Introduction
The phenotype of hair color depends on the levels
and ratio of two types of melanin, namely black-brown eumelanin
(EM) and yellow-red-brown pheomelanin (PM), which are produced by
resident skin melanocytes (1). The
hair color formation includes melanin production in melanocytes and
melanin transport to keratinocytes (2). Several genes and miRNAs are involved
in the pathways of melanin production. microphthalmia-associated
transcription factor (MITF) is an important transcription factor
that activates the key limiting enzymes tyrosinase (TYR),
tyrosinase-related protein 1 (TYRP1), and tyrosinase-related
protein 2 (TYRP2)/DCT for the synthesis and storage of melanin in
melanosomes (3,4). During melanin transportation, the
tripartite complex Rab27a/melanophilin (MLPH)/myosin Va (MYO5A)
plays a crucial role (5) and acts
as a linker in the entire protein complex (6,7). The
complex is required for connecting the melanosome to the actin
cytoskeleton in order to facilitate the normal accumulation of the
organelle in the dendritic tips (8). MYO5A is an actin-based molecular
motor typically involved in the transport of organelles and
vesicles, such as the melanosomes (9,10).
MYO5A can bind to more than one type of cellular structure and its
selectivity is determined by alternate spliced sequences in
melanocytes (11).
Short tandem target mimics (STTMs) are based on
target mimicry (TM) and aim to block the function of small RNA
molecules in animals and plants (12). The expression of the microRNA-143
(miR-143) cluster is required in various types of cells. As
predicted by the TargetScan analysis, miR-143-5p targets MYO5A. In
the present study, the STTM method was used, which is considered a
powerful technology to complement existing small RNA sequestration
(12). An STTM-miR-143-5p plasmid
was constructed to inhibit the function of miR-143-5p in the
regulation of EM and PM production in mouse melanocytes.
Materials and methods
Construction of plasmids
An oligonucleotide containing the STTM-miR-143-5p
sequence was chemically synthesized according to a previously
described method (12). The
oligonucleotide was inserted into the dual-luciferase vector,
pmirGLO (Promega Corporation) in order to construct the expression
plasmid pmirGLO-STTM-miR-143-5p, in which the CMV promoter was used
to induce GFP and STTM-miR-143-5p expression. The null pmirGLO
vector was used as the corresponding negative control (NC). The
luciferase reporter expression plasmid was constructed by cloning
the 3′-UTR sequence of the mouse MYO5A into the dual luciferase
pmirGLO vector (Promega Corporation). The partial sequence of the
mouse MYO5A containing the miR-143-5p binding sites was obtained by
PCR with mouse melanocyte cDNA as the template and the primers that
contained the XhoI and SacI sites (Table I). The PCR product was subsequently
digested with XhoI and SacI and cloned into the
SacI and XhoI restriction sites of the vector in
order to produce the pmirGLO-MYO5A-wt plasmid. In addition, the
MYO5A 3′-UTR with the miR-143-5p binding sites was mutated using a
site-directed gene mutagenesis kit (Beyotime Institute of
Technology) to generate the pmiGLO-MYO5A-mut plasmid. All
constructs were verified by sequencing (Shenzhen Huada Gene Co.,
Ltd.).
 | Table I.Primers used in the present
study. |
Table I.
Primers used in the present
study.
| Primer name | Primer sequence
5′-3′ | Application |
|---|
| MYO5A-F |
AAAATGCTGCGGTTAGGA | RT-qPCR |
| MYO5A-R |
GCTTGGGAGGTATTGTGC | RT-qPCR |
| MYO5A-wt-F |
CGAGCTCAAAATGCTGCGGTTAGG | Luciferase
reporter-wt |
| MYO5A-wt-R |
CCGCTCGAGCCAGTTAAAGAGTTTTGCATAG | Luciferase
reporter-wt |
| MYO5A-mut-F |
TACCTGCAGATGCACCTCTGCAAGTAGCAGACACTGG | Luciferase
reporter-mut |
| MYO5A-mut-R |
CCAGTGTCTGCTACTTGCAGAGGTGCATCTGCAGGTA | Luciferase
reporter-mut |
| miR-143-5p-RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCAGAGA | RT-qPCR |
| miR-143-5p-F |
ACACTCCAGCTGGGGGTGCAGTGCTGCATC | RT-qPCR |
| Common-R |
CGAGCAGTGCAGGGTCCGAGGT | RT-qPCR |
| U6-RT |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCATCT | RT-qPCR |
| U6-F |
CTCGCTTCGGCAGCACA | RT-qPCR |
| MLPH-F |
GGTTTCCTGCTTCTGTTCG | RT-qPCR |
| MLPH-R |
CGTGGCTTATGTTTGTCCC | RT-qPCR |
| Rab27a-F |
TATGGGTTTCCTGCTTCT | RT-qPCR |
| Rab27a-R |
GCCTCCTCCTCTTTCACT | RT-qPCR |
| MITF-F |
TGAGGAGCAGAGCAGGGCAGAGAGT | RT-qPCR |
| MITF-R |
TGGGAAGGTTGGCTGGACAGGAGTT | RT-qPCR |
| TYR-F |
TGAAAATCCTAACTTACTCAGCCCA | RT-qPCR |
| TYR-R |
TCAAACTCAGACAAAATTCCACATC | RT-qPCR |
| TYRP1-F |
CCATTGCTGTAGTGGCTGCGTTGTT | RT-qPCR |
| TYRP1-R |
GGAGAGGCTGGTTGGCTTCATTCTT | RT-qPCR |
| TYRP2-F |
TTGCTCTTGGGGTTGCTGGCTTTTC | RT-qPCR |
| TYRP2-R |
TCCTCCGTGTATCTCTTGCTGCTGA | RT-qPCR |
| β-actin-F |
TTGCTGACAGGATGCAGAAG | RT-qPCR |
| β-actin-R |
ACATCTGCTGGAAGGTGGAC | RT-qPCR |
Melanocyte transfection
The mouse melanocytes used in the present study were
established and maintained in our laboratory (13). The melanocytes were transfected
with the pmirGLO-STTM-miR-143-5p plasmid and/or the pmirGLO null
plasmid (NC) using the Lipofectamine 2000 assay (Invitrogen; Thermo
Fisher Scientific, Inc.) following the manufacturer's instructions.
