|
1
|
Zhu AX, Hong TS, Hezel AF and Kooby DA:
Current management of gallbladder carcinoma. Oncologist.
15:168–181. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miller G and Jarnagin WR: Gallbladder
carcinoma. Eur J Surg Oncol. 34:306–312. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Randi G, Franceschi S and La Vecchia C:
Gallbladder cancer worldwide: Geographical distribution and risk
factors. Int J Cancer. 118:1591–1602. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Misra S, Chaturvedi A, Misra NC and Sharma
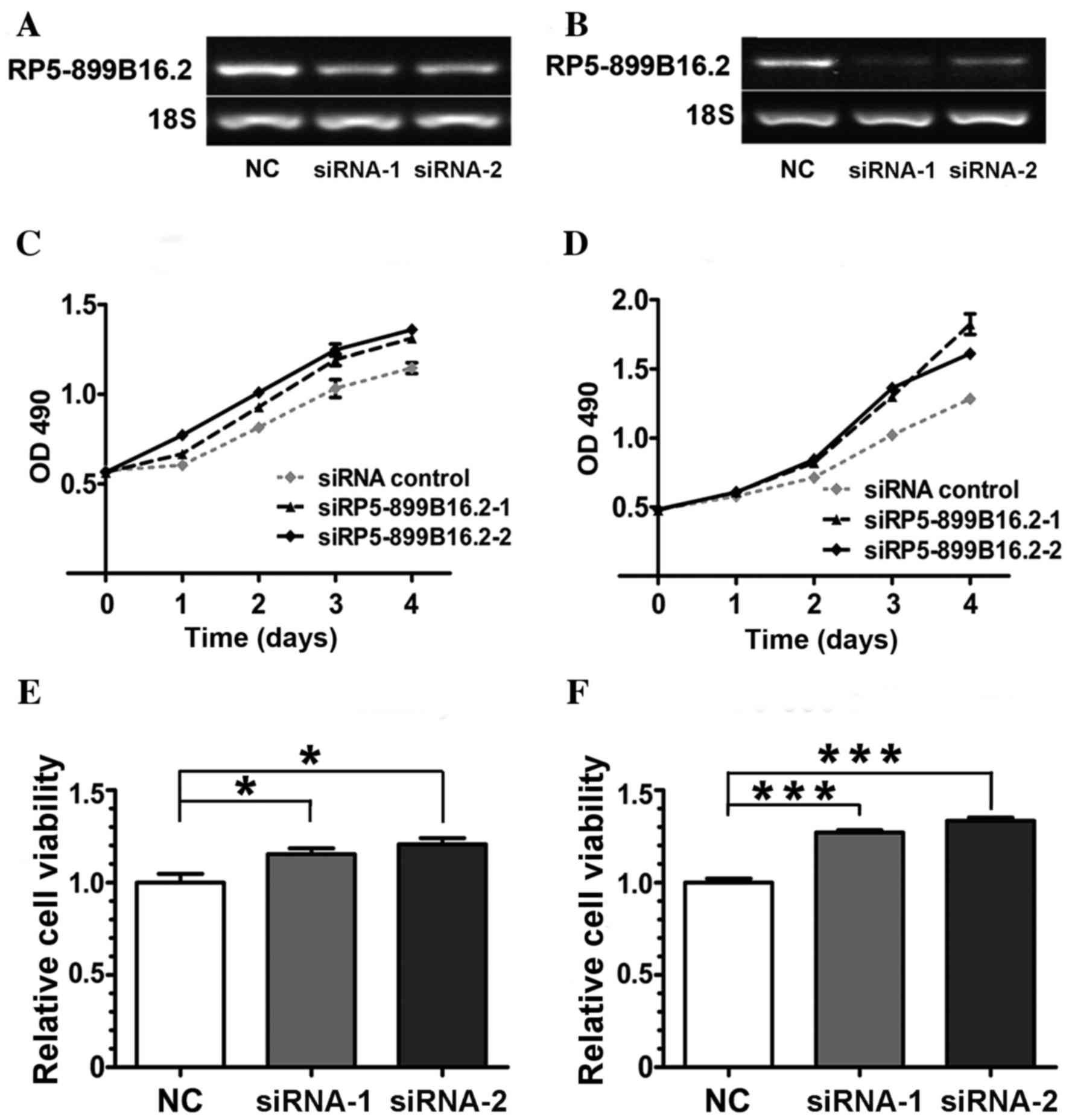
ID: Carcinoma of the gallbladder. Lancet Oncol. 4:167–176. 2003.
