Introduction
Lung cancer is one of the most frequently diagnosed
malignancies, with >14 million new cases and >8 million
mortalities every year (1). The
incidence of lung cancer has been predicted to further increase in
the near future (2). In developing
countries, such as China, lung cancer remains the most common
cancer and the leading cause of cancer-related mortalities
(3). This situation will not
change unless air pollution in China is significantly reduced
(4). With the efforts made on
cancer treatment, such as the development of novel therapeutic
approaches, survival of patients with lung cancer at early stages
has been significantly improved in the past decades (5). However, once metastasis occurs,
treatment outcomes become extremely poor (6). At present, early diagnosis and
treatment are still crucial.
Phospholipase A2 (PLA2) enzymes catalyze the release
of fatty acids from the second carbon group of glycerol. As an
extracellular form of PLA2, secretory phospholipase A2 (sPLA2)
widely participates in the development of different types of human
cancers (7). sPLA2 is considered
to be a promising therapeutic target for cancer treatment (8,9).
sPLA2 has been demonstrated to participate in human disease through
interactions with different signaling proteins (10,11),
whereas its interactions with non-coding RNAs have rarely been
studied. Long non-coding RNA (lncRNA) SRA-like non-coding RNA
(SLNCR1; also known as linc00673) has been proven as an oncogenic
lncRNA in several types of cancers, such as NSCLC and melanoma
(12,13). However, its functionality in NSCLC
remains to be further studied. In the present study, it was
observed that lncRNA SLNCR1 may regulate NSCLC migration, invasion
and stemness through interactions with sPLA2. To evaluate the early
diagnostic value of SLNCR1 for non-small cell lung cancer (NSCLC),
patients with stage I or II NSCLC were included in the study. The
results demonstrated that altered lncRNA SLNCR1 expression may be
used to effectively distinguish patients with early stage NSCLC
from healthy controls.
Materials and methods
Human specimens and cell lines
A total of 66 patients with NSCLC (stage I and II)
who were admitted to Jiangxi Provincial People's Hospital
(Nanchang, China) between January 2016 and January 2018 were
included in the study. Inclusion criteria: i) Diagnosed by biopsy;
ii) stage I or II for early diagnosis analysis. Exclusion criteria:
i) Combined with other disease; ii) received treatment within 6
months prior to admission. Tumor tissues and paired healthy tissues
(within 5 cm from tumors) were collected from three sites. Blood
was extracted from the patients with NSCLC and 42 healthy
volunteers to prepare plasma by centrifuging blood samples in EDTA
tube for 20 min at 1,200 × g at room temperature. The patient group
included 35 males and 31 females, with an age range of 29–68 years
(mean, 48.5±4.7 years). The control group included 22 males and 20
females, with an age range of 28–66 years (mean, 47.1±4.4 years).
The two groups had similar age and sex distributions. The study was
approved by the ethics committee of Jiangxi Provincial People's
Hospital. All participants signed written informed consent.
Two human NSCLC cell lines, H1581 and H1993, were
purchased from American Type Culture Collection (ATCC).
ATCC-formulated RPMI-1640 medium containing 10% FBS (cat. no.
30-2020; ATCC) was used to cultivate cells in a 5% CO2
incubator at 37°C.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA extraction from plasma (0.3 ml) and cells
(106) was performed using RNeasy Mini Kit (Qiagen China
Co., Ltd.) according to manufacturer's instructions. Reverse
transcription was carried out using Applied Biosystems™
High-Capacity cDNA Reverse Transcription kit to synthesize cDNA
following DNase I (Sigma-Aldrich; Thermo Fisher Scientific, Inc.)
digestion. Reverse transcription conditions were 55°C for 20 min
and 80°C for 10 min. SuperScript III Platinum one-Step qRT-PCR SYBR
kit (Thermo Fisher Scientific, Inc.) was used to prepare PCR
reaction systems. The thermocycling conditions were 95°C for 55
sec, followed by 40 cycles of 95°C for 16 sec and 59.5°C for 25
sec. The primers were as follows: Human lncRNA SLNCR1, forward
5′-GGACCCCTTAACGTGGATTAC-3′, reverse 5′-AAATACCTCCAGCTTGGCG-3′;
β-actin, forward 5′-GACCTCTATGCCAACACAGT-3′, reverse
5′-AGTACTTGCGCTCAGGAGGA-3′. The 2−ΔΔCq method (14) was used to normalize expression
levels to β-actin.