The melanocytes were collected two days after, following
transfection for cell lysis and total RNA preparation.
Dual luciferase reporter assay for
miRNA target validation
293T cells were cultured in DMEM (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(FBS) and were transfected with 580 ng of pmirGLO-MYO5A-wt or
pmirGLO-MYO5A-mut plasmids in the presence of 20 ng of
pmirGLO-STTM-miR-143-5p or pmirGLO-NC plasmids using Lipofectamine
2000. These preparations were used for the luciferase assays two
days after co-transfection. Luciferase activity was subsequently
measured using the Dual-Luciferase Reporter Assay kit (Promega
Corporation). The firefly luciferase activity of each test sample
was normalized to that of Renilla. The data were performed
in triplicate and expressed as the mean relative luciferase
activity (mean ± SD).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis of miRNA and mRNA
expression levels
Total RNA from melanocytes was extracted using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's instructions and treated with DNase I
(Sigma-Aldrich; Merck KGaA). For quantitative mRNA expression
analysis, 1 µg of total RNA was reverse-transcribed to cDNA using a
cDNA synthesis kit (Takara Bio, Inc.) as described in the
manufacturer's instructions. The samples were amplified by
quantitative real time PCR using SYBR-Green PCR master mix (Takara
Bio, Inc.). For quantitative miRNA expression analysis, cDNA was
produced using a specific stem-loop RT primer pair, a common
reverse primer (Table I) pair and
a cDNA synthesis kit (Takara Bio, Inc.) according to a previously
described method (14). SYBR-Green
PCR master mix (Takara Bio, Inc.) was used for RT-qPCR reactions
performed on a 7500 Fast Real-Time PCR™ system (Applied Biosystems
Life Technologies; Thermo Fisher Scientific, Inc.). The relative
expression levels of mRNA and miRNA were quantified using the
quantification cycle (Cq) method (15) and the normalization was performed
with regard to the amounts of β-actin and U6 mRNA,
respectively.
Western blot analysis
The cell lysates from melanocytes were obtained
using RIPA cell lysis reagent (Beyotime Institute of Biotechnology)
and western blotting was conducted as previously described
(16). The following primary
antibodies were used: Anti-MYO5A (rabbit resource) at a 1:1,000
dilution (cat. no. 3402; RRID:AB_2148475; Cell Signaling
Technology, Inc.), anti-MLPH (mouse resource) at a 1:1,000 dilution
(cat. no. 66092-1-Ig; RRID:AB_11232039; ProteinTech Group),
anti-RAB27A (mouse resource) at a 1:1,000 dilution (cat. no.
ab55667; RRID:AB_945112), anti-MiTF (rabbit resource) at a 1:1,000
dilution (cat. no. ab20663; RRID:AB_470315), anti-TYR (rabbit
resource) at a 1:1,000 dilution (cat. no. ab61294), anti-TYRP1
(rabbit resource) at a 1:1,000 dilution (cat. no. ab83774;
RRID:AB_2211142), and anti-TYRP2 (rabbit resource) at a 1:1,000
dilution (cat. no. ab74073; AB_1524517; all from Abcam), and
anti-GAPDH (rabbit resource) at a 1:10,000 dilution (cat. no.
AP0063, Bioworld Technology, Inc.). Anti-rabbit IgG secondary
antibodies (cat. no. CW0103) and anti-mouse IgG secondary
antibodies (cat. no. CW0102) were commercially purchased. The
immunoblots were exposed to develop the chemicals and subsequently
scanned using a ChemiDOC™ XRS + imager (Bio-Rad Laboratories,
Inc.). Protein expression was quantified from the band intensity
which was assessed by densitometry using the Image-Pro Plus
software (Olympus Corporation).
Spectrophotometric assay of melanin
content
Following transfection, the harvested melanocytes
were rinsed with phosphate-buffered saline (PBS). The
spectrophotometric assay of the alkali-soluble melanin required
melanocytes that were lysed with l ml of 1 M NaOH at 37°C. The
cells were incubated for 1 h and the absorbance was measured at 475
nm spectrophotometrically. The determination of EM was achieved by
melanocyte hydrolysis at 80°C in 30% hydroiodic acid and 30%
hypophosphoric acid. Following cooling and the addition of 50%
ethanol, the samples were centrifuged at 2,234 × g for 10 min in
order to collect the insoluble eumelanic pigments that were
subsequently solubilized at 80°C in hydrogen peroxide and sodium
hydroxide. Following centrifugation at 10,700 × g for 1 min in a
Sorvall Ultracentrifuge, the supernatant was obtained and its
absorbance was measured at 350 nm. The concentration of PM was
determined in transfected melanocytes that were solubilized in
phosphate buffer (pH 10.5) and centrifuged at 10,700 × g for 10
min. The supernatant was obtained and the absorbance was recorded
at 400 nm. The melanin contents were normalized to the total
numbers of cells. All experiments were performed in triplicate.
