Introduction
Hepatocellular carcinoma (HCC) remains one of the
most prevalent malignant diseases in the world (1). Chronic infection with the hepatitis B
virus (HBV) is the most important risk factor for HCC, accounting
for 55% of global incidences and >80% of cases in Asia and
sub-Saharan Africa (2). Patients
infected with HBV are known to be at an increased risk of
developing HCC over their lifetime (3–5). Previous
studies have reported familial aggregation of HCC (6,7), and
previous meta-analyses have indicated that family history of HCC
increases the risk of HCC in patients with viral hepatitis,
independently of hepatitis (8). A
previous study examined the effect of a family history of HCC on
the incidence of HCC in the entire population who were screened for
HBV seromarkers, and elucidated that family history of HCC
multiplied the risk of HCC at each stage of HBV infection (7). Although numerous molecular studies have
revealed that the HBV-encoded X (HBx) protein performs a critical
role in hepatocarcinogenesis in patients with HBV-associated HCC
(9–12), the mechanisms underlying familial
clustering-associated carcinogenesis remain to be fully
elucidated.
MicroRNAs (miRNAs/miRs) comprise a class of
highly-conserved noncoding RNAs of ~22 nucleotides in length
(13). Mature miRNAs may interact
with the 3′-untranslated regions (UTRs) of target mRNAs to form
RNA-induced silencing complexes, resulting in the inhibition of
translation or mRNA cleavage (14).
Previous studies have revealed that miRNAs may regulate gene
expression at the posttranscriptional level and performs a role in
regulating epigenetic machinery, including DNA methylation and
histone modification (15–18). Since miRNAs perform important roles in
various pivotal biological processes, dysregulated miRNA expression
has been implicated in a variety of human diseases, including
chronic HBV infection and hepatocarcinogenesis (19). An increasing amount of evidence has
indicated that dysregulation of miRNAs has important roles in HBV
infection and HBV-associated HCC (9,20–22). Numerous studies have demonstrated that
the HBx protein is associated with the regulation of miRNAs, which
affects basic tumor processes, including cellular proliferation,
differentiation and metastasis (12,23–27). In
addition, a family history of HCC increases the risk of HCC, even
following adjustment for other risk factors, and in patients
without hepatitis serum markers (28). However, little is known regarding the
association between miRNAs and family aggregation of HCC in
patients with HBV infection.
In the present study, microarray analysis was
performed to study the miRNA profile in the plasma of patients with
HCC with familial aggregation of HCC and HBV infection, compared
with the healthy control. The present study may help improve
understanding of the key roles of miRNAs in the progression of HCC,
and pave the way to future studies on the molecular mechanism of
hepatocarcinogenesis.
Materials and methods
Patients and samples
A total of 3 patients with HCC with a history of
familial aggregation of HCC and HBV infection underwent liver
resection at the Hepatic Surgery Center, Tongji Hospital, Tongji
Medical College, Huazhonog University of Science and Technology
(Wuhan, China) between December 2011 and March 2012. Blood samples
were obtained from these 3 patients prior to liver resection. HCC
was diagnosed based on cytohistological evidence from resected
specimens. These patients were not treated with any
radiochemotherapy prior to blood drawing. Blood collected from a
healthy volunteer was used as a control. The baseline information
of all enrolled subjects is presented in Table I. Written consent for sample
collection was obtained from all the patients, and the protocol was
approved by the Institutional Research Ethics Committee of Tongji
Hospital, Tongji Medical College, Huazhong University of Science
and Technology.
 | Table I.Baseline information of 3 patients
with HCC and 1 volunteer. |
Table I.
Baseline information of 3 patients
with HCC and 1 volunteer.
| Patient | Sex | Age, years | Diagnosis | Edmondson-steiner
grade | Family history of
HCC | Sampling date |
|---|
| 1 | F | 42 | HBV/HCC | II | One sister and one
brother | 2011–12–24 |
| 2 | M | 43 | HBV/HCC | I | Two brothers | 2012-01-28 |
| 3 | M | 38 | HBV/HCC | II | Father and one
brother | 2012-03-17 |
| Control | M | 38 | Healthy | – | None | 2012-03-18 |
Familial aggregation of HCC is defined as having a
first degree relative (including parents, siblings and children)
with HCC (7). The diagnosis of
familial aggregation of HCC of these 3 patients was confirmed with
the help of a standardized questionnaire that was given to all
patients at the baseline visit. Other family members also received
liver resection or medical treatment at Tongji Hospital with the
diagnosis of HCC. Their blood samples are no longer available as
they have passed away.
miRNA microarray analysis
Plasma samples were acquired by high-speed
centrifugation (12,000 × g at 4°C for 10 min) and stored at −80°C.