Cell transfection
Vectors (pcDNA3.1) expressing sPLA2, lncRNA SLNCR1
siRNA (5′-AAGAGGATGGGAAGGACTGAT-3′) and Scrambled siRNA
(5′-UUCUCCGAACGUGUCACGUdTdT-3′) were synthesized by Shanghai
GenePharma Co., Ltd. NSCLC cells were cultured to 70–80%
confluence, and transfection was performed using
Lipofectamine® 3000 Transfection Reagent (Thermo Fisher
Scientific, Inc.) with 10 nM vectors or 30 nM siRNAs. Incubation of
cells with transfection mixtures were performed at 37°C for 6 h.
Cells treated with Lipofectamine® 3000 alone were used
as an untransfected control. Cells transfected with empty vectors
or Scramble siRNAs were used as a negative control. The
transfections were considered successful if the knockdown rate of
lncRNA SLNCR1 reached 50% and the overexpression rate of sPLA2
reached 200%. Three biological replicates were included in all
subsequent experiments. All subsequent experiments were performed
at 24 h post-transfections.
Enzyme-linked immunosorbent assay
(ELISA)
Plasma levels of sPLA2 were measured using a human
sPLA2 ELISA kit (cat. no. MBS265046; MyBioSource, Inc.). Plasma
levels of sPLA2 were expressed as pmol/l.
Transwell migration and Matrigel
invasion assays
RPMI-1640 medium containing 1% FBS was used to
prepare suspensions of transfected cells (5×104
cells/ml). The upper chamber (Corning HTS Transwell 96 well, 8.0 µm
pore, cat. no. CLS3374, Sigma-Aldrich; Merck KGaA) was filled with
100 µl cell suspension, whereas the lower chamber was filled with
RPMI-1640 medium containing 20% FBS. Following 24-h incubation, the
membranes were subjected to staining with 0.5% crystal violet
(Sigma-Aldrich; Merck KGaA) at 25°C for 30 min. Matrigel (cat. no.
356234; EMD Millipore) was used to pre-coat the upper chamber prior
to the invasion assay. Cells in 5 randomly selected visual fields
(magnification, ×40) were counted under Olympus CX22 Microscope
(Olympus Corporation). Cells were counted using Image J v1.46
software (National Institutes of Health). Control group was set to
‘100’. All other groups were normalized to control group.
Flow cytometry
Trypsinization was performed to harvest transfected
cells. Cells (105) were incubated with phycoerythrin
(PE)-conjugated immunoglobulin G (IgG) 1 (cat. no. 130-112-760;
Miltenyi Biotec GmbH) or CD133-PE antibody (cat. no. 130-093-193;
Miltenyi Biotec GmbH) at 4°C for 15 min. Cells were then
resuspended in PBS and signals were detected using FACS Aria system
(BD Immunocytometry Systems) and processed by CellQuest software,
version 5.1 (Becton, Dickinson and Company).
Western blotting
Total protein was extracted from 105
transfected cells using ReadyPrep™ Protein Extraction kit (Bio-Rad
Laboratories, Inc.). Protein samples were quantified using a BCA
kit (Sangon Biotech Co., Ltd.). Different proteins were separated
by 10% SDS-PAGE with 40 µg per lane. Following gel transfer onto
PVDF membranes and blocking in PBS containing 5% non-fat milk for 2
h at room temperature, Membranes were first blotted with rabbit
anti-human primary antibodies sPLA2 (cat. no. ab47105; 1:1,200;
Abcam) and GAPDH (cat. no. ab9485; 1:1,200; Abcam) for 12 h at 4°C,
followed by incubation with secondary goat anti-rabbit horseradish
peroxidase-conjugated IgG antibody (cat. no. MBS435036; 1:1,000;
MyBioSource, Inc.) for 2 h at 24°C. Pierce ECL Western Blotting
Substrate (Thermo Fisher Scientific, Inc.) was used to develop
signals. Densitometric analysis was performed using ImageJ v.1.46
software (National Institutes of Health); expression data were
normalized to GADPH.
Statistical analysis
All in vitro experiments were repeated three
times and data are presented as the mean ± standard deviation.
Comparisons of lncRNA SLNCR1 expression levels in tumor tissues and
adjacent tissues were performed using Student's paired t-test.
Comparisons between patient and control groups were performed using
Student's unpaired t-test. One-way ANOVA followed by Tukey's test
was used for comparisons between cell transfection groups.
Pearson's correlation coefficient was used for correlation
analysis. Diagnostic analysis was performed by receiver operating
characteristics (ROC) curve. P<0.05 was considered to indicate a
statistically significant difference.