Fontana-Masson staining
Following melanocytes transfection with
STTM-miR-143-5p or pmirGLO null vector, the plasmids were washed
thoroughly with PBS (pH 7.4) and the cells were fixed in 4%
formaldehyde for 20 min. The cells were subsequently stained using
Ammoniacal Silver solution from the Fontana-Masson stain kit
(Abcam) at 55°C in the dark for 1 h, and washed with distilled
water thoroughly thereafter. The cells were further stained using
0.2% gold chloride solution and 5% sodium thiosulfate solution for
2 min and counter-stained with Nuclear Fast Red solution for 5 min.
The samples were washed with distilled water following every
staining process. The melanin content was visualized under a light
microscope.
Statistical analysis
The difference in melanin production, the levels of
miRNA, mRNA and protein, and the relative luciferase activities
were analyzed using ANOVA and the protected Fisher's least
significant difference tests. The analysis was conducted using SPSS
11.5 software (SPSS, Inc.) and the differences were determined
using a P-value threshold of 0.05 (P<0.05).
Results
STTM-miR-143 reduces the levels of
miR-143 and its binding to MYO5A in melanocytes
To evaluate whether STTM-miR-143-5p affected miR-143
activity, the expression levels of miR-143 and its predicted target
gene MYO5A were investigated in melanocytes transfected with
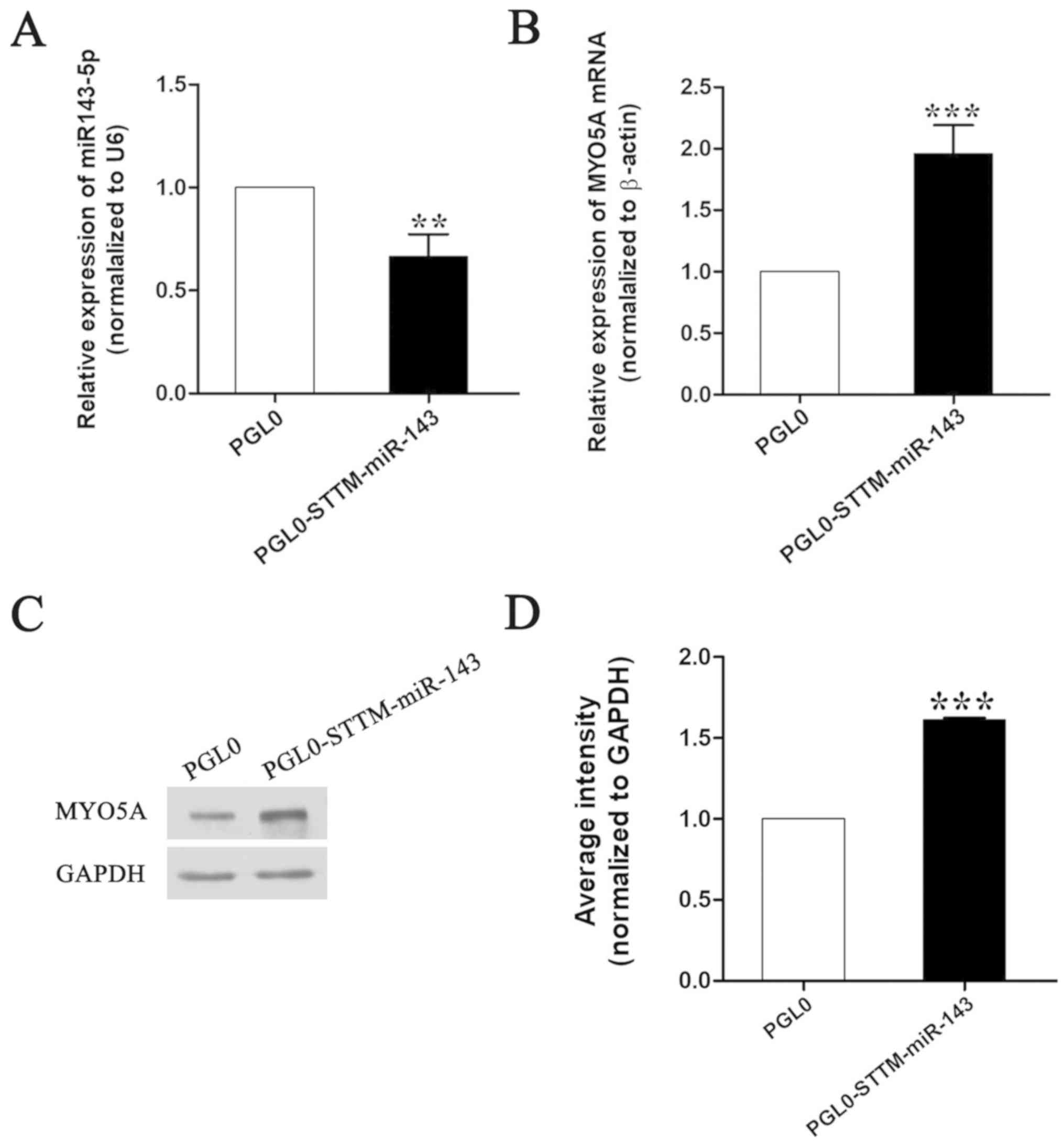
pmirGLO-miR-143 by RT-qPCR. The results indicated that miR-143
expression was significantly decreased and that MYO5A expression
was significantly increased following STTM-miR-143-5p treatment
(Fig. 1).
