Introduction
The incidence of esophageal squamous cell carcinoma
(ESCC) was ~24.4 males and ~4 females per 100,000 individuals in
Japan in 2004; 3.4% of mortalities from all malignant neoplasms are
due to ESCC (1). ESCC has high
biological malignant potential amongst digestive tract cancers due
to the frequent involvement of lymph node metastasis and tumor
invasion of adjacent organs at the early stages (2). Therefore, it is crucial to identify
biomarkers for predicting the malignant potential of ESCC.
DNA copy number alterations, which are associated
with the dysregulation of gene expression, are considered to have a
critical role in the pathogenesis of human cancers (3). The amplification and overexpression of
human epidermal growth factor receptor 2 (HER2) are
associated with poor prognosis in patients with breast cancer, and
HER2 is an effective therapeutic target for trastuzumab
(4). In ESCC, integrative analysis of
copy number and gene expression profiles may facilitate the
identification of genes associated with tumor progression (5–7); the
current study aimed to investigate the genetic and transcriptional
alterations in ESCC.
The caspase-4 (CASP4) gene encodes a protein
involved in immunity and inflammation (8). Previous studies into cell death have
demonstrated that endoplasmic reticulum (ER) stress induces
CASP4-mediated cell apoptosis (9–11).
Although pro-apoptotic caspases are downregulated in certain
cancers (12), few previous clinical
studies have focused on CASP4. Notably, CASP4
expression is suppressed, and is associated with poor prognosis, in
head and neck squamous cell carcinoma (13,14).
Loss-of-function CASP4 mutations are rarely observed in
colorectal cancers (15); however,
the clinical significance and role of CASP4 have yet to be
evaluated in ESCC. The aim of the present study was to elucidate
the clinical role of CASP4 expression in ESCC. The
association between the expression levels and copy number profiles
of CASP4 in tumor samples was investigated. Furthermore,
gene set enrichment analysis (GSEA) (16) was performed to identify the signaling
pathways involved in CASP4 expression in ESCC; the
association between the expression levels of ER stress markers and
CASP4 was also analyzed. Finally, the present study assessed
the significance of CASP4 expression levels in the prognosis
of ESCC.
Materials and methods
Sample collection, DNA extraction and
RNA extraction
ESCC tumor and adjacent tissue samples were
collected from a total of 157 patients diagnosed with ESCC who
underwent surgical resection at the following five institutions:
Juntendo University Hospital (Tokyo, Japan), National Cancer Center
(Tokyo, Japan), Kurume University Hospital (Kurume, Japan), Saitama
Cancer Center (Saitama, Japan), and Kagoshima University Medical
and Dental Hospital (Kagoshima, Japan). The mean age of the
patients was 65.5 years (age range, 40–83 years), with a 137:17
male:female ratio (clinical information was available for 154/157
patients). ESCC tissues samples with tumor stages T1−T4 were
included in the study. The tissue samples were randomly divided
into two sets, with 78 samples included in the discovery set
(clinical information including survival profile was available for
75/78 cases) and 79 samples in the validation set (clinical
information was available for 79 cases); each dataset included the
above-mentioned institutions. In the discovery set, gene expression
profiles were analyzed using human gene expression microarrays
(Whole Human Genome Microarray kit; 4×44; cat. no. G4112F; Agilent
Technologies, Inc., Santa Clara, CA, USA). Array-comparative
genomic hybridization (CGH) was also performed in 57/78 cases in
the discovery set, as genomic DNA was available in these cases. In
the validation set, CASP4 expression levels were analyzed
using reverse transcription quantitative polymerase chain reaction
(RT-qPCR).
Cells were isolated using a laser microdissection
system (LMD; Leica Microsystems GmbH, Wetzlar, Germany) and DNA and
RNA samples were purified and extracted using a QIAamp DNA Micro
kit (Qiagen GmbH, Hilden, Germany) and an RNeasy Micro kit (Qiagen
GmbH). Written informed consent was obtained from all patients and
the study protocol was reviewed and approved by the internal review
board of Kyushu University (Fukuoka, Japan).