Results
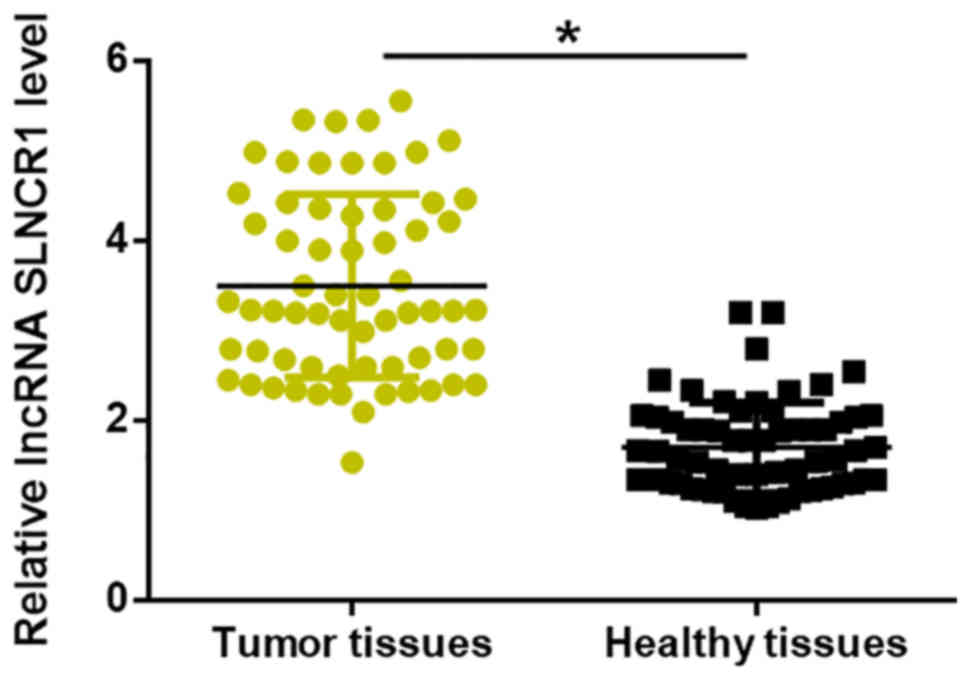
lncRNA SLNCR1 expression is
upregulated in NSCLC tumor tissues
lncRNA SLNCR1 expression in tumor tissues and
adjacent healthy tissues of 66 patients with NSCLC was detected by
RT-qPCR. Compared with adjacent healthy tissues, the expression
levels of lncRNA SLNCR1 were significantly increased in tumor
tissues (P<0.05; Fig. 1).
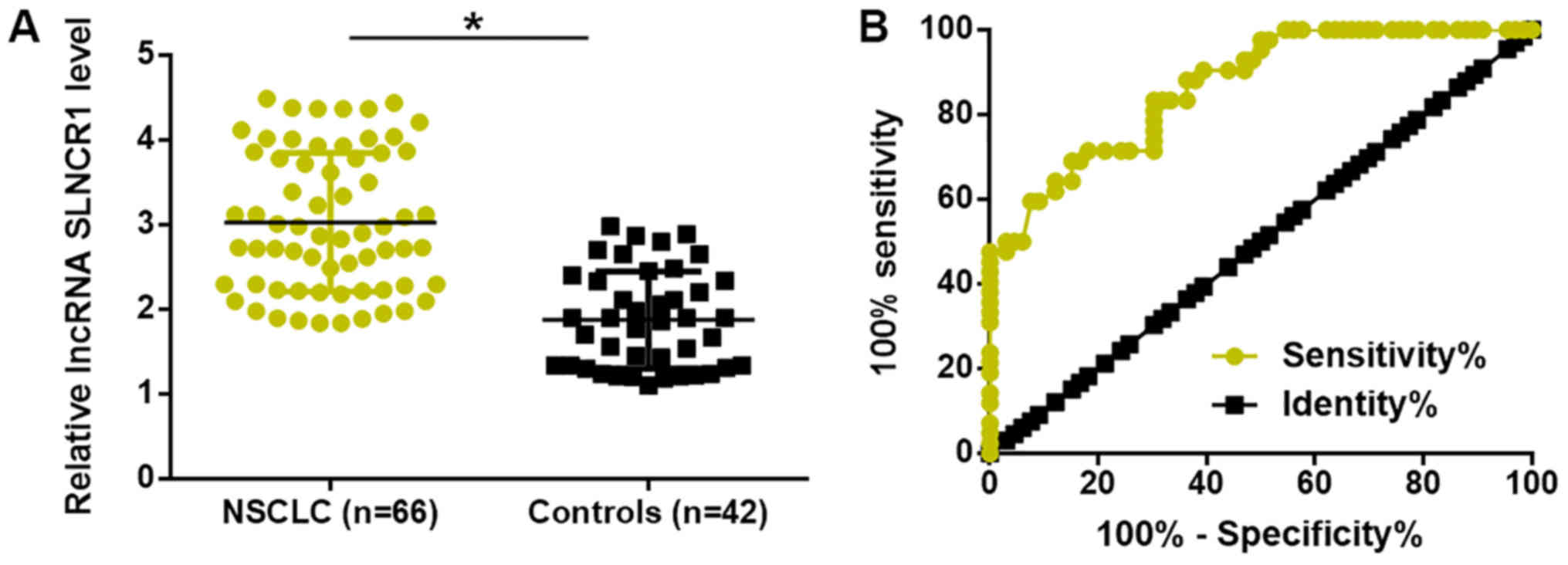
Upregulation of plasma lncRNA SLNCR1
distinguishes patients with NSCLC from healthy controls
Expression levels of lncRNA SLNCR1 in plasma of 66
patients with NSCLC and 44 healthy controls were detected by
RT-qPCR. Compared with healthy controls, expression levels of
plasma lncRNA SLNCR1 were significantly increased in patients with
NSCLC (P<0.05; Fig. 2A). ROC
curve analysis was performed to evaluate the diagnostic value of