Agilent human miRNA V16.0 from Agilent Technologies (Santa Clara,
CA, USA) was used to identify differentiated miRNAs between
familial patients with HCC with HBV infection and the healthy
volunteer. All protocols were performed by the professional
Shanghai Biological Corporation (Shanghai, China).
Data analysis
According to the results of microarray analysis, the
miRNAs that exhibited ≥20-fold increase or decrease in the plasma
of patients with HCC compared with the healthy control were
selected. To evaluate the list of differentially-expressed miRNAs
in the context of the current literature, the names of these
differentially-expressed miRNAs were used as a keyword for online
searches and detailed information was extracted. The following
databases were utilized: PubMed (https://www.ncbi.nlm.nih.gov/pubmed/), Web of Science
(http://www.webofknowledge.com/) and
Google Scholar (http://scholar.google.com) search.
Results
Microarray results of the
differentially-expressed miRNAs in the plasma of patients with
HCC
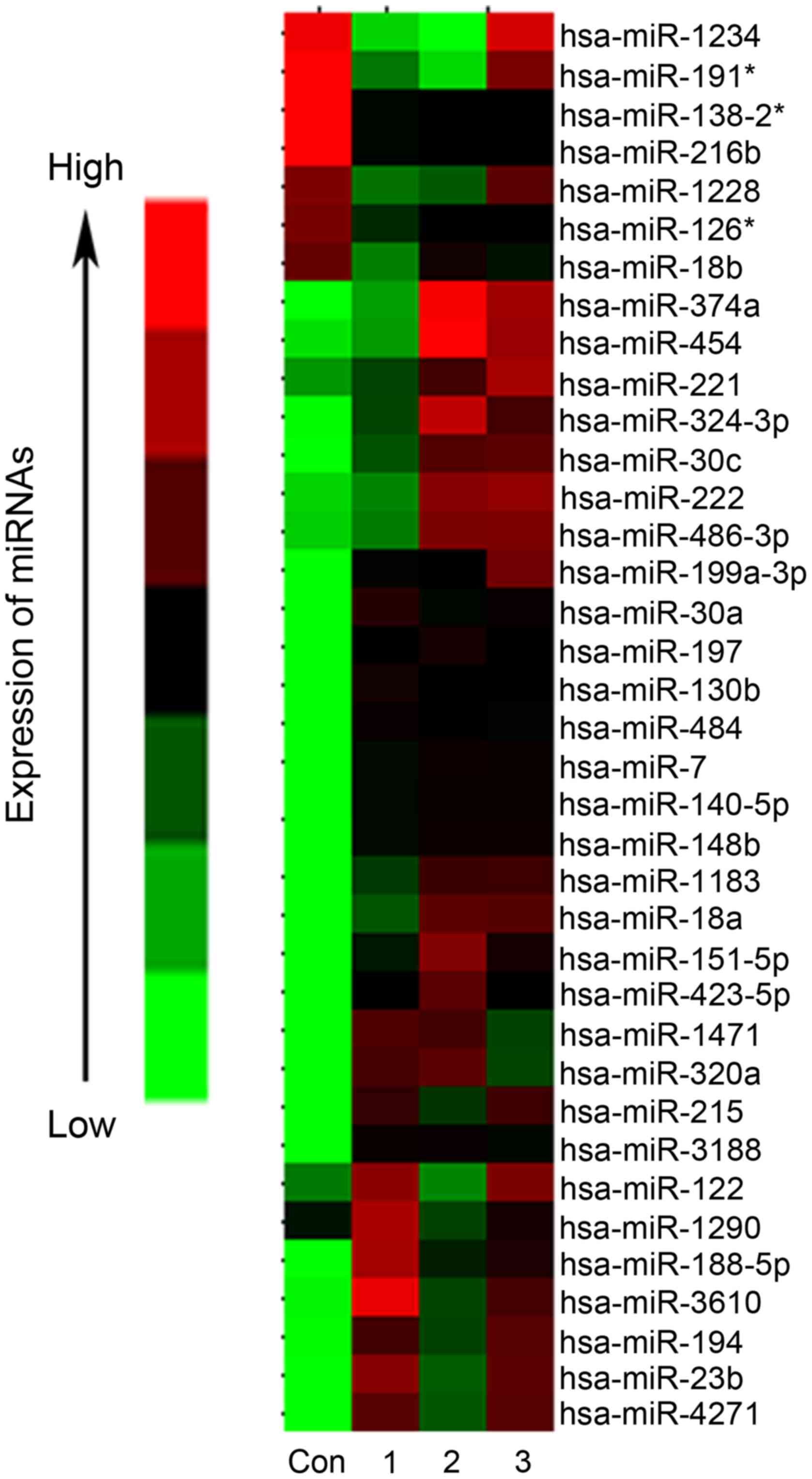
Microarray results identified 37 miRNAs with a
significant difference in expression between the patients with HCC
and the healthy control, including 30 upregulated and 7
downregulated (Fig. 1) (27). Among the differentially expressed
miRNAs, 26 miRNAs exhibited a ≥20-fold increase or decrease in the
plasma of patients with HCC, compared with the healthy control.
By searching the databases of PubMed, Web of Science
and Google Scholar, 15 of these differentially-expressed miRNAs