View Article : Google Scholar : PubMed/NCBI
|
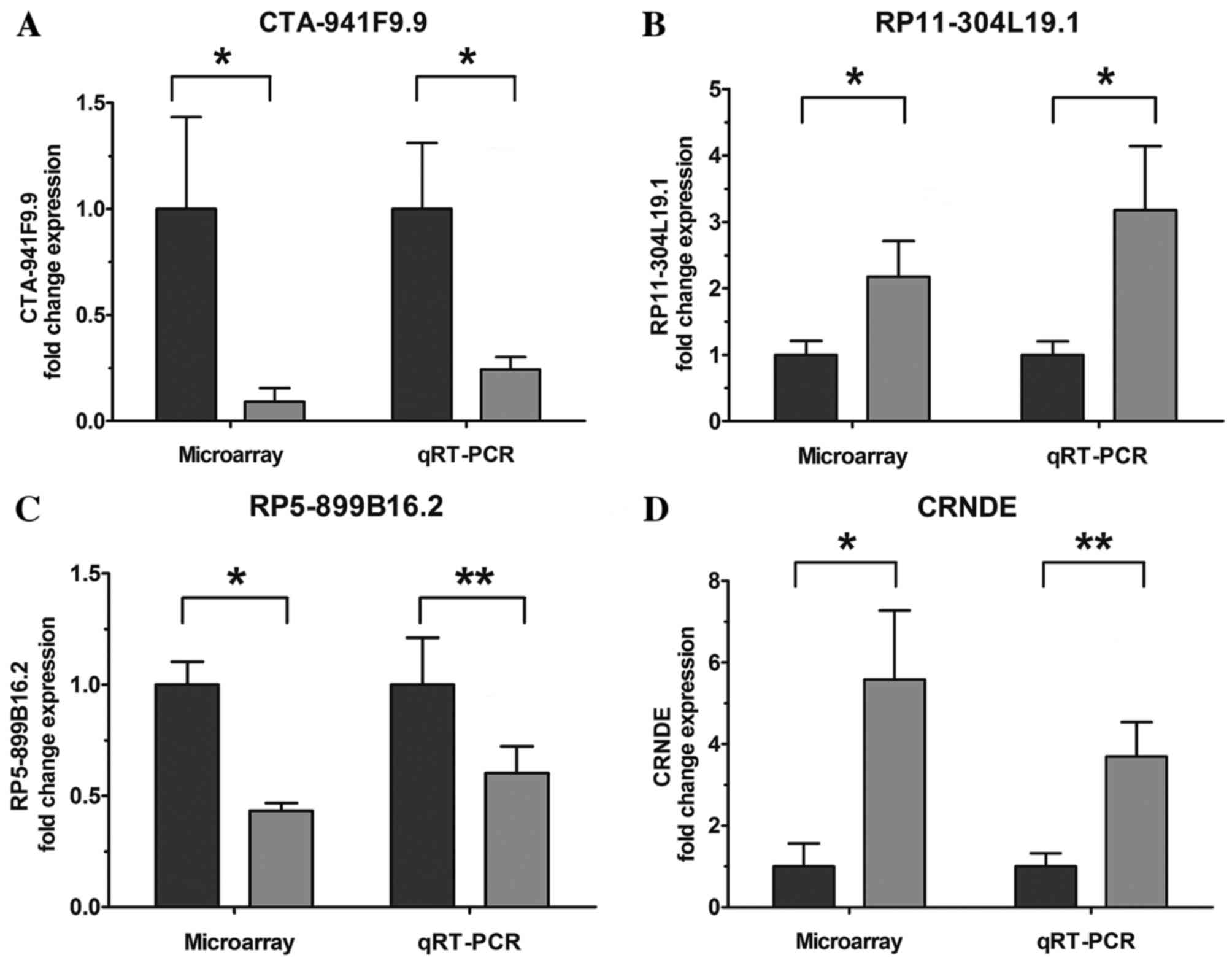
|
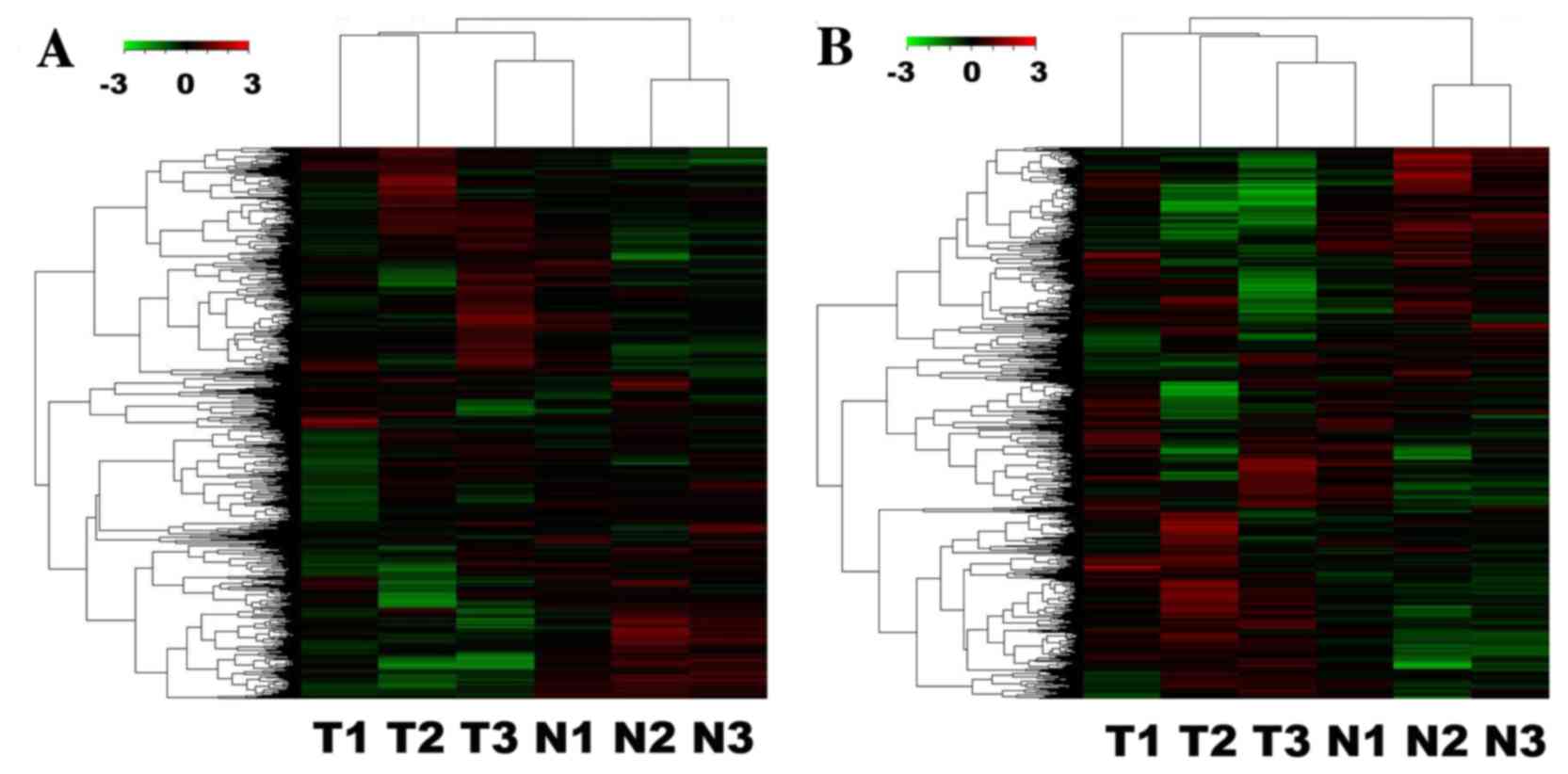