lncRNA SLNCR1 for patients with NSCLC using patients with NSCLC as
true positive cases and healthy controls as true negative cases.
The area under the curve was 0.8660, with standard error of 0.03377
and 95% confidence interval of 0.7998–0.9322 (Fig. 2B).
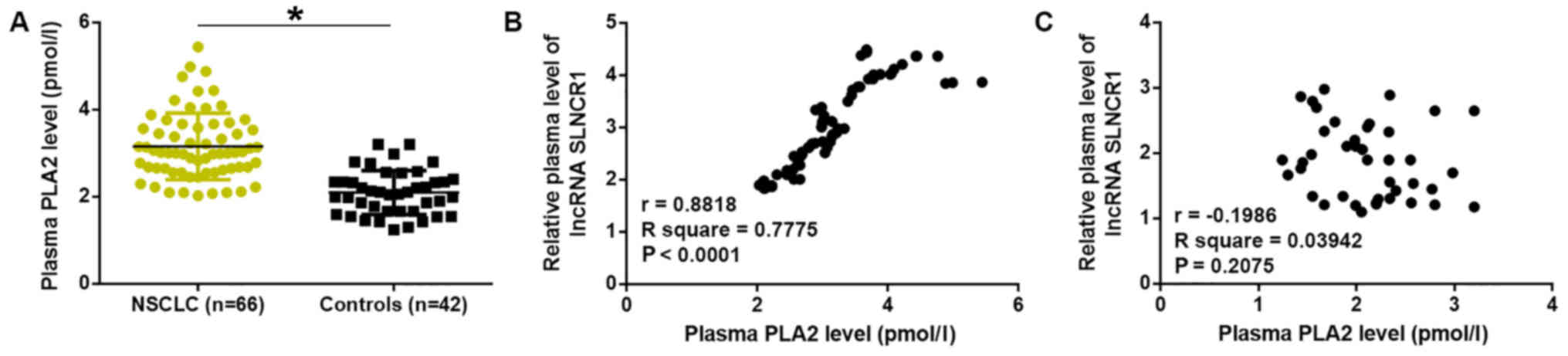
Plasma sPLA2 expression is upregulated
in patients with NSCLC and positively correlates with plasma lncRNA
SLNCR1
ELISA results demonstrated that, compared with
healthy controls, plasma levels of sPLA2 were significantly
upregulated in patients with NSCLC (P<0.05; Fig. 3A). Pearson's correlation analysis
revealed a positive correlation between plasma levels of sPLA2 and
lncRNA SLNCR1 in patients with NSCLC (Fig. 3B). Conversely, no correlation was
observed between plasma levels of sPLA2 and lncRNA SLNCR1 in
healthy controls (Fig. 3C).