have been extensively reported in HCC, and 12 of them have been
fully elucidated for their roles in the progression of
hepatocarcinogenesis (Table II).
Notably, the remaining 11 miRNAs (10 upregulated and 1
downregulated) have never been reported in liver cancer (Table III). Among them, the levels of
plasma miR-3188, miR-374a, miR-4271, miR-1471, miR-188-5p,
miR-1183, miR-3610, miR-454, miR-324-3p and miR-484 were
significantly increased in patients with HCC compared with the
healthy control, corresponding to a fold-change of 124.5, 121.4,
96.8, 96.3, 82.1, 81.9, 64.8, 62.9, 48.7 and 29.4, respectively
(P<0.05). Comparing the patients with HCC with the healthy
control, only plasma miR-216b level was significantly downregulated
with a fold-change of 127.6 (P<0.01). The expression status of
miR-216b was investigated in 150 HCC tissues, and it was observed
that miR-216b expression was downregulated in 90 patients compared
with normal liver tissues. The 5-year overall survival and
disease-free survival rates were significantly improved in HCC
patients with increased miR-216b expression in comparison with the
downregulated miR-216b expression group (P<0.001) (27).
 | Table II.miRNAs have been reported in HCC,
which exhibited at least a 20-fold increase or decrease in plasma
expression of patients with HCC compared with the healthy
control. |
Table II.
miRNAs have been reported in HCC,
which exhibited at least a 20-fold increase or decrease in plasma
expression of patients with HCC compared with the healthy
control.