Array-CGH and copy number
analysis
Genomic DNA samples from 57 tumor specimens were
analyzed using array-CGH. The genomic DNA of 3 samples from the
normal esophageal mucosa was also analyzed and used as a reference
for the array-CGH. DNA labeling and hybridization were performed
using a Genome Microarray kit 244K (Agilent Technologies, Inc.).
The data were evaluated using a microarray scanner (Agilent
Technologies, Inc.) and analyzed using the Feature Extraction
software version 9.1 (Agilent Technologies, Inc.).
Expression array
An expression array was performed using the 78 tumor
samples in the discovery set. The total RNA extracted using the LMD
technique was reverse transcribed to generate double-stranded cDNA.
Amplification was then performed using a T7 RNA polymerase (Agilent
Technologies, Inc.), and the product was converted to
cyanine-labeled cRNA. The labeled cRNA was fragmented and
hybridized to an oligonucleotide microarray (Whole Human Genome
Microarray kit, 4×44; G4112F; Agilent Technologies, Inc.).
Fluorescence intensities were obtained using an Agilent DNA
microarray scanner and subjected to quantile normalization.
GSEA
GSEA was performed using the gene expression data
from the discovery set to investigate how signaling pathways were
differentially regulated depending on the expression levels of
CASP4 in ESCC. The present study used a continuous-type
class file with the CASP4 profile to phenotype labels in
GSEA; the expression level of CASP4 in each sample was used
as an input data in GSEA. The gene sets extracted from the Broad
Institute (Cambridge, MA, USA) database (http://software.broadinstitute.org/gsea/msigdb/collections.jsp)
included gene sets based on the gene ontology pathway
(apoptosis_go, inflammatory_response, and immune_response), gene
sets annotated by the reactome pathway (reactome_apoptosis and
reactome_innate_immune_system) and a gene set based on the biocarta
pathway, biocarta_inflam_pathway.
RT-qPCR
RT-qPCR was performed using a LightCycler system
(Roche Diagnostics, Indianapolis, IN, USA) and a LightCycler 480
Probes Master kit (Roche Diagnostics) according to the
manufacturer's protocol. The following thermal cycling conditions
were used: Initial denaturation at 95°C for 10 min, followed by 45
cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 30
sec, and extension at 40°C for 30 sec. The CASP4 primers
were as follows: Forward, 5′-TTCCTGGCAATTGAAAATGG-3′, and reverse,
5′-TGCAAGCTGTACTAATGAAGGTG-3′. The concentrations of CASP4
in each sample were calculated by plotting their crossing points
against the standard curve from a single experiment. The
CASP4 mRNA level was normalized to the internal standard,
glyceraldehyde 3-phosphate dehydrogenase assessed using the
following primers: GAPDH forward, 5′-AGCCACATCGCTCAGACAC-3′,
and reverse, 5′-GCCCAATACGACCAAATCC-3′.
Statistical analysis
Student's t-tests and Fisher's exact tests were used
to determine significant differences between the groups. The
Kaplan-Meier method was used to evaluate the survival rates and the
survival curves were compared using log-rank tests. Overall
survival was calculated from the date of surgical resection to date
of death/final follow-up. Statistical analysis was performed using
JMP version 5 software (SAS Institute, Buckinghamshire, UK).
P<0.05 was considered to indicate a statistically significant
result.
