Introduction
Uterine leiomyoma is the most common female
reproductive tract benign tumor (1).
This tumor is responsible for severe health problems, including
pain, infertility, repetitive pregnancy loss and, most importantly,
heavy uterine bleeding (2). No
effective medical treatment is currently available, due to the poor
understanding of the exact molecular pathways involved in its
physiopathology (3). Ovarian
hormones, growth factors, retinoic acid, vitamin D, transforming
growth factor-β/Smad and genetic factors are associated with tumor
development (4,5). Leiomyomas are characterized by a
hyperproduction of extracellular matrix (ECM), which is constituted
predominantly by collagens and other proteoglycans (6). The ECM components serve a key role in
the regulation of migration, proliferation and differentiation of
leiomyomas (6).
The interstitial fluid (IF) is an extracellular
fluid bathing and surrounding the tissue cells, and is closely
associated with pathological changes in the tissues. In addition to
nutrients and oxygen, IF contains proteins secreted by tumors via
classical and non-classical secretory pathways, as well as proteins
released by extracellular vesicles (7). Several studies have shown that tumor
growth is not only regulated by the malignancy of the tumor cells,
but is also associated with several factors that are present in the
tumor microenvironment, including cytokines, electrolytes and other
secreted proteins (8). The IF flow
movement may influence the degree of viscosity and elasticity of
the ECM, thus modifying the intercellular communication and
determining an increased mitotic activity (9). Collagen and other proteoglycans such as
lumican (LUM) induce the activation of the integrin signaling
pathway, which serves a key role in cytoskeleton reorganization,
cell motility and proliferation (6).
Early cytogenetic alterations are allegedly insufficient for tumor
development; thus, additional alterations of the MAPK/ERK signaling
pathway appear to be crucial (3).
For the reasons described above, the identification
of changes in protein abundance in leiomyoma IF may be useful for
the understanding of the tumor physiopathology. Proteomic studies
on leiomyomas, carried out to identify dysregulated proteins, have
been conducted in two-dimensional electrophoresis (2-DE) coupled
with mass spectrometry (MS) (10) and
gel electrophoresis liquid chromatography-MS (11). Our previous proteomic studies on
leiomyoma tissue identified several dysregulated proteins involved
in metabolic processes, oxidative stress and cell migration
(12,13). In a previous study, seven proteins
with changes in abundance were identified in the IF of leiomyomas
(14). To date, no other proteomic
study has been conducted on the IF of leiomyomas.
The aim of the present study was to analyze the
proteome of leiomyoma and myometrium IF in order to identify
changes in the abundance of proteins potentially associated with
tumor growth. A comparative proteomic approach identified 17
proteins associated with cellular migration and proliferation with
different abundance in leiomyoma IF.
Materials and methods
Samples
Tissue samples were obtained from eight
premenopausal female patients who underwent hysterectomy for
symptomatic uterine leiomyoma. Samples were collected between
February 2015 and June 2015 at the Institute for Maternal and Child
Health, IRCCS ‘Burlo Garofolo’ (Trieste, Italy). The procedures
complied with the Declaration of Helsinki and were approved by the
Technical and Scientific Committee of the Institute for Maternal
and Child Health, IRCCS ‘Burlo Garofolo’ of Trieste, Italy
(approval no. RC 19/08). All subjects signed a written informed
consent. The patients did not receive any medication (including
hormonal therapy) during the three months prior to surgery. All
samples were obtained during the proliferative phase. Two samples
were collected from each patient: One from the central area of the
leiomyoma and one from the unaffected myometrium. The median age of
the patients was 46 years (range, 42–52 years).
Preparation of IF
The IF was obtained as previously described
(14,15). Two samples (300 mg each) were
collected from each patient: One from the unaffected myometrium and
one from the central area of the leiomyoma. All leiomyomas were
confirmed histologically as benign ordinary leiomyomas. Leiomyoma
and myometrium tissues were dissected into 0.1–0.3-cm3 pieces and
rinsed three times with sterile PBS containing a protease inhibitor
mixture (2 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 1 mM
EDTA and 10 mM NaF). The samples were incubated with PBS in a
humidified CO2 incubator for 1 h at 37°C. The
supernatant was centrifuged at 5,000 × g for 20 min at 4°C,
followed by centrifugation at 15,000 × g for 30 min at 4°C to
remove cell debris. Approximately 5 ml of supernatant was desalted
and concentrated using an Ultrafree-4 Centrifugal Filter Unit (EMD
Millipore, Billerica, MA, USA) with a molecular mass cut-off of 10
kDa at 4,000 × g at 25°C until the remaining volume reached 200 µl.
The obtained supernatant was designated as the IF, and the protein
content of the supernatant was determined by Bradford assay.
2-DE and image analysis
For 2-DE, 700 µg of proteins from each IF sample
were denatured in 350 µl of dissolution buffer [7 M urea, 2 M
thiourea, 4% 3-[(3-cholamidopropyl)
dimethylammonio]-1-propanesulfonate, 40 mM Tris, 65 mM
dithiothreitol (DTT) and 0.24% Bio-Lyte 3/10 (Bio-Rad Laboratories,
Inc., Hercules, CA, USA)] and 7 µg bromophenol blue. ReadyStrip™ pH
3–10 non-linear (NL) 18-cm immobilized pH gradient (IPG) strips
(Bio-Rad Laboratories, Inc.) were rehydrated in a dissolution
buffer at 50 V for 12 h at 20°C, and isoelectric focusing (IEF) was
performed in a PROTEAN IEF Cell (Bio-Rad Laboratories, Inc.) using
the following protocol: 150 V for 1 h, 250 V for 2 h, increase from
250 V to 10,000 V in 2 h, 10,000 V for 10 h and 10,000 V for 6 h,
for a total of 170,000 V/h. Serial incubations were performed
following IEF. First, the IPG strips were equilibrated for 20 min
in an equilibration buffer [6 M urea, 2% SDS, 50 mM Tris-HCl (pH
8.8), 30% glycerol and 1% DTT] and then in another equilibration
buffer containing 4% iodoacetamide instead of DTT. For the second
dimension, the equilibrated IPG strips were transferred to a 12%
polyacrylamide gel (19×20 cm). The SDS gels were subjected to
electrophoresis on a Protean II XL Cell (Bio-Rad Laboratories,
Inc.) at 200 V constant voltage, until the bromophenol blue band
reached the bottom of the gel. Upon electrophoresis, the gels were
fixed in 40% methanol and 10% acetic acid, and then stained for 48
h with colloidal Coomassie Brilliant Blue. Triple experimental
replicates were performed per sample. Molecular masses were
determined by comparison with Precision Plus Protein™ Prestained
Standards (Bio-Rad Laboratories, Inc.), covering a range between 10
and 250 kDa. 2-DE gels were scanned with a Molecular Imager
PharosFX system and analyzed using the Proteomweaver software
version 4.0 (both from Bio-Rad Laboratories, Inc.).