| miRNA name | Sequence | Fold-change | Targets | Published year | Chr | HBx regulation |
|---|
| hsa-miR-215 |
GTCTGTCAATTCATAGGTCAT | 277.3 | PTPRT | 2014 | chr1 | Yes |
| hsa-miR-3188 | CCCCGTATCCGCA | 124.5 | mTOR | 2016 | chr19 | No |
| hsa-miR-23b |
GGTAATCCCTGGCAATG | 102.9 | c-Met | 2009 | chr9 | No |
| hsa-miR-18a |
CTATCTGCACTAGATGCA | 97.3 | ER-α | 2009 | chr13 | Yes/no |
| hsa-miR-320a | TCGCCCTCTCAAC | 88.3 | GNAI1 | 2012 | chr8 | No |
|
hsa-miR-199a-3p |
TAACCAATGTGCAGACTACT | 63.7 | mTOR/c-Met | 2010 | chr1 | No |
| hsa-miR-151-5p |
ACTAGACTGTGAGCTCC | 61.2 | RhoGDIA | 2010 | chr8 | No |
| hsa-miR-140-5p |
CTACCATAGGGTAAAACCACT | 38.2 | TGF-β | 2013 | chr16 | No |
| hsa-miR-130b |
ATGCCCTTTCATCATTGC | 34.9 | PPAR-γ | 2014 | chr22 | No |
| hsa-miR-197 |
GCTGGGTGGAGAAGGTG | 33.8 | CD82 | 2014 | chr1 | No |
| hsa-miR-148b |
ACAAAGTTCTGTGATGCAC | 33.4 | NA | 2014 | chr12 | No |
| hsa-miR-30a |
CTTCCAGTCGAGGATG | 31.1 | SNAI1 | 2014 | chr6 | No |
| hsa-miR-7 |
ACAACAAAATCACTAGTCTTCC | 24.5 | CCNE1 | 2013 | chr9 | Yes |
| hsa-miR-30c |
GCTGAGAGTGTAGGATGT | 24.4 | NA | 2014 | chr1 | No |
| hsa-miR-194 |
TCCACATGGAGTTGCT | 22.2 | NA | 2014 | chr1 | No |
| hsa-miR-138-2* |
AACCCTGGTGTCGTGA | −106.3 | Cyclin D3 | 2012 | chr16 | Yes/no |
 | Table III.miRNAs have never been reported in
HCC, which exhibited at least a 20-fold increase or decrease in
expression of patients with HCC compared with the healthy
control. |
Table III.
miRNAs have never been reported in
HCC, which exhibited at least a 20-fold increase or decrease in
expression of patients with HCC compared with the healthy
control.
| miRNA name | Sequence | Fold-change | Chr |
|---|
| hsa-miR-374a |
CACTTATCAGGTTGTATTATAA | 121.4 | chrX |
| hsa-miR-4271 |
CCCCACCTTTTCTTCC | 96.8 | chr3 |
| hsa-miR-1471 |
ACACCTGGCTCCACA | 96.3 | chr2 |
| hsa-miR-188-5p | CCCTCCACCATGC | 82.1 | chrX |
| hsa-miR-1183 |
TGCCCACTCTCACCA | 81.9 | chr7 |
| hsa-miR-3610 | CGGCGCCTCCTT | 64.8 | chr8 |
| hsa-miR-454 |
ACCCTATAAGCAATATTGCAC | 62.9 | chr17 |
| hsa-miR-324-3p |
CCAGCAGCACCTGGGG | 48.7 | chr17 |
| hsa-miR-484 |
ATCGGGAGGGGACTGA | 29.4 | chr16 |
| hsa-miR-216b |
TCACATTTGCCTGCAG | −127.6 | chr2 |
Discussion
Dysregulation of miRNAs is involved in the
initiation and progression of various cancers, including HCC
(19). Previous studies revealed that
common and specific mechanisms exist at the miRNA level during
HBV-induced hepatocarcinogenesis (29), and miRNAs may function as potential
biomarkers for HBV-associated HCC (20,30). In
addition, family history of HCC may improve the risk of HCC at each
stage of HBV infection (7). The
present study aimed to identify the plasma miRNAs that are
differentially expressed in patients with HCC with familial
aggregation of HCC and HBV infection.
In the present study, by virtue of microarray
analysis, specific groups of miRNAs were identified, whose
expression is significantly altered in the plasma of patients with
HCC with familial aggregation of HCC and HBV infection. A total of
37 differently-expressed miRNAs were identified, and among them, 26
miRNAs exhibited ≥20-fold-changes in the plasma of patients with
HCC compared with the healthy controls. By reviewing the available
literature published in previous years, among these 37
differentially expressed miRNAs, 15 miRNAs have been reported in
HCC, and 12 miRNAs of them have been elaborated for the detailed
molecular mechanism in the progression of HCC (Table II). The remaining 11 miRNAs have
never been studied in liver cancer (Table III). Among the 11 differentially
dysregulated miRNAs, the function of miR-374a, miR-188-5p,
miR-1183, miR-454, miR-324-3p, miR-484, miR-3188, and miR-216b have
been studied in human cancers (27,31–43);
however, the exact function of miR-4271, miR-1471 and miR-3610
remains unclear.