5
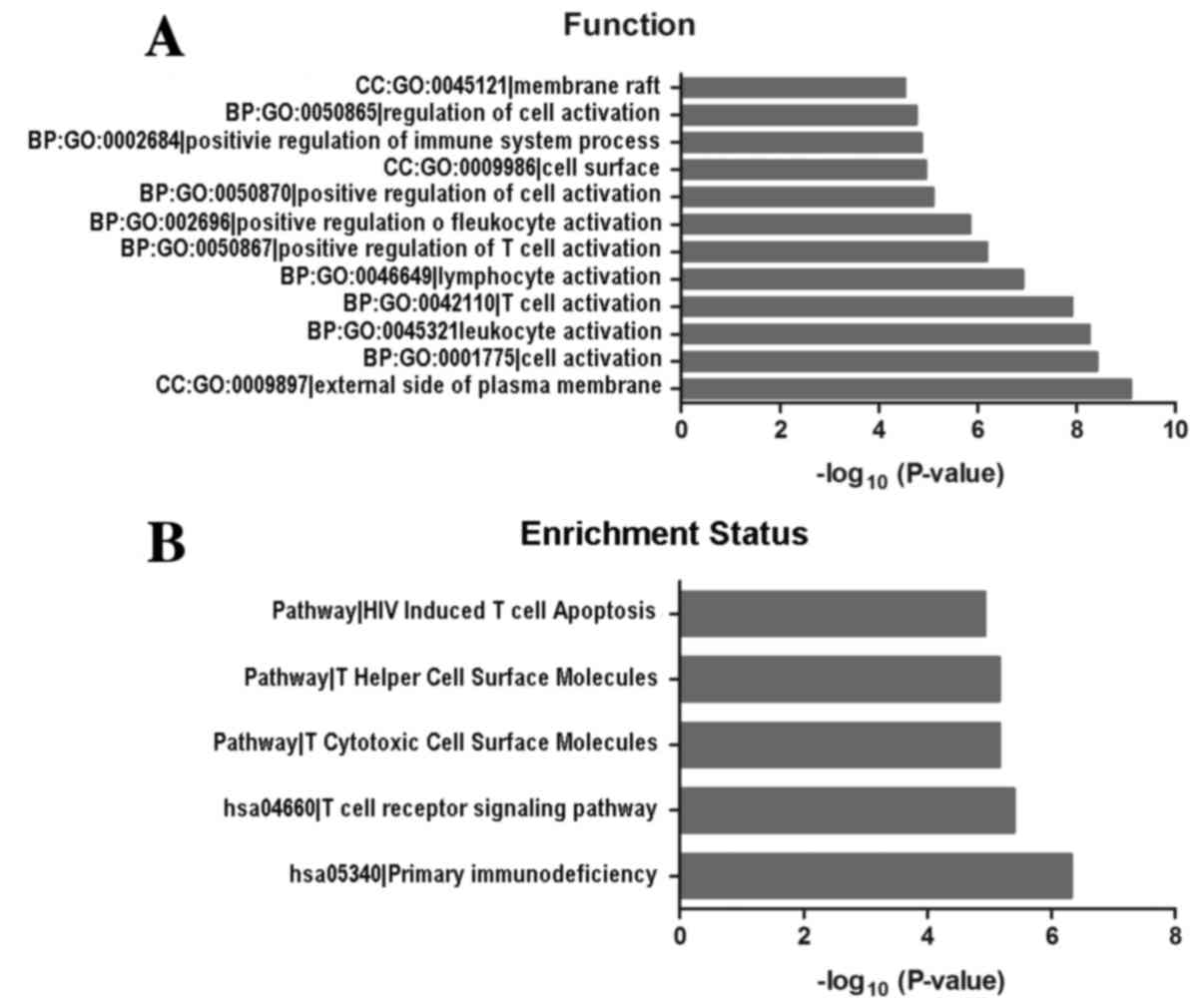
|
Yamaguchi K, Chijiiwa K, Saiki S,
Nishihara K, Takashima M, Kawakami K and Tanaka M: Retrospective
analysis of 70 operations for gallbladder carcinoma. Br J Surg.