Quantification of spot levels
Spot quantification was performed as previously
described (12). 2-DE image analysis
was performed using the Proteomweaver 4 software. The analysis was
performed by matching all gels from eight myometrial IFs and eight
leiomyoma IFs. Differences were considered to be significant when
the ratio of the mean percentage of relative volume (%V),
determined as %V=Vsingle spot/Vtotal spot,
was 1.5-fold for upregulated and 0.6-fold for downregulated
proteins, and satisfied the non-parametric Wilcoxon signed-rank
test for matched samples (P<0.05). Fold change was calculated as
the ratio between the mean %V of the uterine leiomyoma and that of
the normal myometrium.
Trypsin digestion and matrix-assisted
laser desorption/ionization (MALDI)-time-of-flight (TOF)/TOF
analysis
Spots from 2-DE gels were thoroughly washed with
alternating changes of 50 mM NH4HCO3 and
acetonitrile (ACN), and then dried under vacuum. A total of 5 µl of
sequencing-grade modified trypsin (12.5 ng/ml in 50 mM
NH4HCO3; Promega Corporation, Madison, WI,
USA) were added to each gel spot, and samples were kept in ice for
30 min to allow the gel to completely rehydrate. A total of 50 mM
NH4HCO3 was added to each sample until gel
pieces were completely covered, and digestion was carried out
overnight at 37°C. Peptides were extracted with three changes of
50% ACN/0.1% trifluoroacetic acid (TFA; Honeywell Riedel-de Haën,
Seelze, Germany), dried under vacuum and then dissolved in 10 µl of
0.1% TFA. One µl of each sample was mixed with an equal volume of
matrix [α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany), 5 mg/ml in 70% ACN/0.1% TFA], and 0.8 µl of
the resulting solution was spotted onto a stainless steel MALDI
target plate (AB SCIEX, Framingham, MA, USA). Mass spectrometry
(MS)/MS analysis was performed on a 4800 MALDI-TOF/TOF Analyzer (AB
SCIEX) in a data-dependent mode: For each sample, a full MS scan
was acquired, followed by MS/MS spectra of the 10 most intense
signals.
Data were converted into Mascot generic format files
and analyzed with Proteome Discoverer software (version 1.4; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) interfaced to a Mascot
search engine (version 2.2.4; Matrix Science Ltd., London, UK).
Data were searched against the human section of the UniProt
database (www.uniprot.org; version 20,150,401;
90,411 sequences). Enzyme specificity was set to trypsin with ≤1
missed cleavage. Tolerance was set to 50 ppm and 0.3 Da for parent
and fragment ions, respectively. Carbamidomethylation of cysteines
was set as a fixed modification, while methionine oxidation was set
as a variable modification. The software calculates the false
discovery rate based on the search against a randomized database.
Proteins were considered as positively identified if ≥2 unique
peptides were identified for each protein with high confidence
(q<0.01).
Prediction of secreted protein
candidates
SecretomeP 2.0 (Center for Biological Sequence
Analysis, Technical University of Denmark, Lyngby, Denmark) was
used to classify proteins as following a classical (proteins with
signal peptide) or non-classical (proteins without signal peptide
but whose neural network score exceeded the normal threshold of
0.5) secretory pathway. To verify the possible release of proteins
by exosomes and microvesicles, manual annotation on the Exocarta
(exocarta.org) and Vesiclepedia (microvesicles.org/index.html) databases was
used.
Western blotting
Western blot analysis was performed as previously
described (16). An equal amount of
protein to that used for 2-DE analysis (30 µg) was separated by 12%
SDS-PAGE and then transferred to a nitrocellulose membrane. The
residual binding sites on the membrane were blocked by treatment
with 5% skim milk in TBS containing 0.05% Tween-20 (TBST) for 2 h
at room temperature. Next, the membrane was incubated overnight at
4°C with 1:500-diluted primary rabbit polyclonal antibody against
cellular retinoic acid binding protein 2 (CRABP2; cat. no.
SAB2100477), 1:300-diluted primary rabbit polyclonal antibody
against phosphoglycerate mutase 1 (PGAM1; cat. no. WH0005223M1),
1:800-diluted primary rabbit polyclonal antibody against aldolase A
(ALDOA; cat. no. SAB1410385), 1:700-diluted primary rabbit
polyclonal antibody against LUM (cat. no. SAB1401232),
1:500-diluted primary rabbit polyclonal antibody against annexin A2
(ANXA2; cat. no. SAB1410612), 1:1,000-diluted primary rabbit
polyclonal antibody against calreticulin (CALR; cat. no. AV30114)
and 1:200-diluted primary rabbit polyclonal antibody against
peroxiredoxin (PRDX1; cat. no. HPA007730) (all antibodies were
supplied by Sigma-Aldrich; Merck KGaA). The membrane was washed
three times in TBST for 10 min, and then incubated for 2 h at 4°C
with a horseradish peroxidase-conjugated anti-rabbit immunoglobulin
G antibody (cat. no. A8275) (Sigma-Aldrich; Merck KGaA) at 1:3,000
dilution. Protein expression was visualized by chemiluminescence
(SuperSignal West Pico Chemiluminescent Substrate; Thermo Fisher
Scientific, Inc.), and the intensity of the signals was quantified
by VersaDoc Imaging System (Bio-Rad Laboratories, Inc.). The
intensities of the immunostained bands were normalized with the
protein intensities measured by Red Ponceau S reagent
(Sigma-Aldrich; Merck KGaA) from the same blot.