Results
Copy number loss inhibits CASP4
expression in ESCC
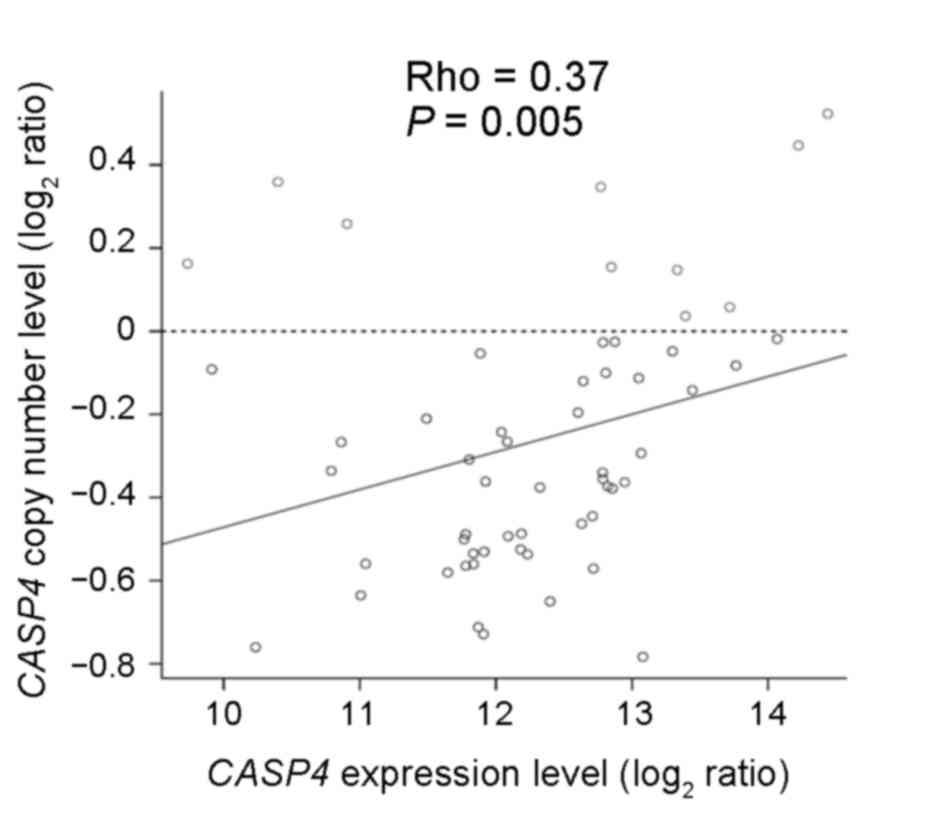
The copy number alterations accompanying changes in
CASP4 gene expression were analyzed using the array-CGH and
the expression array data from the ESCC discovery set. The present
study demonstrated that CASP4 copy number loss occurred in
47/57 ESCC samples and that there was a significant association
between the copy number and expression levels (Spearman's
correlation, rho=0.37; P=0.005; Fig.
1). In ESCC, copy number loss appeared to suppress CASP4
gene expression, suggesting that CASP4 may act as a
tumor-suppressor gene.
Higher CASP4 expression levels were
significantly associated with the signaling pathways involved in