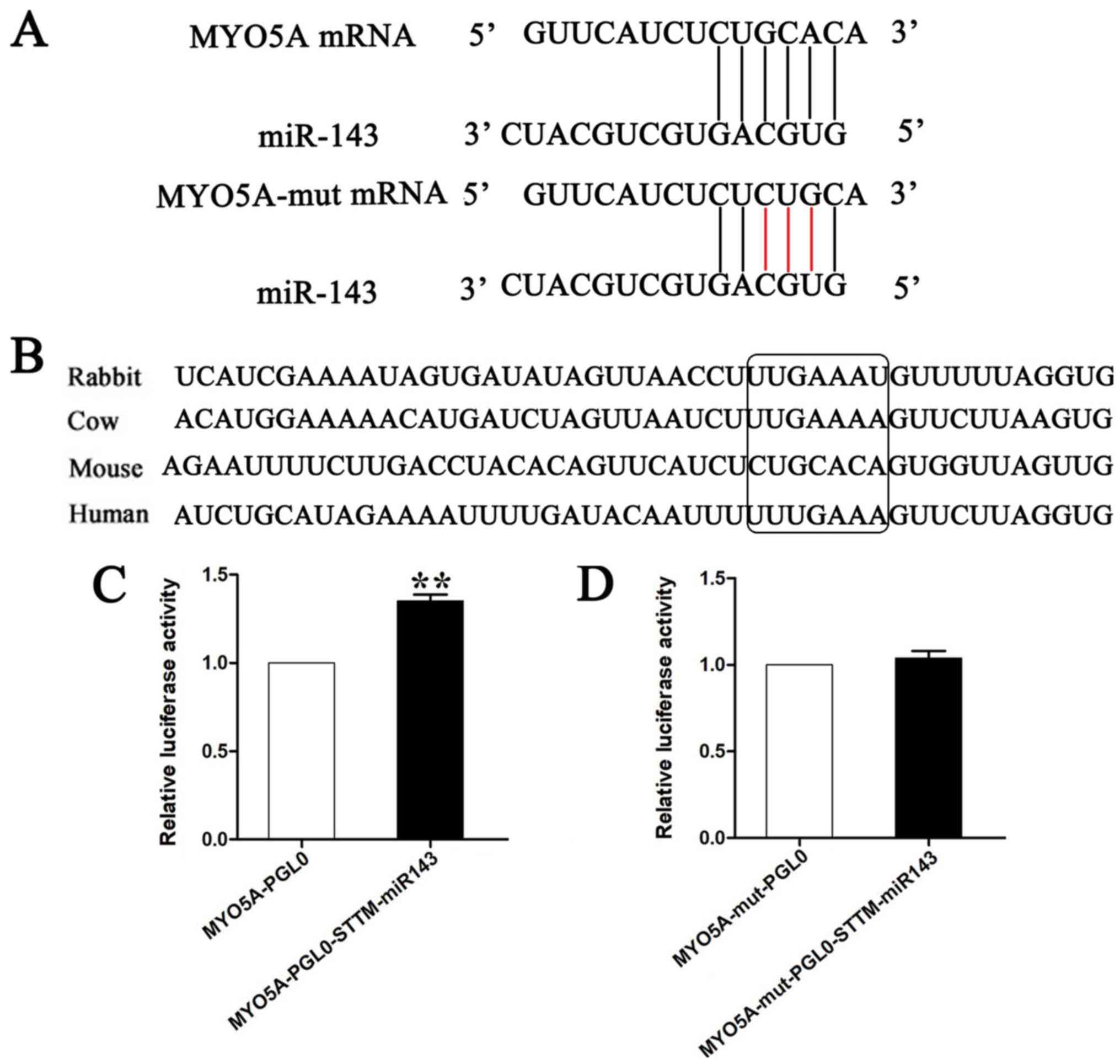
To determine whether binding to conserved target
sites could cause a reduction in the expression levels of MYO5A
(Fig. 2A and B), the plasmids
containing the 3′-UTR of MYO5A were co-transfected into melanocytes
with pmirGLO-STTM-miR-143 or the corresponding pmirGLO null vector.
Co-transfection of the pmirGLO-STTM-miR-143 and the
pmirGLO-MYO5A-wt plasmids or of the empty vector and the
pmirGLO-MYO5A-wt plasmids into the cells indicated that
pmirGLO-STTM-miR-143 significantly increased the luciferase
activity of wild-type of MYO5A. Furthermore, luciferase reporter
assays indicated that point mutations in the sequences of
pmirGLO-MYO5A-mut abolished the effects of endogenous miR-143 by
reducing complementarity between miR-143 and the target sites of
MYO5A (Fig. 2C and D). These data
indicated that miR-143 could bind to the 3′-UTR of MYO5A and
significantly inhibit its expression.
Effect of STTM-miR-143 overexpression
on the melanogenic gene expression
The melanocytes that were transfected with the
pmirGLO-STTM-miR-143 and negative control plasmids were harvested
in order to examine the levels of the melanogenic genes, namely
TYR, TYRP1 and TYRP2. In addition, the levels of the
transcription factor MITF, and of the melanin transport genes
MLPH, and Rab27a were examined. RT-qPCR and western
blotting analyses demonstrated that overexpression of
pmirGLO-STTM-miR-143 in melanocytes significantly increased the
mRNA and protein levels of MLPH, Rab27a, MITF, TYRP1 and TYR.
Conversely, overexpression of pmirGLO-STTM-miR-143 in melanocytes
significantly decreased the mRNA and protein levels of Tyrp2
(Fig. 3).