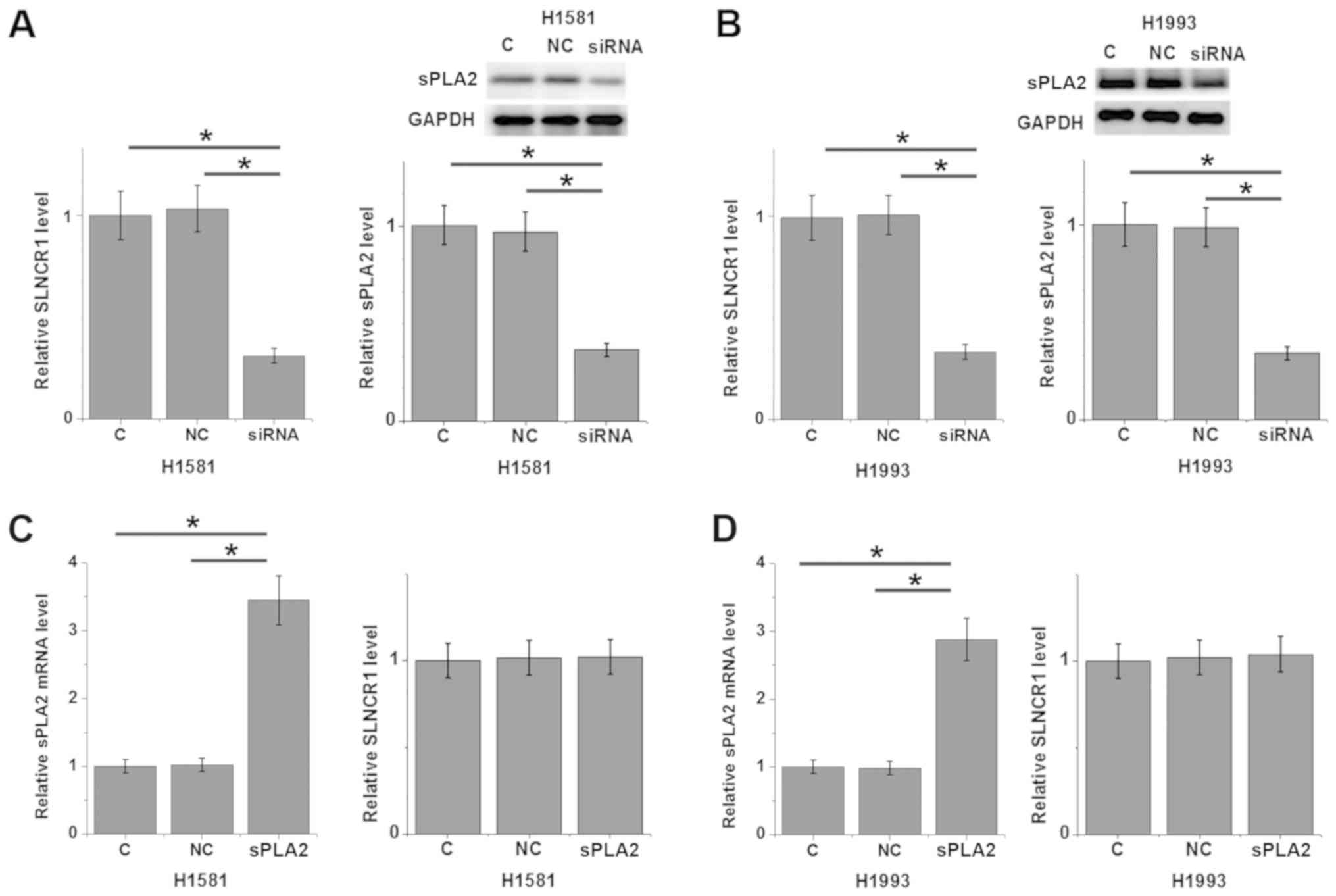
lncRNA SLNCR1 siRNA silencing leads to
downregulation of sPLA2 in NSCLC cell lines H1581 and H1993
Compared with untransfected control cells and
negative control cells, transfection with lncRNA SLNCR1 siRNA
significantly reduced the expression levels of lncRNA SLNCR1 and
sPLA2 in NSCLC cell lines H1581 (P<0.05; Fig. 4A) and H1993 (P<0.05; Fig. 4B). By contrast, sPLA2
overexpression exhibited no significant effects on lncRNA SLNCR1
expression in cells of H1581 (Fig.
4C) and H1993 (Fig. 4D) cell
lines.
lncRNA SLNCR1 regulates NSCLC cell
migration, invasion and stemness through sPLA2
Compared with the control groups, lncRNA SLNCR1
siRNA silencing and sPLA2 overexpression clearly affected the
expression of sPLA2 protein (Fig.