Ingenuity Pathway Analysis (IPA)
Changes in protein abundance identified by MS in
leiomyoma IF were analyzed by IPA (Qiagen GmbH, Hilden, Germany) as
previously described (16). Selected
genes were incorporated in canonical pathways and bio-functions,
and were used to generate biological networks. For the filter
summary, only associations where confidence was high (predicted) or
that had been experimentally observed were considered. As cut-off,
and adjusted P<0.05 value was used. The network score was based
on the hypergeometric distribution, and was calculated with a
two-tailed Fisher's exact test. Since the majority of proteins
identified participated in multiple processes, only the most
relevant ones were reported.
Statistical analysis
Statistical analyses were carried out with the
non-parametric Wilcoxon signed-rank test for matched samples for
both 2-DE and western blot data. P<0.05 was considered to
indicate a statistically significant difference. All analyses were
conducted with Stata/IC 14.1 for Windows (StataCorp LP, College
Station, TX, USA).
Results
2-DE and MS
In the present study, the IF of eight leiomyomas and
myometria from eight premenopausal women were resolved using 2-DE.
Approximately 2,000 protein spots were detected in the 2-DE gels
for the two types of IFs. Correlation of gel-pairs performed with
an average matching efficiency of ~75%. The analysis revealed 25
protein spots with different abundance in the IF of leiomyoma
compared with that of the IF of the myometrium. Only protein spots
with fold change in %V ≥1.5 or ≤0.6 in intensity that were
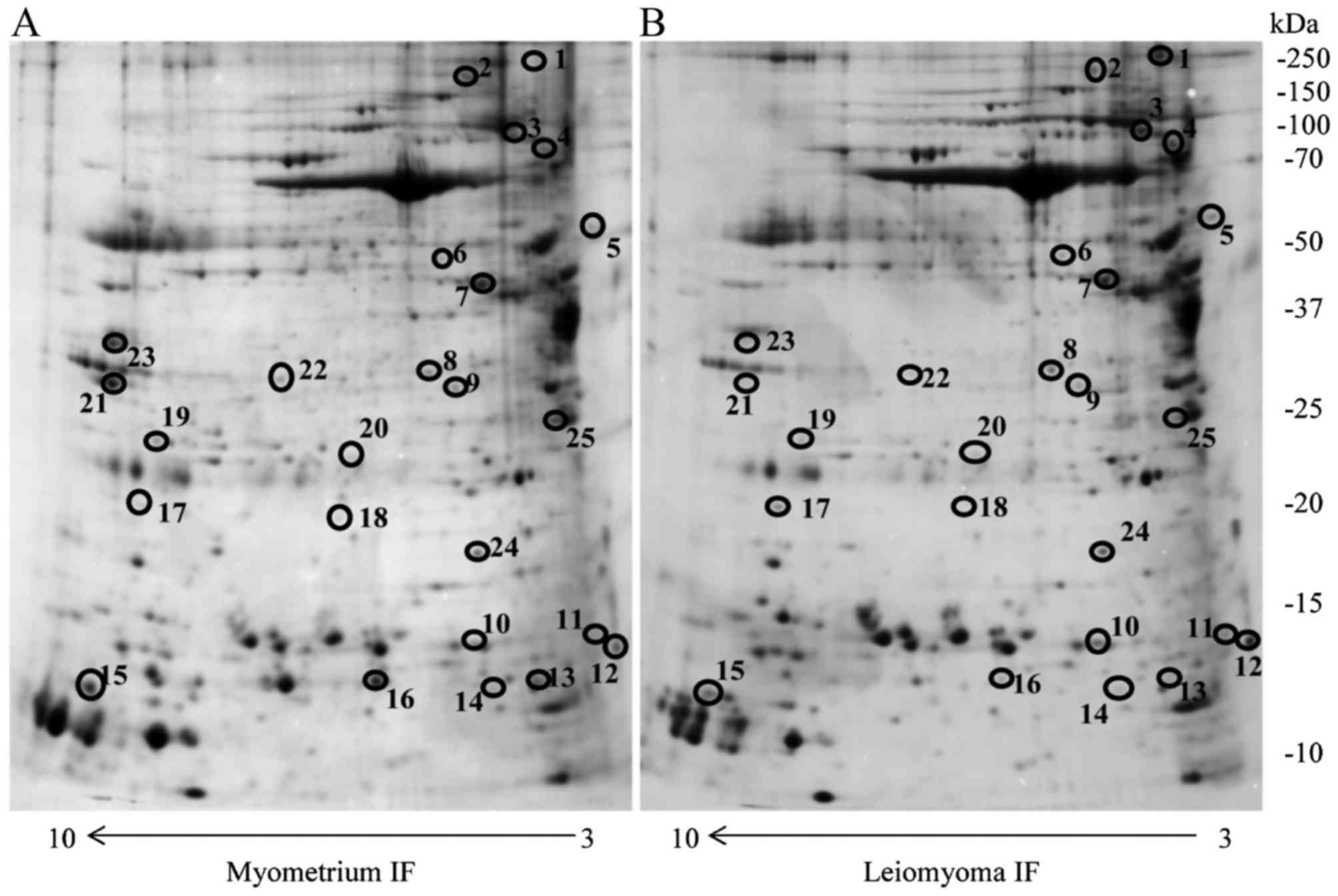
significant (P<0.05) were considered (Fig. 1). These proteins were identified by
MALDI-TOF/TOF and database search against the human section of the
UniProt database (version 20150401; 90,411 sequences) (Table I).
 | Table I.List of proteins with significantly
different abundance in the interstitial fluid of the leiomyoma
compared with that of the normal myometrium, as identified by
matrix-assisted laser desorption/ionization-TOF/TOF mass
spectrometry, and classified by their corresponding molecular and
cellular functions and subcellular localization. Only the most
relevant molecular and cellular functions are reported. |
Table I.