apoptosis, inflammatory responses, and immune responses in
ESCC
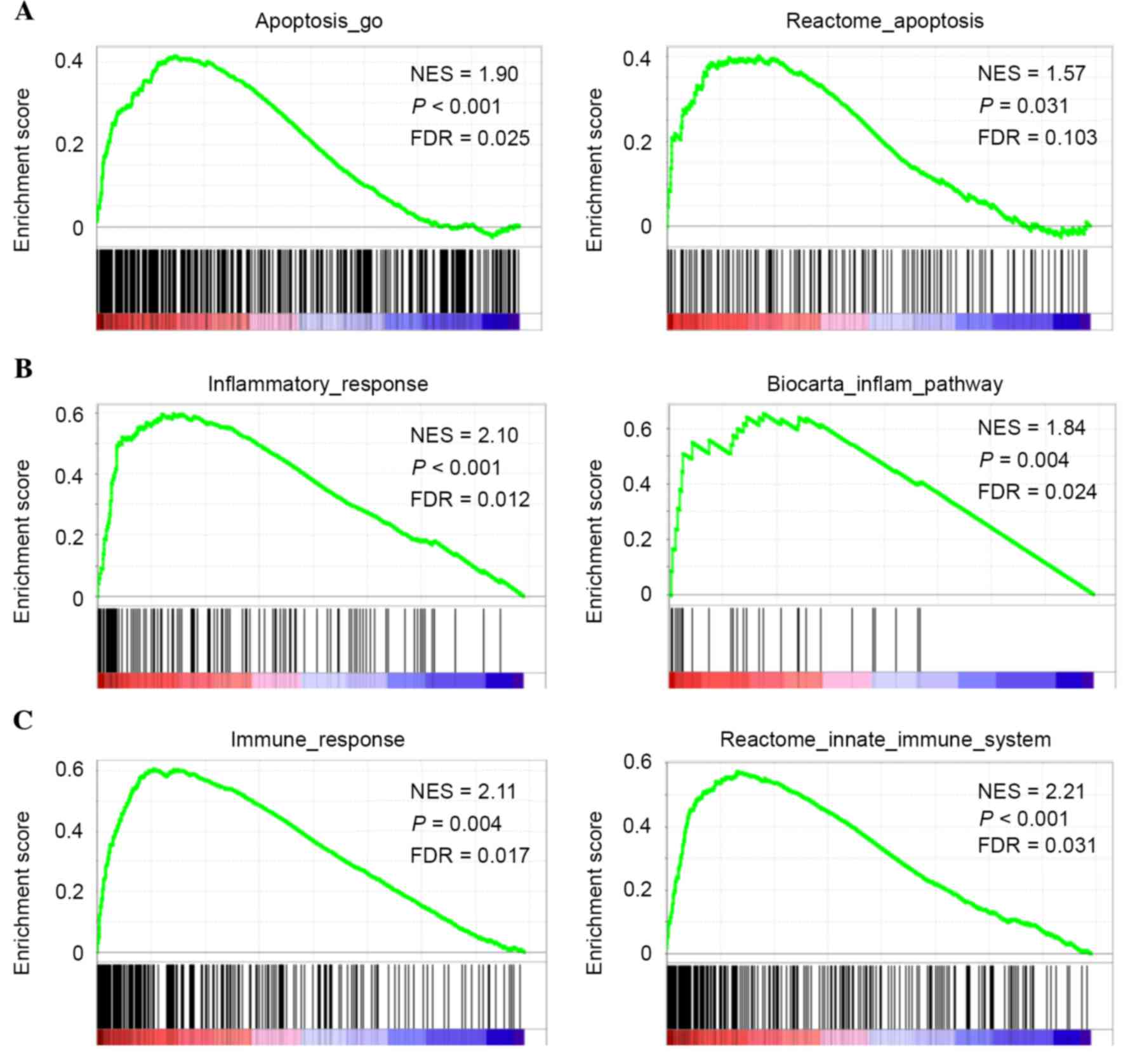
GSEA of the 78 ESCC samples from the discovery set
revealed that certain gene sets involved in the apoptotic signaling
pathway were significantly upregulated in ESCCs expressing high
levels of CASP4 as follows: Apoptosis_go, P<0.001, false
discovery rate (FDR)=0.025; reactome_apoptosis P=0.031, FDR=0.103
(Fig. 2A). The present study also
demonstrated that higher CASP4 expression was significantly
associated with the enrichment of gene sets involved in the
inflammatory response, including inflammatory_response (P<0.001;
FDR=0.012) and biocarta_inflam_pathway (P=0.004; FDR=0.024;
Fig. 2B), and of gene sets involved