5A). In addition, lncRNA SLNCR1 siRNA silencing significantly
reduced, whereas sPLA2 overexpression significantly increased the
migratory ability (Fig. 5B),
invasive ability (Fig. 5C) and
stemness (Fig. 5D) of H1581 and
H1993 cells. sPLA2 overexpression partially attenuated the
inhibitory effects of lncRNA SLNCR1 siRNA silencing on cell
migration (Fig. 5B), invasion
(Fig. 5C) and stemness (Fig. 5D) (P<0.05). Notably, the changes
in sPLA2 expression followed a similar pattern to the changes of
cell migration, invasion and stemness (Fig. 5).
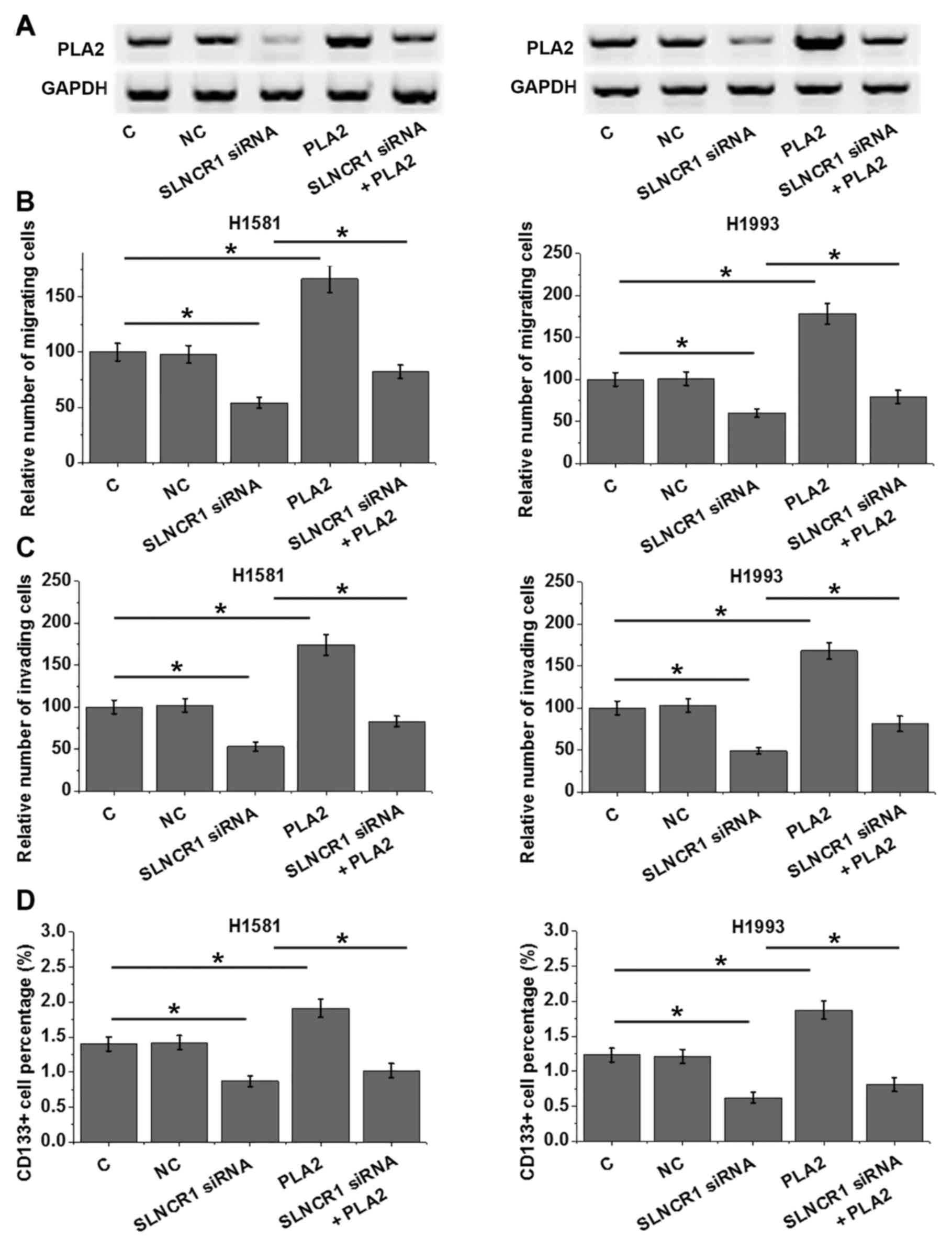 | Figure 5.lncRNA SLNCR1 regulates NSCLC cell
migration, invasion and stemness through sPLA2. (A) lncRNA SLNCR1
siRNA silencing and sPLA2 overexpression affected the expression of
sPLA2 protein in H1581 and H1993 cells. (B-D) lncRNA SLNCR1 siRNA
silencing reduced, whereas sPLA2 overexpression significantly
increased the migratory ability (B), invasive ability (C) and
stemness (D) of H1581 and H1993 cells vs. control and NC. In
addition, sPLA2 overexpression attenuated the inhibitory effects of
lncRNA SLNCR1 siRNA silencing on cell migration, invasion and
stemness; *P<0.05. C, untransfected control; lncRNA, long
non-coding RNA; NC, negative control; NSCLC, non-small cell lung
cancer; siRNA, small interfering RNA; sPLA2, secretory
phospholipase A2; SLNCR1, SRA-like non-coding RNA. |
Discussion
The present study found that lncRNA SLNCR1 is an
oncogenic lncRNA in NSCLC, which is the major pathological type of
lung cancer. The data also revealed that lncRNA SLNCR1 may regulate
NSCLC migration, invasion and stemness through interactions with
sPLA2.