List of proteins with significantly
different abundance in the interstitial fluid of the leiomyoma
compared with that of the normal myometrium, as identified by
matrix-assisted laser desorption/ionization-TOF/TOF mass
spectrometry, and classified by their corresponding molecular and
cellular functions and subcellular localization. Only the most
relevant molecular and cellular functions are reported.
| Accession
no.a | Spot no. | Protein
description | Gene symbol | Protein score | Fold
changeb | Molecular and
cellular functions | Secretome P
2.0 | Exocarta | Veciclepedia | P-value |
|---|
| P51884 | 4 | Lumican | LUM | 122.78 | 6.0 | Cell migration | SignalP |
|
| 0.0139 |
| Q5HY54 | 1 | Filamin A | FLNA | 362.86 | 4.8 | Cellular assembly
and organization | 0.45 | Yes | Yes | 0.0193 |
| P29373 | 14 | Cellular retinoic
acid | CRABP2 | 142.69 | 3.0 | Cell
proliferation | 0.59 |
|
| 0.0140 |
|
|
| binding protein
2 |
| Q06830 | 17 | Peroxiredoxin
1 | PRDX1 | 394.64 | 3.0 | Cell migration | 0.53 |
|
| 0.0173 |
| P05387 | 11 | Ribosomal protein
lateral | RPLP2 | 288.99 | 2.7 | Not known | 0.27 | Yes | Yes | 0.0296 |
|
|
| stalk subunit
P2 |
| P27797 | 5 | Calreticulin | CALR | 642.74 | 2.5 | Cell migration | SignalP |
|
| 0.0173 |
| P32119 | 24 | Peroxiredoxin
2 | PRDX2 | 161.45 | 2.0 | Cell
proliferation | 0.52 |
|
| 0.0117 |
| A0A087WV45 | 10 | Transthyretin | TTR | 100.29 | 1.80 | Not known | SignalP |
|
| 0.0295 |
| P18669 | 20 | Phosphoglycerate
mutase 1 | PGAM1 | 450.56 | 1.80 | Cell
proliferation | 0.41 | Yes | Yes | 0.0251 |
| P55072 | 3 | Valosin-containing
protein | VCP | 345.34 | 1.70 | Cell migration | 0.16 | Yes | Yes | 0.0296 |
| P60709 | 9 | β-actin | ACTB | 316.16 | 1.70 | Cell migration | 0.50 | Yes | Yes | 0.0418 |
| B4DXW1 | 6 | Actin-related
protein 3 | ACTR3 | 400.00 | 1.60 | Cell migration | 0.44 | Yes | Yes | 0.0296 |
| P62158 | 12 | Calmodulin 1 | CALM1 | 283.75 | 1.60 | Cell migration | 0.45 | Yes | Yes | 0.0497 |
| P07195 | 8 | L-lactate
dehydrogenase | LDHB | 457.75 | 1.50 | Cell
proliferation | 0.57 |
|
| 0.0249 |
|
|
| B chain |
| K7EJ44 | 15 | Profilin 1 | PFN1 | 263.83 | 0.60 | Cellular assembly
and | 0.47 | Yes | Yes | 0.0296 |
|
|
|
|
|
|
| organization |
| P04083 | 21 | Annexin 1 | ANXA1 | 671.52 | 0.60 | Cell
proliferation | 0.51 |
|
| 0.0296 |
| P12277 | 7 | Creatine kinase
B | CKB | 466.67 | 0.48 | Cellular
assembly | 0.26 | Yes | Yes | 0.0357 |
|
|
|
|
|
|
| and
organization |
|
| Q9Y490 | 2 | Talin 1 | TLN1 | 156.87 | 0.30 | Cellular
assembly | 0.23 | Yes | Yes | 0.0193 |
|
|
|
|
|
|
| and
organization |
| P07355 | 22 | Annexin 2 | ANXA2 | 442.58 | 0.19 | Cell
proliferation | 0.75 |
|
| 0.0117 |
| E7EP94 | 25 | Heat shock protein
family | HSPA1A | 60.00 | 0.16 | Cell migration | 0.28 | Yes | Yes | 0.0193 |
|
|
| A (Hsp70) member
1A |
| Q01995 | 16 | Transgelin | TAGLN | 280.64 | 0.16 | Cellular
function | 0.56 |
|
| 0.0117 |
|
|
|
|
|
|
| and
maintenance |
| H3BQN4 | 23 | Aldolase A | ALDOA | 370.58 | 0.10 | Cell migration | 0.36 | Yes | Yes | 0.0192 |
| Q5T0R7 | 19 | Adenylyl
cyclase-associated | CAP1 | 374.82 | 0.06 | Cell migration | 0.44 | Yes | Yes | 0.0293 |
In addition, proteins reported in our previous
study, including desmin (DES), transgelin (TAGLN), serpin family A
member 1 (SERPINA1), lamin A/C (LMNA), actinin α1 (ACTN1), PRDX2
and heat shock protein family A (Hsp70) member 1A (HSPA1A), were
also identified (14). Of these
proteins, TAGLN, PRDX2 and HSPA1A were significant, while DES,
SERPINA1 (more abundant in the leiomyoma compared with in the
myometrium), LMNA and ACTN1 (less abundant in the leiomyoma
compared with in the myometrium) were not significant, and thus
were not investigated further.
In total, 22 of the 25 proteins with different
abundance reported in the present study have not been previously
identified in the leiomyoma IF. Furthermore, among the identified
proteins, one (namely PGAM1) has never been previously associated
with leiomyoma.
Prediction of protein secretion and
functional analysis of the leiomyoma proteome
To define if the 25 identified proteins were
secreted by the classical or non-classical secretory pathway, the
SignalIP version 4.1 (www.cbs.dtu.dk/services/SignalP) and SecretomeP 2.0
(www.cbs.dtu.dk/services/SecretomeP) prediction servers
were used. In addition, the Vesiclepedia and Exocarta databases,
which contain proteins identified in extracellular vesicles and
exosomes released by non-classical secretory pathways respectively,
were used. Out of the 25 proteins, three were assigned to the
classical secretory pathway predicted by their signal peptide: LUM,
CALR and transthyretin. The remaining proteins were predicted to be
secreted by non-classical pathways to the extracellular space
through various mechanisms (Table
I).
IPA
Proteins with significantly different abundance in
the IF of leiomyoma compared with that of the myometrium were used
in the core analysis with IPA software. The analysis revealed that
the identified proteins were associated with cellular migration,
proliferation, assembly, organization, function and
maintenance.
A total of 17 proteins were observed to be
associated with cellular migration and proliferation according to
the IPA prediction (Table I).