in the immune response, including immune_response (P=0.004;
FDR=0.017) and reactome_innate_immune_system (P<0.001;
FDR=0.031; Fig. 2C). These data were
consistent with CASP4 being an inflammatory caspase and a
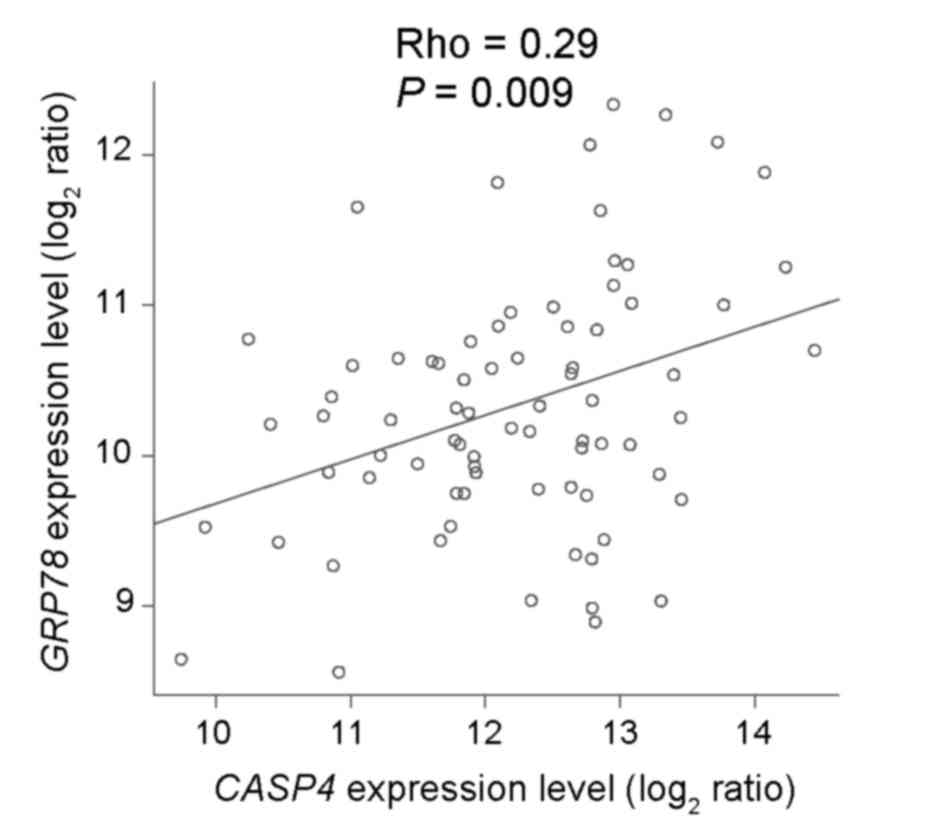
critical mediator of the innate immune response (8). The current study subsequently evaluated
the association between the expression levels of CASP4 and
the ER chaperone glucose-regulated protein (GRP) 78, which
is used as an ER stress marker (9–11,17). CASP4 expression levels were
positively associated with GRP78 expression levels
(Spearman's correlation, rho=0.29; P=0.009; Fig. 3), demonstrating that CASP4 has
a role in regulating ER stress-induced cell death in ESCC.
