J:
5-lipoxygenase deficiency reduces hepatic inflammation and tumor
necrosis factor α-induced hepatocyte damage in hyperlipidemia-prone
ApoE-null mice. Hepatology. 51:817–827. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li SQ, Zhu S, Wan XD, Xu ZS and Ma Z:
Neutralization of ADAM8 ameliorates liver injury and accelerates
liver repair in carbon tetrachloride-induced acute liver injury. J
Toxicol Sci. 39:339–351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Higuchi Y, Yasui A, Matsuura K and
Yamamoto S: CD156 transgenic mice. Different responses between
inflammatory types. Pathobiology. 70:47–54. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li X, Liu R, Luo L, Yu L, Chen X, Sun L,
Wang T, Hylemon PB, Zhou H, Jiang Z and Zhang L: Role of
AMP-activated protein kinase α1 in 17α-ethinylestradiol-induced
cholestasis in rats. Arch Toxicol. 91:481–494. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li X, Liu R, Zhang L and Jiang Z: The
emerging role of AMP-activated protein kinase in cholestatic liver
diseases. Pharmacol Res. 125:105–113. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Moustafa T, Fickert P, Magnes C, Guelly C,
Thueringer A, Frank S, Kratky D, Sattler W, Reicher H, Sinner F, et
al: Alterations in lipid metabolism mediate inflammation, fibrosis,
and proliferation in a mouse model of chronic cholestatic liver
injury. Gastroenterology. 142:140–151.e12. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Leuenberger N, Pradervand S and Wahli W:
Sumoylated PPARalpha mediates sex-specific gene repression and
protects the liver from estrogen-induced toxicity in mice. J Clin
Invest. 119:3138–3148. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li T and Chiang JY: Nuclear receptors in
bile acid metabolism. Drug Metab Rev. 45:145–155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen P, Li D, Chen Y, Sun J, Fu K, Guan L,
Zhang H, Jiang Y, Li X, Zeng X, et al: p53-mediated regulation of
bile acid disposition attenuates cholic acid-induced cholestasis in
mice. Br J Pharmacol. 174:4345–4361. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang H, Li TW, Ko KS, Xia M and Lu SC:
Switch from Mnt-Max to Myc-Max induces p53 and cyclin D1 expression
and apoptosis during cholestasis in mouse and human hepatocytes.
Hepatology. 49:860–870. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wilkins BJ, Lorent K, Matthews RP and Pack
M: p53-mediated biliary defects caused by knockdown of cirh1a, the
zebrafish homolog of the gene responsible for North American Indian
Childhood Cirrhosis. PLoS One. 8:e776702013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
So J, Khaliq M, Evason K, Ninov N, Martin
BL, Stainier D and Shin D: Wnt/β-catenin signaling controls
intrahepatic biliary network formation in zebrafish by regulating
notch activity. Hepatology. 67:2352–2366. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Okabe H, Yang J, Sylakowski K, Yovchev M,
Miyagawa Y, Nagarajan S, Chikina M, Thompson M, Oertel M, Baba H,
et al: Wnt signaling regulates hepatobiliary repair following
cholestatic liver injury in mice. Hepatology. 64:1652–1666. 2016.
View Article : Google Scholar : PubMed/NCBI
|