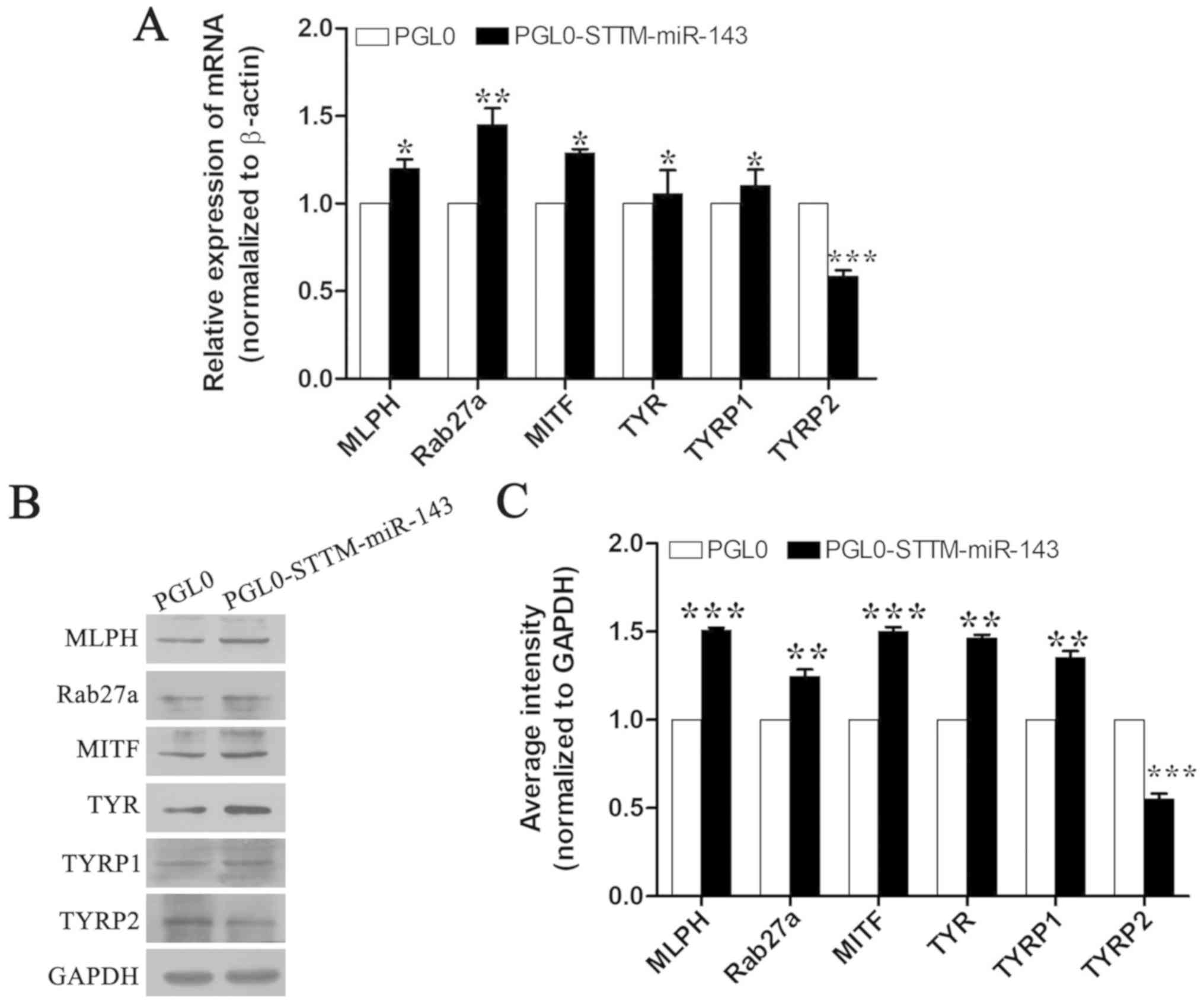 | Figure 3.Effects of STTM-miR-143-5p on the
expression of the melanogenic genes in melanocytes (A) mRNA
expression of the genes MLPH, Rab27a, MITF, TYR, TYRP1, and
TYRP2 in mouse melanocytes transfected with the
STTM-miR-143-5p expression plasmid. (B and C) Analysis of MLPH,
Rab27a, MITF, TYR, TYRP1, and TYRP2 protein expression in
melanocytes transfected with the STTM-miR-143-5p expression
plasmids. The data were normalized to β-actin levels and expressed
as the relative fold change. The data are expressed as the mean ±
SD (n=3). *P<0.05; **P<0.01; ***P<0.001 vs. respective
PGL0. STTM, short tandem target mimic; MLPH, melanophilin;
MITF, microphthalmia-associated transcription factor; TYR,
tyrosinase; TYRP1, tyrosinase-related protein 1;
TYRP2, tyrosinase-related protein 2. |
Effects of STTM-miR-143 overexpression
on melanin production
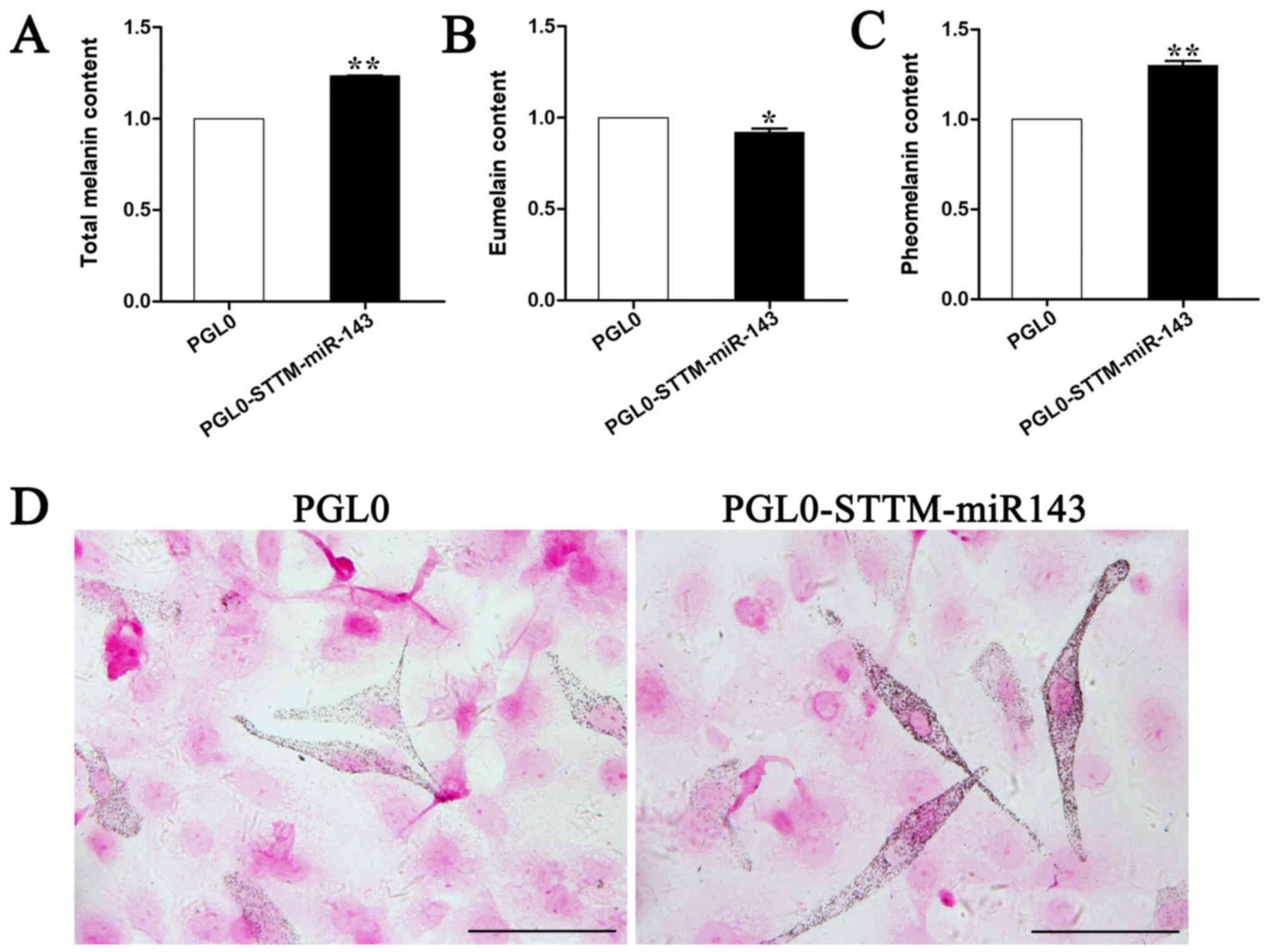
To determine whether pmirGLO-STTM-miR-143
overexpression could affect melanin production, melanocytes
transfected with pmirGLO-STTM-miR-143 plasmid were harvested and
total alkali-soluble melanogenesis (ASM), pheomelanogenesis (PM),
and eumelanogenesis (EM) were measured. The results indicated that
pmirGLO-STTM-miR-143 overexpression in mouse melanocytes increased
ASM and PM by 30% (P<0.01; Fig. 4A
and C), while it decreased EM by approximately 20% (P<0.01;
Fig. 4B). Fontana-Masson staining
confirmed the localization of melanin in the melanocytes.
Overexpression of the pmirGLO-STTM-miR-143-5p plasmids in
melanocytes significantly increased the melanin content and
distribution compared with those noted in the negative control
(Fig. 4D).
Discussion