sPLA2 has oncogenic functions in cancer biology and
usually exhibits upregulated expression in certain types of human
cancers, including lung cancer (15,16).
Consistent with previous studies (15,16),
in the present study plasma sPLA2 was significantly upregulated in
patients with NSCLC compared with healthy controls. overexpression
of sPLA2 not only promotes cancer cell migration and invasion, but
also participates in the maintenance of prostate cancer cell
stemness (17,18). CD133 is a widely used marker for
cancer stem cells (19). In the
present study, increased NSCLC cell migration, invasion and
stemness were also observed following sPLA2 overexpression. The
results further supported a potential oncogenic role for sPLA2 in
lung cancer and provided new insights to its functionality in this
disease.
lncRNA SLNCR1 is an oncogenic lncRNA in many
different types of cancers, such as NSCLC and melanoma (12,13);
the present study demonstrated an upregulation of lncRNA SLNCR1
expression levels in tumor tissues compared with adjacent healthy
tissues of patients with NSCLC. Significantly higher plasma levels
of lncRNA SLNCR1 were also observed in patients with NSCLC compared
with healthy controls, which indicated a potential oncogenic role
for lncRNA SLNCR1 in NSCLC. lncRNA SLNCR1 has been previously
demonstrated to mediate melanoma invasion (13). In another study, SLNCR1 was proved
to serve roles in regulating multiple cell behaviors in NSCLC, such
as invasion, migration and proliferation (12). Consistent with previous studies
(12,13), the present study also demonstrated
that lncRNA SLNCR1 may be involved in the regulation of cancer cell
migration in NSCLC. The present study is the first to report the
role of SLNCRA in regulating cancer cell stemness in NSCLC.
Therefore, the inhibition of lncRNA SLNCR1 expression may serve as
a potential therapeutic target for NSCLC.
Notably, a significantly positive correlation
between plasma lncRNA SLNCR1 and sPLA2 was identified in patients
with NSCLC. In vitro experiments using NSCLC cell lines also
demonstrated that lncRNA SLNCR1 may be an upstream activator of
sPLA2 in the regulation of migration, invasion and stemness of
NSCLC cells. To the best of our knowledge, this is the first
reported interaction between sPLA2 and lncRNAs. However, the
interaction between lncRNA SLNCR1 and sPLA2 is unlikely to be
direct owing to the lack of correlation between plasma lncRNA
SLNCR1 and sPLA2 in healthy controls. The development and
progression of NSCLC are associated with pathological factors,
which may mediate the interaction between lncRNA SLNCR1 and sPLA2.
However, the present study failed to identify these pathological
mediators. Future studies are needed to identify the mediators
involved in this process.
In conclusion, lncRNA SLNCR1 and secretory sPLA2
were both upregulated in NSCLC. lncRNA SLNCR1 may regulate cancer
cell migration, invasion and stemness in NSCLC through interactions
with secretory sPLA2.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
WX designed the experiments. WX, QX, DK and ZW
performed the experiments. QLu, QLi, HW and LC assisted with the
experiments and analyzed data. WX drafted the manuscript. All
authors approved the manuscript.