CASP4 expression levels predict the
prognosis of patients with ESCC
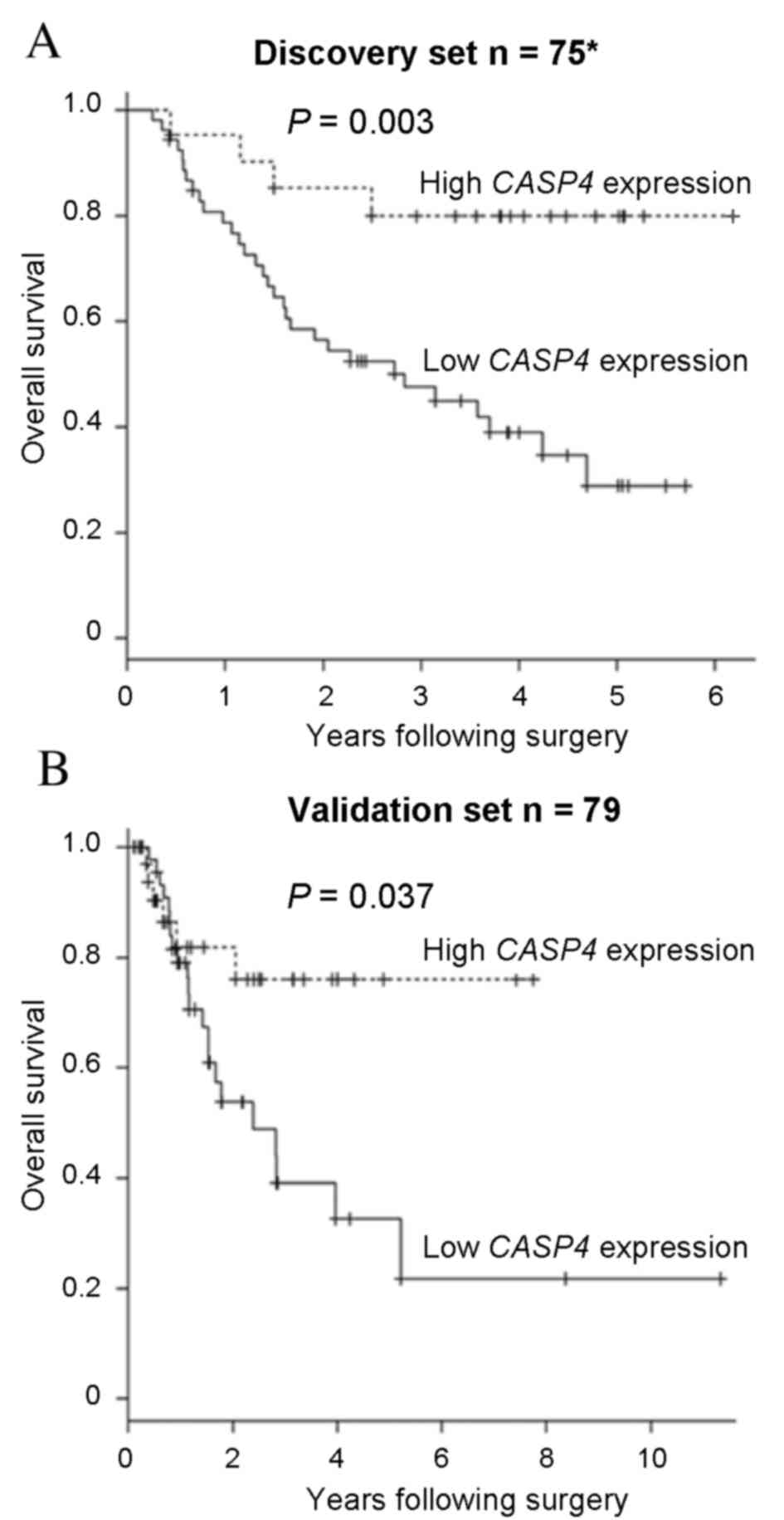
To examine the clinical significance of CASP4
expression in ESCC, a survival analysis was performed. The present
study identified that low CASP4 expression levels were
significantly associated with a poor overall survival rate in the
discovery set (P=0.003; Fig. 4A). To
investigate the clinical significance of CASP4 expression
levels in ESCC, the validation set was also analyzed. Low
CASP4 expression levels were observed to be significantly
associated with lymphatic invasion (P=0.003; Table I). The overall survival rate for the
validation set was also analyzed, and revealed no significant
differences in the overall survival rate between the discovery and
validation sets (P=0.805). Concordant with the discovery set data,
the CASP4 low expression group had a significantly poorer
overall survival rate, compared with the CASP4 high
expression group, in the validation set (P=0.037; Fig. 4B). These results suggest that
CASP4 may have a tumor suppressor role in ESCC.
 | Table I.CASP4 mRNA expression and
clinicopathological factors in the validation set. |
Table I.