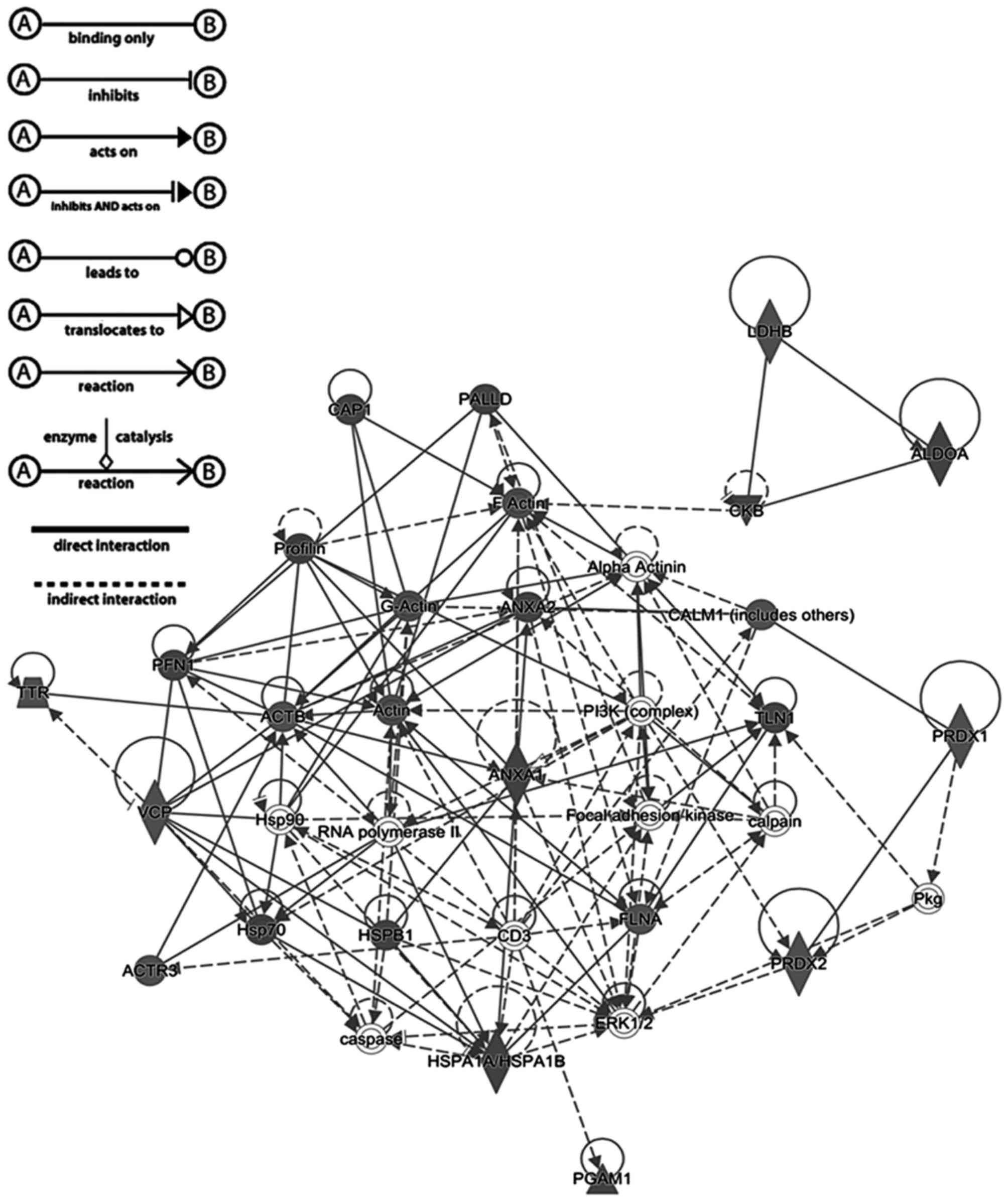
Fig. 2 shows a graphical
representation of the predicted interaction between the proteins
with significantly different abundance identified in the present
study. This network (score, 56) includes 25 identified proteins
involved in neurological diseases, cellular migration, and skeletal
and muscular disorders. The representative interaction network
suggests that the proteins with changes in abundance identified in
the present study may influence the activity of other proteins
within the network.
Immunohistochemical study of protein
expression
Proteins with different abundance in the leiomyoma
IF (namely LUM, PRDX, CALR, CRABP2, ANXA2, ALDOA and PGAM1) were
further validated by western blotting in five leiomyomas (IF) vs.
normal myometrium (IF) (Fig. 3). The
abundance of LUM, PRDX1, CALR, CRABP2 and PGAM1 was significantly
higher in leiomyomas than in the myometrium, while for ANXA2 and
ALDOA, their abundance was significantly lower in leiomyomas with
respect to that in the myometrium, thus confirming the results
obtained from the 2-DE analysis. To normalize the results of the
western blot analysis, the total protein content of each sample, as
determined by Red Ponceau S staining, was used. According to our
previous results (12), proteins
encoded by housekeeping genes (i.e., β-actin and tubulin) are
usually upregulated in leiomyomas and, therefore, not adequate to
be used as controls for normalization.