Genomic analysis has been previously used to
investigate several aspects of pigment cell biology, including
melanocyte- and melanosome-associated functions, pigment disorders,
normal pigmentation variation and even melanoma (17). miRNAs are regulators of cellular
events, such as differentiation, proliferation, and reprogramming
(18), and have often been
implicated in the regulation of melanogenesis for the determination
of hair color (19). Our previous
study identified miR-143 as a negative regulator of proliferation,
migration and melanogenesis in mouse melanocytes by direct
targeting of the TGF-β-activated kinase 1 (TAK1) (20). However, the effects of miR-143
knockdown on melanogenesis have not been previously identified.
STTM is a powerful technology used to block miRNA function in
animals (12). Our previous
studies utilized STTM technology in order to inhibit miRNA-508-3p
and demonstrated its efficacy in upregulating melanogenesis
(21). In the present study, the
same methodology was used to inhibit miR-143 expression in
melanocytes.
Melanocytes located in the skin are derived directly
from the neural crest cells and the embryonic cells that are
present in the skin by the dorsolateral migration pathway (22). They are under the modulation of
intrinsic factors and are controlled by extracellular signals
(22). miR-143 plays a key role in
embryonic stem cell (ESC) pluripotency (23), by targeting the sex-determining
region Y-box 2 (SOX2), the Krüppel-like factor 4 (KLF4), and the
octamer binding transcription factor 4 (OCT4) (20). These functions indicate that
miR-143 promotes ESC differentiation into different cell lineages,
such as neural crest cells (20).