84:200–204. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grobmyer SR, Lieberman MD and Daly JM:
Gallbladder cancer in the twentieth century: Single institution's
experience. World J Surg. 28:47–49. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu XS, Shi LB, Li ML, Ding Q, Weng H, Wu
WG, Cao Y, Bao RF, Shu YJ, Ding QC, et al: Evaluation of two
inflammation-based prognostic scores in patients with resectable
gallbladder carcinoma. Ann Surg Oncol. 21:449–457. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartlett DL, Fong Y, Fortner JG, Brennan
MF and Blumgart LH: Long-term results after resection for
gallbladder cancer. Implications for staging and management. Ann
Surg. 224:639–646. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Butte JM, Matsuo K, Gönen M, D'Angelica
MI, Waugh E, Allen PJ, Fong Y, DeMatteo RP, Blumgart L, Endo I, et
al: Gallbladder cancer: Differences in presentation, surgical
treatment, and survival in patients treated at centers in three
countries. J Am Coll Surg. 212:50–61. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cziupka K, Partecke LI, Mirow L, Heidecke
CD, Emde C, Hoffmann W, Siewert U, van den Berg N, von Bernstorff W
and Stier A: Outcomes and prognostic factors in gallbladder cancer:
A single-centre experience. Langenbecks Arch Surg. 397:899–907.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ooi A, Suzuki S, Nakazawa K, Itakura J,
Imoto I, Nakamura H and Dobashi Y: Gene amplification of Myc and
its coamplification with ERBB2 and EGFR in gallbladder
adenocarcinoma. Anticancer Res. 29:19–26. 2009.PubMed/NCBI
|
|
12
|
Nagahashi M, Ajioka Y, Lang I, Szentirmay
Z, Kasler M, Nakadaira H, Yokoyama N, Watanabe G, Nishikura K,
Wakai T, et al: Genetic changes of p53, K-ras, and microsatellite
instability in gallbladder carcinoma in high-incidence areas of
Japan and Hungary. World J Gastroenterol. 14:70–75. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
An integrated encyclopedia of DNA elements
in the human genome. Nature. 489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Faghihi MA, Modarresi F, Khalil AM, Wood
DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ,
Wahlestedt C, et al: Expression of a noncoding RNA is elevated in
Alzheimer's disease and drives rapid feed-forward regulation of
beta-secretase. Nat Med. 14:723–730. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Johnson R: Long non-coding RNAs in
Huntington's disease neurodegeneration. Neurobiol Dis. 46:245–254.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang J, Zhang P, Wang L, Piao HL and Ma
L: Long non-coding RNA HOTAIR in carcinogenesis and metastasis.
Acta Biochim Biophys Sin (Shanghai). 46:1–5. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vikram R, Ramachandran R and Abdul KS:
Functional significance of long non-coding RNAs in breast cancer.