CASP4 mRNA expression and
clinicopathological factors in the validation set.
| Factor | High CASP4
expression n=39 | Low CASP4
expression n=40 | P-value |
|---|
| Age (mean ± SD) | 65.92±7.87 | 63.38±8.43 | 0.169a |
| Gender |
|
|
|
| Male | 35 | 36 | 0.973b |
|
Female | 4 | 4 |
|
| Tumor
differentiation |
|
|
|
| Well | 15 | 14 | 0.818b |
| Mod or
poor | 24 | 26 |
|
| Depth |
|
|
|
| T1 | 4 | 5 | 1.000b |
| T2-4 | 35 | 35 |
| Lymph node
metastasis |
|
|
|
|
Negative | 12 | 10 | 0.622b |
|
Positive | 27 | 30 |
|
| Lymphatic
invasion |
|
|
|
|
Negative | 12 | 2 | 0.003b,c |
|
Positive | 27 | 38 |
|
| Venous
invasion |
|
|
|
|
Negative | 7 | 4 | 0.348b |
|
Positive | 32 | 36 |
|
Discussion
The current study demonstrated that the
downregulation of CASP4 expression levels is associated with
ESCC progression. The gene expression and copy number profiles of
clinical tissue samples were analyzed using a bioinformatics
approach and demonstrated that the copy number loss of CASP4
was associated with the decreased expression levels of CASP4
observed in ESCC. The CASP4 low expression group had a poor
prognosis, compared with the CASP4 high expression group;
these results were reproducible in each clinical ESCC cohort. These
results suggest that CASP4 may function as a
tumor-suppressor gene and may be a useful biomarker for predicting
the prognosis in ESCC.
CASP4 has been previously reported to induce
cell death (9–11). A few studies have identified that
CASP4 is involved in ER stress-induced apoptosis in
neurodegenerative disorders (9),
muscular dystrophy (10) and retinal
pigment epithelial cells (11).
Concordantly, the present study demonstrated that CASP4
expression levels were significantly associated with the apoptotic
signaling pathway and the expression levels of GRP78 in
ESCC. Furthermore, CASP4 encodes a protein involved in
inflammation and immune responses (8). Kobayashi et al (18) reported that CASP4 is involved
in the innate immune response and inflammatory cell death in
bacterial infection. Similarly, the current study indicated that
CASP4 was significantly associated with the inflammatory and
immune responses that may contribute to the inhibition of ESCC
progression.
The data demonstrates that low CASP4
expression is significantly associated with lymphatic invasion. We
hypothesized that CASP4 expression may contribute to the
early phase of ESCC progression, as cell death, including
apoptosis, is more frequent during the early phase of tumor
progression, and lymphatic invasion occurs earlier than lymph node
metastasis. Therefore, it is possible that CASP4 expression
levels may predict not only the prognosis, but also early phase
tumor progression in ESCC.
In conclusion, the current study indicated that
CASP4 may be a tumor-suppressor gene associated with the
signaling pathways underlying apoptosis, inflammatory responses and
immune responses in ESCC. The results also suggested that ER stress
induces CASP4-mediated apoptosis. CASP4 expression
may therefore be a useful clinical biomarker for predicting the
prognosis of patients with ESCC.
Acknowledgements
This study used the supercomputing resources
provided by the Human Genome Center, Institute of Medical Science,
University of Tokyo. This study was supported by the following
foundations: Grants-in-Aid for Scientific Research (grant nos.
26861003, 15H04921, 25461953 and 21229015); Funding Program for
Next Generation World-Leading Researchers (grant no. LS094).
References
|
1
|
Kuwano H, Nishimura Y, Oyama T, Kato H,
Kitagawa Y, Kusano M, Shimada H, Takiuchi H, Toh Y, Doki Y, et al:
Guidelines for diagnosis and treatment of carcinoma of the
esophagus April 2012 edited by the Japan Esophageal Society.