In mammalian skin, melanocytes produce melanin and form mature
melanosomes that complete their pigmentation through their transfer
to adjacent keratinocytes. These structures can also grow hair
shafts at the hair bulb following transportation from the Golgi
region to the dendrite tips (24,25).
Melanocytes respond to environmental stimuli from surrounding
keratinocytes to control differentiation and pigmentation, thereby
determining hair and skin colors (26).
MITF is one of the key regulators of
melanogenesis-limiting enzymes of the tyrosinase family, which
include TYR, TYRP1, and TYRP2/DCT (27), The expression of MITF is modulated
at the transcriptional level by specific transcription factors or
at the post-transcriptional level by miRNAs (28). miR-143-5p can regulate MITF levels
by targeting TAK1 (20). In the
present study, it was demonstrated that MITF was upregulated
in the presence of STTM-miR-143 in mouse melanocytes. MITF
functions via a ‘lineage addiction’ mechanism to control specific
aspects of the phenotypic expression of the melanocytic lineage and
facilitate the transition of cells to a non-invasive stage. TYR and
TYRP1 levels were decreased following miR-143 knockdown by STTM.
However, DCT levels were increased following STTM-miR-143
overexpression. Therefore, it was hypothesized that MITF was not
the only transcription factor involved in the regulation of
TYRP2. In addition, the data were in agreement with those
from a similar study demonstrating that TYR and TYRP1 levels were
markedly decreased (27). The
present study further demonstrated that TYRP2 expression was
increased in MITF-knockdown melanocytes (MITF-KD) (27). Furthermore, TYRP2 expression levels
were not regulated by the content of cellular melanin or by the
levels of other regulatory enzymes in the melanogenic pathway
(29). The mechanism of TYRP2
regulation has not been investigated to date. However, although
MITF has been reported to be essential, it is not sufficient to
induce the expression of melanogenic enzymes (30). In addition, it is a critical
suppressor of innate immunity and can cause innate immune
dysregulation of pigmentation, with implications in vitiligo, which
is an autoimmune depigmentation disease (31).
During melanin transportation, melanosomes are
transported across actin filaments via the association of the motor
protein myosin Va with RAB27a and MLPH (32). RAB27a is a member of the Rab family
of small GTPases, which are important regulators of membrane
transport (33). Myosin Va
requires simultaneous interaction with multiple components of a
complex containing RAB27a and MLPH on the melanosomes (34). The C-terminus of MLPH can directly
interact with actin, which is important for proper transportation
and distribution of the melanosome to the dendrite tips (35). A positive correlation has been
noted between the ability of MLPH to recruit MYO5A to melanosomes
and the promotion of melanosomes in the peripheral retention
(25). The accumulation of the
pigment around the nucleus of the melanocytes and the abnormal
transfer of melanin from melanocyte dendrites to keratinocytes can
lead to albinism (36). miR-143
has been revealed to target MYO5A, and the present study
demonstrated that STTM-miR-143 increased MYO5A levels and
consequently MLPH and Rab27a levels, thus suggesting a stable
interaction among MLPH, Rab27a and MYO5A. The absence of any one of
these components may result in the failure of MYO5A to bind to
melanosomes (34). In summary,
STTM-miR-143 is an artificial biological tool used to control the
melanogenesis by decreasing the expression of miR-143. This process
regulates the levels of MYO5A, and in turn affects melanin
secretion in melanocytes or in melanoma cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Shanxi
Scholarship Council of China (2017–072) and The Young Sanjin
Scholars Distinguished Professor Program (RF).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SQ and BL performed plasmid construction, cell
culture and transfection, and gene expression experiments. JZ and
XL performed melanin production, and analyzed and interpreted the
data. CD and RF contributed to the conception and design of the
experiments, and critically revised the manuscript for important
intellectual content. SQ contributed to drafting the manuscript.
All authors have read and approved the final manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ito S and Wakamatsu K: Chemistry of mixed
melanogenesis-pivotal roles of dopaquinone. Photochem Photobiol.
84:582–592. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vachtenheim J and Borovansky J:
‘Transcription physiology’ of pigment formation in melanocytes:
Central role of MITF. Exp Dermatol. 19:617–627. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Levy C, Khaled M and Fisher DE: MITF:
Master regulator of melanocyte development and melanoma oncogene.
Trends Mol Med. 12:406–414. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bertolotto C, Busca R, Abbe P, Bille K,
Aberdam E, Ortonne JP and Ballotti R: Different cis-acting elements
are involved in the regulation of TRP1 and TRP2 promoter activities
by cyclic AMP: Pivotal role of M boxes (GTCATGTGCT) and of
microphthalmia. Mol Cell Biol. 18:694–702. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hume AN and Seabra MC: Melanosomes on the
move: A model to understand organelle dynamics. Biochem Soc Trans.
39:1191–1196. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Matesic L, Yip R, Reuss AE, Swing DA,
O'Sullivan TN, Fletcher CF, Copeland NG and Jenkins NA: Mutations
in Mlph, encoding a member of the Rab effector family, cause the
melanosome transport defects observed in leaden mice. Proc Natl
Acad Sci USA. 98:10238–10243. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kuroda TS, Ariga H and Fukuda M: The
actin-binding domain of Slac2-a/melanophilin is required for
melanosome distribution in melanocytes. Mol Cell Biol.
23:5245–5255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu X, Sakamoto T, Zhang F, Sell JR and
Hammer JA III: In vitro reconstitution of a transport complex
containing Rab27a, melanophilin and myosin Va. FEBS Lett.