Of these 11 miRNAs, miR-216b exhibited a 127.6-fold
decrease in expression in the plasma of patients with HCC compared
with that of the healthy control. Deng et al (44) firstly observed miR-216b expression in
nasopharyngeal carcinoma and indicated that miR-216b was
downregulated in nasopharyngeal carcinoma cell lines and specimens.
miR-216b may suppress tumor growth and invasion by targeting KRAS,
which performs an important role in the initiation and progression
of HBV-associated HCC in mice models (45). Other studies have also revealed that
miR-216b functions as a tumor-suppressor gene in colorectal and
pancreatic cancers (46,47). The function of miR-216b in the
pathogenesis of HCC was then studied. The present results indicated
that miR-216b expression was significantly lower (P<0.001) in
tumor tissues compared with adjacent liver tissues, and its
expression was associated with tumor size, HBV infection, HBV-DNA
quantity and incidence of portal vein tumor thrombosis (27). Prognostic analysis revealed that the
5-year overall survival and disease-free survival rates were
significantly improved in patients with HCC with increased miR-216b
expression in comparison with the downregulated miR-216b expression
group (P<0.001). In an in vitro study, miR-216b inhibited
cellular proliferation, migration and invasion of HCC by directly
targeting insulin-like growth factor mRNA binding protein 2, and
was downregulated by HBx (27). When
a miR-216b-specific inhibitor was used to block miR-216b expression
in SMMC-7721 cells, in vitro and in vivo assays
revealed a significant increase in proliferation, migration and
invasion, compared with control SMMC-7721 cells (27).
Cai et al (31)
demonstrated that miR-374a may activate Wnt/β-catenin signaling to
promote breast cancer metastasis and may perform as a therapeutic
target for early metastatic breast cancer. Wnt/β-catenin signaling
has been demonstrated to perform an important role in the
development and promotion of liver cancer metastasis (48), and the HBx protein is essential for
the activation of Wnt/β-catenin signaling in hepatoma cells
(49). The expression of miR-374a was
investigated in tumor tissues and adjacent normal tissues in the
present study, and it was identified that no difference was
observed between HCC and para-HCC tissue. The function of miR-374a
in HBV-associated HCC requires additional study. A previous study
indicated that miR-1183 and miR-188-5p may represent specific
predictors of response to chemoradiotherapy in rectal cancer
(32). miR-188-5p is involved in the
process of prostate cancer by regulating lysosomal protein
transmembrane 4 β (LAPTM4B) (33).
The downregulation of miR-188-5p is an independent prognostic
factor for poor overall and biochemical recurrence-free survival
rates, restoration of miR-188-5p in prostate cancer (PCa) cells
significantly suppresses proliferation, migration and invasion
in vitro and inhibits tumor growth and metastasis in
vivo (33); Overexpression of
miR-188-5p in PC-3 cells may significantly enhance the
chemosensitivity of the cells to adriamycin (33). The
miR-188-5p/LAPTM4B/phosphoinositide-3 kinase/protein kinase B
regulatory network performs an important role in PCa progression
and chemotherapeutic drug sensitivity (33). Recently, Fang et al (50) indicated that miR-188-5p was
significantly decreased in HCC tissue, and that overexpression of
miR-188-5p suppresses tumor cellular proliferation and metastasis
by directly targeting fibroblastic growth factor 5 in HCC. However,
in the present study, miR-188-5p exhibited an 82.1-fold increase in
the plasma of patients with HCC compared with the healthy control.
This demonstrated that the expression of miRNAs in the serum cannot
completely reflect its expression in the tissue.
Hu et al (34)
detected the serum miRNA profiling in patients with breast cancer.