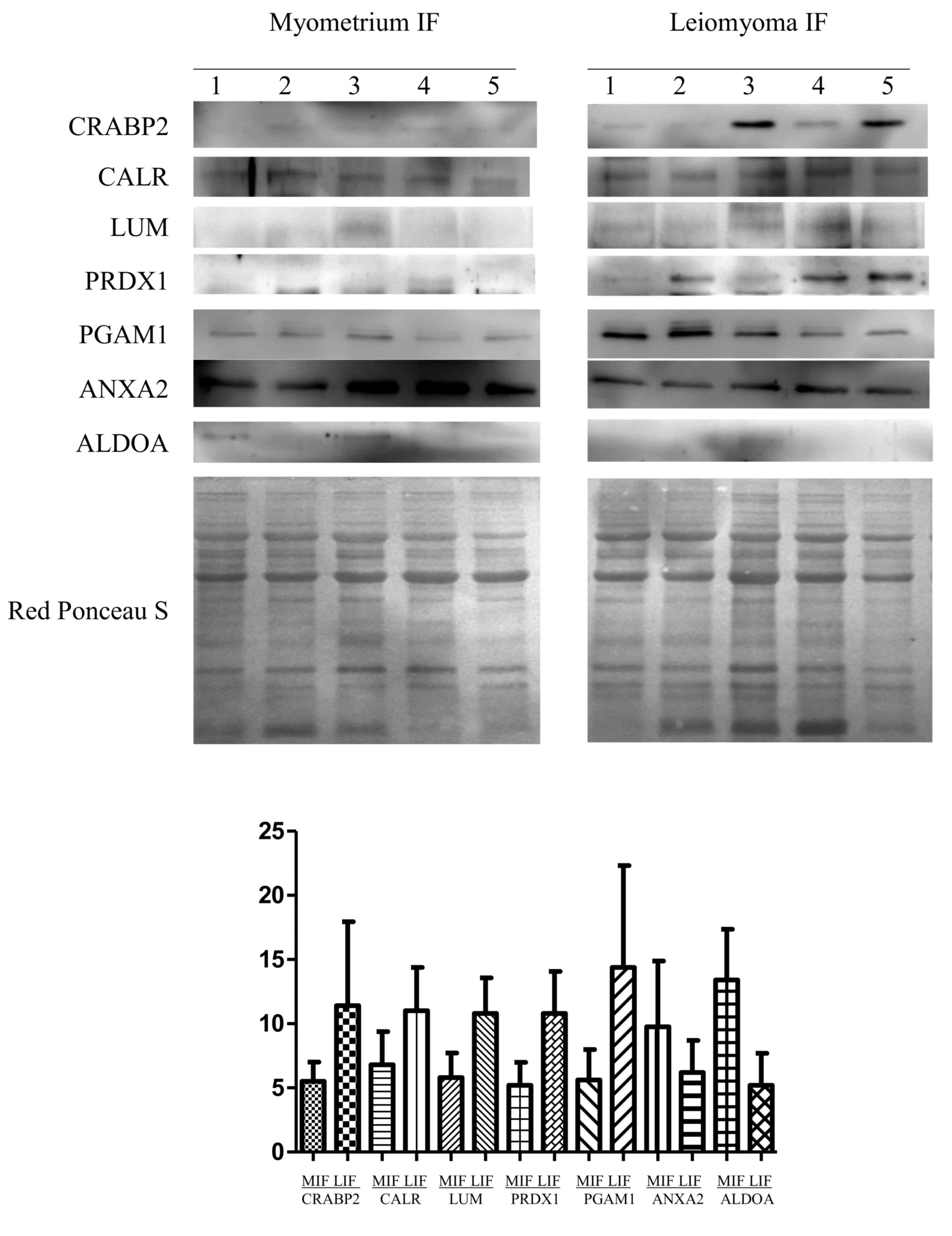 | Figure 3.Western blot analysis of CRABP2,
CALR, LUM, PRDX1, PGAM1, ANXA2 and ALDOA in paired MIF and LIF. The
intensity of the immunostained bands was normalized against the
total protein intensities measured from the same blot stained with
Red Ponceau S. The bar graph shows the relative expression (band
density) of proteins in the MIF and LIF. The results are shown as a
histogram (mean) with error bars representing the standard
deviation. All differences were observed to be significant
(Wilcoxon signed-rank test for matched samples, P<0.05). MIF,
myometrium IF; LIF, leiomyoma IF; IF, interstitial fluid; CRABP2,
cellular retinoic acid binding protein 2; CALR, calreticulin; LUM,
lumican; PRDX, peroxiredoxin; PGAM1, phosphoglycerate mutase 1;
ANXA2; annexin A2; ALDOA, aldolase A. |
Discussion
In tumor diseases, secreted proteins create
conditions favorable to the disorder, promoting tumor progression
and growth (17). Proteomic studies
are essential for the acquisition of knowledge about the
qualitative and quantitative composition of the IF of the cell, and
may be crucial to understand the biology of cell interaction,
proliferation, differentiation, communication and migration. In a
previous study, seven proteins that were differently expressed in
the IF of leiomyomas compared with that of the normal myometrium
were identified (13). The present
study continued to investigate different abundant proteins likely
to be associated with tumor development in the IF.
The adoption of 18-cm pH 3–10 NL strips with
polyacrylamide gels of 19×20 cm in size instead of 7-cm pH 3–10
strips with polyacrylamide gels of 8×8 cm in size markedly
increased the spot number. This allowed the identification of 22
proteins that had not been previously identified in the leiomyoma
IF, while PGAM1 had never previously been associated with
leiomyoma. IPA analysis predicted that 17 out of the 25 proteins
are associated with cellular migration and proliferation. In
addition, the abundance of the proteins identified in our previous
study (DES, TAGLN, SERPINA1, LMNA, ACTN1, PRDX2 and HSPA1A) is in
line with the results of the present study on the leiomyoma IF.
Furthermore, the levels of CRABP2, CALM1, LDHB, SERPINA1 and DES
were confirmed to be upregulated, while those of TAGLN, LMNA and
ACTN1 were downregulated, both in the proteome (12,13) and in
the IF. Seven of these proteins (LUM, PRDX1, CRABP2, CALR, ANXA2,
ALDOA and PGAM1) were validated by western blotting, thus
confirming the 2-DE data.
An inadequate vascularization during tumor growth
results in microenvironmental stress, which induces the activation
of a range of stress response pathways, including the unfolding
protein response (UPR) (18). UPR
activation is required for tumor cell growth under hypoxic
conditions (18). CALR is a protein
involved in the UPR pathway. It participates in several biological
processes, including cell adhesion, gene expression, RNA stability
and regulation of Ca2+ homeostasis (19,20). Cell
stress increases the concentration of nitric oxide, inducing the
release of CALR from cells in the extracellular space (21). This may be associated with alterations
in the viscosity and elasticity of the ECM.
The presence of adenylyl cyclase-associated protein
1 (CAP1) in the extracellular environment was documented in the
urine of patients with systemic autoimmune diseases, and was
identified as an efficient substrate of gelatinase B/matrix
metallopeptidase (MMP)-9 (22).
Degradation of CAP1 by MMP-9 is important for tumor proliferation
(22), as it may accelerate ECM
accumulation, thereby promoting tumor growth.
Inflammatory and pro-inflammatory factors serve a
key role in cancer development with the involvement of the
transcription factor nuclear factor (NF)-κB, which also has a
crucial role in cell growth, tumorigenesis and apoptosis (23). Several pro-inflammatory factors,
including NF-κB, interleukin-1β, tumor necrosis factor-α and
cyclooxygenase-2, are involved in the cellular proliferation and
growth of leiomyomas (23). PRDX1 and
PRDX2 are released by cells in the presence of pro-inflammatory
factors. Their release stimulates the production of inflammatory
cytokines, thus increasing the proliferative effect, which serves a
role in matrix production (24,25).