580:5863–5868. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rudolf R, Bittins CM and Gerdes HH: The
role of myosin V in exocytosis and synaptic plasticity. J
Neurochem. 116:177–191. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alves CP, Yokoyama S, Goedert L, Pontes
CLS, Sousa JF, Fisher DE and Espreafico EM: MYO5A gene is a target
of MITF in melanocytes. J Invest Dermatol. 137:985–989. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Au SY and Huang JD: A tissue-specific exon
of myosin Va is responsible for selective cargo binding in
melanocytes. Cell Motility Cytoskeleton. 53:89–102. 2002.
View Article : Google Scholar
|
|
12
|
Tang G, Yan J, Gu Y, Qiao M, Fan R, Mao Y
and Tang X: Construction of short tandem target mimic (STTM) to
block the functions of plant and animal microRNAs. Methods.
58:118–125. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bai R, Sen A, Yu Z, Yang G, Wang H, Fan R,
Lv L, Lee KB, Smith GW and Dong C: Validation of methods for
isolation and culture of alpaca melanocytes: A novel tool for in
vitro studies of mechanisms controlling coat color. Asian Austral J
Anim. 23:430–436. 2010. View Article : Google Scholar
|
|
14
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong C, Wang H, Xue L, Dong Y, Yang L, Fan
R, Yu X, Tian X, Ma S and Smith GW: Coat color determination by
miR-137 mediated down-regulation ofmicrophthalmia-associated
transcription factor in a mouse model. RNA. 18:1679–1686. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Loftus SK: The next generation of
melanocyte data: Genetic, epigenetic, and transcriptional resource
datasets and analysis tools. Pigment Cell Melanoma Res. 31:442–447.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cordes KR, Sheehy NT, White MP, Berry EC,
Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN and Srivastava D:
miR-145 and miR-143 regulate smooth muscle cell fate and
plasticity. Nature. 460:705–710. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim KH, Lee TR and Cho EG: SH3BP4, a novel
pigmentation gene, is inversely regulated by miR-125b and MITF. Exp
Mol Med. 49:e3672017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ji K, Zhang P, Zhang J, Fan R, Liu Y, Yang
S, Hu S, Liu X and Dong C: MicroRNA 143-5p regulates alpaca
melanocyte migration, proliferation, and melanogenesis. Exp
Dermatol. 27:166–171. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu B, Zhang J, Yang S, Ji K, Liu X, Du B,
Jia Q, Qi S, Li X and Fan R: Effect of silencing microrna-508 by
STTM on melanogenesis in alpaca (Vicugna pacos). Gene. 678:343–348.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sommer L: Generation of melanocytes from
neural crest cells. Pigment Cell Melanoma Res. 24:411–421. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tian X, Jiang J, Fan R, Wang H, Meng X, He
X, He J, Li H, Geng J, Yu X, et al: Identification and
characterization of microRNAs in white and brown alpaca skin. BMC
Genomics. 13:5552012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jackson IJ: Molecular and developmental
genetics of mouse coat color. Annu Rev Genet. 28:189–217. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hume AN, Tarafder AK, Ramalho JS,
Sviderskaya EV and Seabra MC: A coiled-coil domain of melanophilin
is essential for Myosin Va recruitment and melanosome transport in
melanocytes. Mol Biol Cell. 17:4720–4735. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cichorek M, Wachulska M, Stasiewicz A and
Tymińska A: Skin melanocytes: Biology and development. Postepy
Dermatol Alergol. 30:30–41. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang P, Li Y, Hong W, Zhen J, Ren J, Li Z
and Xu A: The changes of microRNA expression profiles and
tyrosinase related proteins in MITF knocked down melanocytes. Mol
Biosyst. 8:2924–2931. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hartman ML and Czyz M: MITF in melanoma:
Mechanisms behind its expression and activity. Cell Mol Life Sci.
72:1249–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pak BJ, Li Q, Kerbel RS and Ben-David Y:
TYRP2-mediated resistance to cis-diamminedichloroplatinum (II) in
human melanoma cells is independent of tyrosinase and TYRP1
expression and melanin content. Melanoma Res. 10:499–505. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gaggioli C, Buscà R, Abbe P, Ortonne JP
and Ballotti R: Microphthalmia-associated transcription factor
(MITF) is required but is not sufficient to induce the expression
of melanogenic genes. Pigment Cell Res. 16:374–382. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Harris ML, Fufa TD, Palmer JW, Joshi SS,
Larson DM, Incao A, Gildea DE, Trivedi NS, Lee AN, Day CP, et al: A
direct link between MITF, innate immunity, and hair graying. PLoS
Biol. 16:e20036482018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chang H, Choi H, Joo KM, Kim D and Lee TR:
Manassantin B inhibits melanosome transport in melanocytes by
disrupting the melanophilin-myosin Va interaction. Pigment Cell
Melanoma Res. 25:765–772. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zerial M and McBride H: Rab proteins as
membrane organizers. Nat Rev Mol Cell Biol. 2:107–117. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hume AN, Collinson LM, Hopkins CR, Strom
M, Barral DC, Bossi G, Griffiths GM and Seabra MC: The leaden gene
product is required with Rab27a to recruit myosin Va to melanosomes
in melanocytes. Traffic. 3:193–202. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kuroda TS, Itoh T and Fukuda M: Functional
analysis of slac2-a/melanophilin as a linker protein between Rab27A
and myosin Va in melanosome transport. Methods Enzymol.
403:419–431. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Marks MS and Seabra MC: The melanosome:
Membrane dynamics in black and white. Nat Rev Mol Cell Biol.
2:738–748. 2001. View
Article : Google Scholar : PubMed/NCBI
|