Normalized by the two endogenous control miRNAs, the authors
revealed that the serum miR-324-3p expression may act as a
non-invasive prediction biomarker for breast cancer. Macconi et
al (35) indicated that
miR-324-3p promotes renal fibrosis by targeting prolyl
endopeptidase. In addition, a previous study revealed that
miR-324-3p targets the promoter of RelA, commonly known as p65, a
subunit of nuclear factor-kB, and significantly induced the
endogenous RelA mRNA and protein expression in PC12 cells (36). The present results also demonstrated
that miR-324-3p exhibited a 48.7-fold increase in the plasma of
patients with HCC compared with the control, which may be
considered as a biomarker in HCC.
miR-484 has been identified to be increased in the
serum of patients with breast and pancreatic cancer (37). The present results revealed that
miR-484 exhibited a 29.7-fold-change in expression in patients with
HCC compared with the healthy control. These findings indicated
that miR-484 may be produced as a result of processes common to
these three cancers. The study by Wang et al (38) also indicated that miR-484 may function
as an oncogene, suppress translation of mitochondrial fission
protein (Fis1) and inhibit Fis1-mediated fission and apoptosis.
Previous studies have demonstrated that miR-454 acts
as an oncogene or tumor suppressor in cancer (39–41).
miR-454 has been reported to be decreased in tissue of patients
with esophageal cancer compared with normal tissue, which may
perform as novel molecular markers of esophageal cancer (39). Niu et al (40) demonstrated that miR-454 expression was
downregulated in osteosarcoma tissues, acting as a tumor suppressor
gene in osteosarcoma. The miR-454 expression was increased in
colorectal cancer (CRC) tissues and CRC cells. Overexpression of
miR-454 promoted the proliferation and anchorage-independent growth
of CRC cells and its oncogenic effect was mediated chiefly through
direct suppression of cylindromatosis expression (41). In the present study, miR-454 exhibited
a 62.9-fold increase in the plasma of patients with HCC in
comparison with the healthy control, demonstrating that miR-454 may
function as an oncogene. A total of 110 pairs of HCC tissues and
para-cancer tissues were compared, and the results revealed that
miR-454 was significantly increased in 83 HCC tissues compared with
para-cancer tissues. The 5-year overall survival and disease-free
survival rates were significantly improved in patients with low
expression of miR-454 (data unpublished). Until recently, Yu et
al (42) indicated that miR-454
expression was upregulated in HCC cell lines and tissues. Knockdown
of miR-454 inhibited HCC cellular proliferation, invasion and
epithelial mesenchymal transition (EMT), whereas overexpression of
miR-454 promoted HCC cellular proliferation and invasion and EMT,
which was consistent with the present results. miR-3188 directly
targets mTOR to stimulate its own expression, and participates in
FOXO1-mediated repression of cell growth, tumorigenesis and
nasopharyngeal carcinoma chemotherapy resistance (43). However, miR-3188 has never been
studied in HCC, and the aim of the present study was to investigate
the function of miR-3188 in the pathogenesis of HCC.
In conclusion, the present study (in a small sample
size) identified a list of differentially-expressed miRNA
candidates in the plasma of patients with familial HCC with HBV
infection compared with the healthy control. A total of 11 of these
miRNAs have not previously been reported in the molecular
pathogenesis of hepatocarcinogenesis. These differentially
expressed miRNAs in the plasma of patients with HCC with familial
aggregation of HCC and HBV infection may lay the foundation for
additional studies on the role of miRNAs in the pathogenesis of
HCC, and provide the basal work for future study of the molecular
mechanisms of HCC. Additional studies are necessary in order to
understand the regulatory mechanisms of these miRNAs in the process
of hepatocarcinogenesis.
References
|
1
|
Zhang EL, Liang BY, Chen XP and Huang ZY:
Severity of liver cirrhosis: A key role in the selection of
surgical modality for Child-Pugh A hepatocellular carcinoma. World
J Surg Oncol. 13:1482015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kew MC: Epidemiology of chronic hepatitis
B virus infection, hepatocellular carcinoma, and hepatitis B
virus-induced hepatocellular carcinoma. Pathol Biol (Paris).
58:273–277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang HI, Lu SN, Liaw YF, You SL, Sun CA,
Wang LY, Hsiao CK, Chen PJ, Chen DS and Chen CJ; Taiwan
Community-Based Cancer Screening Project Group, : Hepatitis B e
antigen and the risk of hepatocellular carcinoma. N Engl J Med.