Esophagus. 12:1–30. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Solomon E, Borrow J and Goddard AD:
Chromosome aberrations and cancer. Science. 254:1153–1160. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yan M, Parker BA, Schwab R and Kurzrock R:
HER2 aberrations in cancer: Implications for therapy. Cancer Treat
Rev. 40:770–780. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sawada G, Niida A, Hirata H, Komatsu H,
Uchi R, Shimamura T, Takahashi Y, Kurashige J, Matsumura T, Ueo H,
et al: An integrative analysis to identify driver genes in
esophageal squamous cell carcinoma. PLoS One. 10:e01398082015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu N, Wang C, Ng D, Clifford R, Yang HH,
Tang ZZ, Wang QH, Han XY, Giffen C, Goldstein AM, et al: Genomic
characterization of esophageal squamous cell carcinoma from a
high-risk population in China. Cancer Res. 69:5908–5917. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi ZZ, Shang L, Jiang YY, Hao JJ, Zhang
Y, Zhang TT, Lin DC, Liu SG, Wang BS, Gong T, et al: Consistent and
differential genetic aberrations between esophageal dysplasia and
squamous cell carcinoma detected by array comparative genomic
hybridization. Clin Cancer Res. 19:5867–5878. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McIlwain DR, Berger T and Mak TW: Caspase
functions in cell death and disease. Cold Spring Harb Perspect
Biol. 5:a0086562013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hitomi J, Katayama T, Eguchi Y, Kudo T,
Taniguchi M, Koyama Y, Manabe T, Yamagishi S, Bando Y, Imaizumi K,
et al: Involvement of caspase-4 in endoplasmic reticulum
stress-induced apoptosis and Abeta-induced cell death. J Cell Biol.
165:347–356. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Moorwood C and Barton ER: Caspase-12
ablation preserves muscle function in the mdx mouse. Hum Mol Genet.
23:5325–5341. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bian ZM, Elner SG and Elner VM: Dual
involvement of caspase-4 in inflammatory and ER stress-induced
apoptotic responses in human retinal pigment epithelial cells.
Invest Ophthalmol Vis Sci. 50:6006–6014. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Philchenkov A, Zavelevich M, Kroczak TJ
and Los M: Caspases and cancer: Mechanisms of inactivation and new
treatment modalities. Exp Oncol. 26:82–97. 2004.PubMed/NCBI
|
|
13
|
Li H, Wang J, Zeng Z, Fu X and Zhang W:
Expression and correlation of apoptosis-related gene c-IAP2 and
caspase-4 in head and cervical undifferentiation squamous cell
carcinoma. Lin Chuang Er Bi Yan Hou Ke Za Zhi. 17:739–741. 2003.(In
Chinese). PubMed/NCBI
|
|
14
|
Li H, Zeng ZH, Wang LH and Wang JQ:
Expression and correlation of apoptosis-related gene c-IAP2 and
caspase-4 in sinonasal squamous carcinoma. Zhonghua Er Bi Yan Hou
Ke Za Zhi. 39:324–327. 2004.(In Chinese). PubMed/NCBI
|
|
15
|
Soung YH, Jeong EG, Ahn CH, Kim SS, Song
SY, Yoo NJ and Lee SH: Mutational analysis of caspase 1, 4, and 5
genes in common human cancers. Hum Pathol. 39:895–900. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moenner M, Pluquet O, Bouchecareilh M and
Chevet E: Integrated endoplasmic reticulum stress responses in
cancer. Cancer Res. 67:10631–10634. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kobayashi T, Ogawa M, Sanada T, Mimuro H,
Kim M, Ashida H, Akakura R, Yoshida M, Kawalec M, Reichhart JM, et
al: The Shigella OspC3 effector inhibits caspase-4, antagonizes
inflammatory cell death, and promotes epithelial infection. Cell
Host Microbe. 13:570–583. 2013. View Article : Google Scholar : PubMed/NCBI
|