Breast Cancer. 21:515–521. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tu ZQ, Li RJ, Mei JZ and Li XH:
Down-regulation of long non-coding RNA GAS5 is associated with the
prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol.
7:4303–4309. 2014.PubMed/NCBI
|
|
22
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding
Q, Weng H, Shu YJ, Liu TY, Jiang L, et al: MALAT1 promotes the
proliferation and metastasis of gallbladder cancer cells by
activating the ERK/MAPK pathway. Cancer Biol Ther. 15:806–814.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma M, Kong X, Weng M, Zhang MD, Qin YY,
Gong W, Zhang WJ and Quan ZW: Long non-coding RNA-LET is a positive
prognostic factor and exhibits tumor-suppressive activity in
gallbladder cancer. Mol Carcinogen. 54:1397–1406. 2015. View Article : Google Scholar
|
|
25
|
Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD,
Qin YY, Gong W and Quan ZW: Long non-coding RNA HOTAIR, a c-Myc
activated driver of malignancy, negatively regulates miRNA-130a in
gallbladder cancer. Mol Cancer. 13:1562014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY,
Gong W and Quan ZW: Long non-coding RNA CCAT1 promotes gallbladder
cancer development via negative modulation of miRNA-218-5p. Cell
Death Dis. 6:e15832015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta DeltaC(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Patterson TA, Lobenhofer EK,
Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W, Fang H, Kawasaki ES,
Hager J, Tikhonova IR, et al: Performance comparison of one-color
and two-color platforms within the MicroArray Quality Control
(MAQC) project. Nat Biotechnol. 24:1140–1150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Khachane AN and Harrison PM: Mining
mammalian transcript data for functional long non-coding RNAs. PLoS
One. 5:e103162010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pan YF, Feng L, Zhang XQ, Song LJ, Liang
HX, Li ZQ and Tao FB: Role of long non-coding RNAs in gene
regulation and oncogenesis. Chin Med J (Engl). 124:2378–2383.
2011.PubMed/NCBI
|
|
31
|
Cheng Y, Jutooru I, Chadalapaka G, Corton
JC and Safe S: The long non-coding RNA HOTTIP enhances pancreatic
cancer cell proliferation, survival and migration. Oncotarget.
6:10840–10852. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heubach J, Monsior J, Deenen R, Niegisch
G, Szarvas T, Niedworok C, Schulz WA and Hoffmann MJ: The long
noncoding RNA HOTAIR has tissue and cell type-dependent effects on
HOX gene expression and phenotype of urothelial cancer cells. Mol
Cancer. 14:1082015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang X, Zhou Y, Mehta KR, Danila DC,
Scolavino S, Johnson SR and Klibanski A: A pituitary-derived MEG3
isoform functions as a growth suppressor in tumor cells. J Clin
Endocrinol Metab. 88:5119–5126. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Graham LD, Pedersen SK, Brown GS, Ho T,
Kassir Z, Moynihan AT, Vizgoft EK, Dunne R, Pimlott L, Young GP, et
al: Colorectal neoplasia differentially expressed (CRNDE), a novel
gene with elevated expression in colorectal adenomas and
adenocarcinomas. Genes Cancer. 2:829–840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ellis BC, Molloy PL and Graham LD: CRNDE:
A long non-coding RNA involved in CanceR, neurobiology and
DEvelopment. Front Genet. 3:2702012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Taft RJ, Pang KC, Mercer TR, Dinger M and
Mattick JS: Non-coding RNAs: Regulators of disease. J Pathol.
220:126–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Goto Y, Hayashi R, Muramatsu T, Ogawa H,
Eguchi I, Oshida Y, Ohtani K and Yoshida K: JPO1/CDCA7, a novel
transcription factor E2F1-induced protein, possesses intrinsic
transcriptional regulator activity. Biochim Biophys Acta.
1759:60–68. 2006. View Article : Google Scholar : PubMed/NCBI
|