Ethics approval and consent to
participate
The study was approved by the ethics committee of
Jiangxi Provincial People's Hospital. All participants signed
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rahib L, Smith BD, Aizenberg R, Rosenzweig
AB, Fleshman JM and Matrisian LM: Projecting cancer incidence and
deaths to 2030: The unexpected burden of thyroid, liver, and
pancreas cancers in the United States. Cancer Res. 74:2913–2921.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guo Y, Zeng H, Zheng R, Li S, Barnett AG,
Zhang S, Zou X, Huxley R, Chen W and Williams G: The association
between lung cancer incidence and ambient air pollution in China: A
spatiotemporal analysis. Environ Res. 144:60–65. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng X, Schipper M, Kidwell K, Lin J,
Reddy R, Ren Y, Chang A, Lv F, Orringer M and Spring Kong FM:
Survival outcome after stereotactic body radiation therapy and
surgery for stage I non-small cell lung cancer: A meta-analysis.
Int J Radiat oncol Biol Phys. 90:603–611. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sperduto PW, Yang TJ, Beal K, Pan H, Brown
PD, Bangdiwala A, Shanley R, Yeh N, Gaspar LE, Braunstein S, et al:
Estimating survival in patients with lung cancer and brain
metastases: An update of the graded prognostic assessment for lung
cancer using molecular markers (Lung-molGPA). JAMA Oncol.
3:827–831. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schewe M, Franken PF, Sacchetti A, Schmitt
M, Joosten R, Böttcher R, van Royen ME, Jeammet L, Payré C, Scott
PM, et al: Secreted phospholipases A2 are intestinal stem cell
niche factors with distinct roles in homeostasis, inflammation and
cancer. Cell Stem Cell. 19:38–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Quach ND, Arnold RD and Cummings BS:
Secretory phospholipase A2 enzymes as pharmacological targets for
treatment of disease. Biochem Pharmacol. 90:338–348. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pedada SR, Yarla NS, Tambade PJ,
Dhananjaya BL, Bishayee A, Arunasree KM, Philip GH, Dharmapuri G,
Aliev G, Putta S and Rangaiah G: Synthesis of new secretory
phospholipase A2-inhibitory indole containing isoxazole derivatives
as anti-inflammatory and anticancer agents. Eur J Med Chem.
112:289–297. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rubio JM, Rodríguez JP, Gil-de-Gómez L,
Guijas C, Balboa MA and Balsinde J: Group V secreted phospholipase
A2 is upregulated by IL-4 in human macrophages and mediates
phagocytosis via hydrolysis of ethanolamine phospholipids. J
Immunol. 194:3327–3339. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu Y, Li Y, Shang M, Jian Y, Wang C,
Bardeesi AS, Li Z, Chen T, Zhao L, Zhou L, et al: Secreted
phospholipase A2 of Clonorchis sinensis activates hepatic stellate
cells through a pathway involving JNK signalling. Parasit Vectors.
10:1472017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H,
Kong J, Ding K, Shen HM, Wu H, et al: Long non-coding RNA linc00673
regulated non-small cell lung cancer proliferation, migration,
invasion and epithelial mesenchymal transition by sponging
miR-150-5p. Mol Cancer. 16:1182017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schmidt K, Joyce CE, Buquicchio F, Brown
A, Ritz J, Distel RJ, Yoon CH and Novina CD: The lncRNA SLNCR1
mediates melanoma invasion through a conserved SRA1-like region.
Cell Rep. 15:2025–2037. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hulstaert E, Brochez L, Volders PJ,
Vandesompele J and Mestdagh P: Long non-coding RNAs in cutaneous
melanoma: Clinical perspectives. Oncotarget. 8:43470–43480. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu S and Dong Z: overexpression of
secretory phospholipase A2-IIa supports cancer stem cell phenotype
via HER/ERBB-elicited signaling in lung and prostate cancer cells.
Int J Oncol. 50:2113–2122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Brglez V, Pucer A, Pungerčar J, Lambeau G
and Petan T: Secreted phospholipases A2 are
differentially expressed and epigenetically silenced in human
breast cancer cells. Biochem Biophys Res Commun. 445:230–235. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Choi YA, Lim HK, Kim JR, Lee CH, Kim YJ,
Kang SS and Baek SH: Group IB secretory phospholipase A2 promotes
matrix metalloproteinase-2-mediated cell migration via the
phosphatidylinositol-3 kinase and Akt pathway. J Biol Chem.
279:36579–36585. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu Y and Wu PY: CD133 as a marker for
cancer stem cells: Progresses and concerns. Stem Cells Dev.
18:1127–1134. 2009. View Article : Google Scholar : PubMed/NCBI
|