CRABP2 is a key protein for the binding and
transport of retinoic acid (26).
Increasing levels of retinoic acid induce the overexpression of
MMP-11, which degrades insulin-like growth factor-binding proteins,
and may result in cell proliferation, decreased apoptosis and
acceleration of ECM accumulation, thereby promoting the growth of
uterine leiomyomas (27).
The present study identified two glycolytic enzymes
(ALDOA and PGAM1) with different abundance in the leiomyoma IF.
This further confirms that glycolytic enzymes can be secreted by
cells, attest metabolic dysregulation in leiomyomas (12) and perform non-enzymatic activities
such as structural or regulatory functions at their extracellular
localization (28).
ANXA2 is a Ca2+-dependent phospholipid
binding protein able to bind plasminogen and mediate the localized
generation of plasmin (29). The
present study has shown that the IF of leiomyomas is characterized
by a lower abundance compared with the myometrium, of ALDOA and
ANXA2, which could be associated with a decreased proteolytic
degradation of the ECM in leiomyomas.
HSPA1A is a member of the Hsp70 family (30). In cooperation with other chaperones,
HSPA1A mediates the folding of newly translated polypeptides in the
cytosol, as well as within organelles (31). Once secreted, HSPA1A participates in
paracrine or autocrine interactions with adjacent cell surfaces,
controlling the expression of several genes associated with
apoptosis and cell migration (32,33).
It is well known that leiomyomas are characterized
by an abnormal ECM production (6).
Proteoglycan components of the ECM such as LUM bind to integrin,
activating the integrin signaling pathway, and leading to
cytoskeletal rearrangement and cell motility (34). Physiologically, LUM is involved in
several biological processes, including the glycosaminoglycan
metabolic process, angiogenesis, cell proliferation, migration,
adhesion and ECM organization (35).
Leiomyoma growth may be closely associated with secreted LUM, which
is in turn associated with cell motility and growth.
IPA revealed remarkable data on the interaction
among proteins. Several proteins identified in the present study
(including PRDX2, ANXA1, ANXA2, HSPA1A, filamin Aα, CALR,
calmodulin 1 and talin 1) interact substantially with extracellular
signal-regulated kinase-1/2, platelet-derived growth factor-BB and
calpain, which serve a key role in cell growth, adhesion, survival
and differentiation (36,37).
In conclusion, the comparative proteomic approach
conducted in the present study identified a number of proteins
associated with cellular migration and proliferation with different
abundance in the IF. Secreted proteins may be involved in
mechanisms associated with leiomyoma development. Further studies
will be required to investigate the role of these proteins in the
leiomyoma IF and their association with tumor development, in order
to identify novel therapeutic targets and develop new
pharmacological approaches for the treatment of leiomyoma.
Acknowledgements
The authors acknowledge the help of Dr Emmanouil
Athanasakis (Medical Genetics Department, Institute for Maternal
and Child Health-IRCCS ‘Burlo Garofolo’, Trieste, Italy) for data
acquisition. The present study was internally funded by the
Institute for Maternal and Child Health-IRCCS ‘Burlo Garofolo’ of
Trieste, Italy (grant no. RC 19/08).
References
|
1
|
Hashimoto K, Azuma C, Kamiura S, Kimura T,
Nobunaga T, Kanai T, Sawada M, Noguchi S and Saji F: Clonal
determination of uterine leiomyomas by analyzing differential
inactivation of the X-chromosome-linked phosphoglycerokinase gene.
Gynecol Obstet Invest. 40:204–208. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haney AF: Clinical decision making
regarding leiomyomata: What we need in the next millennium. Environ
Health Perspect. 108:(Suppl 5). S835–S839. 2000. View Article : Google Scholar
|
|
3
|
Borahay MA, Al-Hendy A, Kilic GS and
Boehning D: Signaling pathways in leiomyoma: Understanding
pathobiology and implications for therapy. Mol Med. 21:242–256.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Catherino WH and Malik M: Uterine
leiomyomas express a molecular pattern that lowers retinoic acid
exposure. Fertil Steril. 87:1388–1398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arslan AA, Gold LI, Mittal K, Suen TC,
Belitskaya-Levy I, Tang MS and Toniolo P: Gene expression studies
provide clues to the pathogenesis of uterine leiomyoma: New
evidence and a systematic review. Hum Reprod. 20:852–863. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Malik M and Catherino WH: Novel method to
characterize primary cultures of leiomyoma and myometrium with the
use of confirmatory biomarker gene arrays. Fertil Steril.
87:1166–1172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gromov P, Gromova I, Olsen CJ,
Timmermans-Wielenga V, Talman ML, Serizawa RR and Moreira JM: Tumor
interstitial fluid-a treasure trove of cancer biomarkers. Biochim
Biophys Acta. 1834:2259–2270. 2103. View Article : Google Scholar
|
|
8
|
Spano D and Zollo M: Tumor
microenvironment: A main actor in the metastasis process. Clin Exp
Metastasis. 29:381–395. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Norian JM, Owen CM, Taboas J, Korecki C,
Tuan R, Malik M, Catherino WH and Segars JH: Characterization of
tissue biomechanics and mechanical signaling in uterine leiomyoma.
Matrix Biol. 31:57–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lv J, Zhu X, Dong K, Lin Y, Hu Y and Zhu
C: Reduced expression of 14-3-3 gamma in uterine leiomyoma as
identified by proteomics. Fertil Steril. 90:1892–1898. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lemeer S, Gholami AM, Wu Z and Kuster B:
Quantitative proteome profiling of human myoma and myometrium
tissue reveals kinase expression signatures with potential for
therapeutic intervention. Proteomics. 15:356–364. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ura B, Scrimin F, Arrigoni G, Franchin C,
Monasta L and Ricci G: A proteomic approach for the identification
of up-regulated proteins involved in the metabolic process of the
leiomyoma. Int J Mol Sci. 17:5402016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ura B, Scrimin F, Arrigoni G, Athanasakis
E, Aloisio M, Monasta L and Ricci G: Abnormal expression of
leiomyoma cytoskeletal proteins involved in cell migration. Oncol
Rep. 35:3094–3100. 2016.PubMed/NCBI
|
|
14
|
Ura B, Scrimin F, Zanconati F, Arrigoni G,
Monasta L, Romano A, Banco R, Zweyer M, Milani D and Ricci G:
Two-dimensional gel electrophoresis analysis of the leiomyoma
interstitial fluid reveals altered protein expression with a
possible involvement in pathogenesis. Oncol Rep. 33:2219–2226.
2015.PubMed/NCBI
|
|
15
|
Gromov P, Gromova I, Bunkenborg J, Cabezon
T, Moreira JM, Timmermans-Wielenga V, Roepstorff P, Rank F and
Celis JE: Up-regulated proteins in the fluid bathing the tumour
cell microenvironment as potential serological markers for early
detection of cancer of the breast. Mol Oncol. 4:65–89. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mischiati C, Ura B, Roncoroni L, Elli L,
Cervellati C, Squerzanti M, Conte D, Doneda L, de Polverino Laureto
P, de Franceschi G, et al: Changes in protein expression in two
cholangiocarcinoma cell lines undergoing formation of multicellular
tumor spheroids in vitro. PLoS One. 10:e01189062015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li S and McLachlan JA: Estrogen-associated
genes in uterine leiomyoma. Ann N Y Acad Sci. 948:112–120. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Desiniotis A and Kyprianou N: Significance
of talin in cancer progression and metastasis. Int Rev Cell Mol
Biol. 289:117–147. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vandewynckel YP, Laukens D, Geerts A,
Bogaerts E, Paridaens A, Verhelst X, Janssens S, Heindryckx F and
Van Vlierberghe H: The paradox of the unfolded protein response in
cancer. Anticancer Res. 33:4683–4694. 2013.PubMed/NCBI
|
|
20
|
Totary-Jain H, Naveh-Many T, Riahi Y,
Kaiser N, Eckel J and Sasson S: Calreticulin destabilizes glucose
transporter-1 mRNA in vascular endothelial and smooth muscle cells
under high-glucose conditions. Circ Res. 97:1001–1008. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tarr JM, Young PJ, Morse R, Shaw DJ, Haigh
R, Petrov PG, Johnson SJ, Winyard PG and Eggleton P: A mechanism of
release of calreticulin from cells during apoptosis. J Mol Biol.
401:799–812. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cauwe B, Martens E, Van den Steen PE,
Proost P, Van Aelst I, Blockmans D and Opdenakker G: Adenylyl
cyclase-associated protein-1/CAP1 as a biological target substrate
of gelatinase B/MMP-9. Exp Cell Res. 314:2739–2749. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Plewka A, Madej P, Plewka D, Kowalczyk A,
Miskiewicz A, Wittek P, Leks T and Bilski R: Immunohistochemical
localization of selected pro-inflammatory factors in uterine myomas
and myometrium in women of various ages. Folia Histochem Cytobiol.
51:73–83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mullen L, Hanschmann EM, Lillig CH,
Herzenberg LA and Ghezzi P: Cysteine oxidation targets
peroxiredoxins 1 and 2 for exosomal release through a novel
mechanism of redox-dependent secretion. Mol Med. 13:98–108.
2015.
|
|
25
|
Kubota D, Mukaihara K, Yoshida A, Tsuda H,
Kawai A and Kondo T: Proteomics study of open biopsy samples
identifies peroxiredoxin 2 as a predictive biomarker of response to
induction chemotherapy in osteosarcoma. J Proteomics. 91:393–404.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aström A, Pettersson U and Voorhees JJ:
Structure of the human cellular retinoic acid-binding protein II
gene. Early transcriptional regulation by retinoic acid. J Biol
Chem. 267:25251–25255. 1992.PubMed/NCBI
|
|
27
|
Zaitseva M, Vollenhoven BJ and Rogers PA:
Retinoic acid pathway genes show significantly altered expression
in uterine fibroids when compared with normal myometrium. Mol Hum
Reprod. 13:577–585. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Capello M, Ferri-Borgogno S, Cappello P
and Novelli F: α-Enolase: A promising therapeutic and diagnostic
tumor target. FEBS J. 278:1064–1074. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Valapala M, Maji S, Borejdo J and
Vishwanatha JK: Cell surface translocation of annexin A2
facilitates glutamate-induced extracellular proteolysis. J Biol
Chem. 289:15915–15926. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hageman J, Vos MJ, van Waarde MA and
Kampinga HH: Comparison of intra-organellar chaperone capacity for
dealing with stress-induced protein unfolding. J Biol Chem.
282:34334–34345. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen Z, Barbi J, Bu S, Yang HY, Li Z, Gao
Y, Jinasena D, Fu J, Lin F, Chen C, et al: The ubiquitin ligase
Stub1 negatively modulates regulatory T cell suppressive activity
by promoting degradation of the transcription factor Foxp3.
Immunity. 39:272–285. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mambula SS, Stevenson MA, Ogawa K and
Calderwood SK: Mechanisms for Hsp70 secretion: Crossing membranes
without a leader. Methods. 43:168–175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Daniels GA, Sanchez-Perez L, Diaz RM,
Kottke T, Thompson J, Lai M, Gough M, Karim M, Bushell A and Chong
H: A simple method to cure established tumors by inflammatory
killing of normal cells. Nat Biotechnol. 22:1125–1132. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Martin San S, Soto-Suazo M, De Oliveira
SF, Aplin JD, Abrahamsohn P and Zorn TM: Small leucine-rich
proteoglycans (SLRPs) in uterine tissues during pregnancy in mice.
Reproduction. 125:585–595. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nikitovic D, Papoutsidakis A, Karamanos NK
and Tzanakakis GN: Lumican affects tumor cell functions, tumor-ECM
interactions, angiogenesis and inflammatory response. Matrix Biol.
35:206–214. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lorenz K, Schmitt JP, Schmitteckert EM and
Lohse MJ: A new type of ERK1/2 autophosphorylation causes cardiac
hypertrophy. Nat Med. 15:75–83. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Richard I, Broux O, Allamand V,
Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C,
Pasturaud P, Roudaut C, et al: Mutations in the proteolytic enzyme
calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell.
81:27–40. 1995. View Article : Google Scholar : PubMed/NCBI
|