347:168–174. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lok AS: Prevention of hepatitis B
virus-related hepatocellular carcinoma. Gastroenterology. 127 Suppl
1:S303–S309. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mair RD, Valenzuela A, Ha NB, Ayoub WS,
Daugherty T, Lutchman GA, Garcia G, Ahmed A and Nguyen MH:
Incidence of hepatocellular carcinoma among US patients with
cirrhosis of viral or nonviral etiologies. Clin Gastroenterol
Hepatol. 10:1412–1417. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu MW, Chang HC, Liaw YF, Lin SM, Lee SD,
Liu CJ, Chen PJ, Hsiao TJ, Lee PH and Chen CJ: Familial risk of
hepatocellular carcinoma among chronic hepatitis B carriers and
their relatives. J Natl Cancer Inst. 92:1159–1164. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Loomba R, Liu J, Yang HI, Lee MH, Lu SN,
Wang LY, Iloeje UH, You SL, Brenner D and Chen CJ; REVEAL-HBV Study
Group, : Synergistic effects of family history of hepatocellular
carcinoma and hepatitis B virus infection on risk for incident
hepatocellular carcinoma. Clin Gastroenterol Hepatol.
11:1636–1645.e1-3. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Turati F, Edefonti V, Talamini R,
Ferraroni M, Malvezzi M, Bravi F, Franceschi S, Montella M, Polesel
J, Zucchetto A, et al: Family history of liver cancer and
hepatocellular carcinoma. Hepatology. 55:1416–1425. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xie KL, Zhang YG, Liu J, Zeng Y and Wu H:
MicroRNAs associated with HBV infection and HBV-related HCC.
Theranostics. 4:1176–1192. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao Z, Hu Y, Shen X, Lao Y, Zhang L, Qiu
X, Hu J, Gong P, Cui H, Lu S, et al: HBx represses RIZ1 expression
by DNA methyltransferase 1 involvement in decreased miR-152 in
hepatocellular carcinoma. Oncol Rep. 37:2811–2818. 2017.PubMed/NCBI
|
|
11
|
Wu Y, Zhang J, Zhang H and Zhai Y:
Hepatitis B virus X protein mediates yes-associated protein 1
upregulation in hepatocellular carcinoma. Oncol Lett. 12:1971–1974.
2016.PubMed/NCBI
|
|
12
|
Xu C, Zhou W, Wang Y and Qiao L: Hepatitis
B virus-induced hepatocellular carcinoma. Cancer Lett. 345:216–222.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang S and He X: The role of microRNAs in
liver cancer progression. Br J Cancer. 104:235–240. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Braconi C, Huang N and Patel T:
MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor
suppressor gene expression by interleukin-6 in human malignant
cholangiocytes. Hepatology. 51:881–890. 2010.PubMed/NCBI
|
|
16
|
Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X
and Wu Z: miR-101 is down-regulated by the hepatitis B virus ×
protein and induces aberrant DNA methylation by targeting DNA
methyltransferase 3A. Cell Signal. 25:439–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu
Z, Liu R and Wu Z: Epigenetic repression of miR-132 expression by
the hepatitis B virus × protein in hepatitis B virus-related
hepatocellular carcinoma. Cell Signal. 25:1037–1043. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dong Y and Wang A: Aberrant DNA
methylation in hepatocellular carcinoma tumor suppression (Review).
Oncol Lett. 8:963–968. 2014.PubMed/NCBI
|
|
19
|
Liu WH, Yeh SH and Chen PJ: Role of
microRNAs in hepatitis B virus replication and pathogenesis.
Biochim Biophys Acta. 1809:678–685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z,
Wang JF, Zhang Z, Lu S, Huang X, et al: Plasma microRNA panel to
diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin
Oncol. 29:4781–4788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fu Y, Wei X, Tang C, Li J, Liu R, Shen A
and Wu Z: Circulating microRNA-101 as a potential biomarker for
hepatitis B virus-related hepatocellular carcinoma. Oncol Lett.
6:1811–1815. 2013.PubMed/NCBI
|
|
22
|
Katayama Y, Maeda M, Miyaguchi K, Nemoto
S, Yasen M, Tanaka S, Mizushima H, Fukuoka Y, Arii S and Tanaka H:
Identification of pathogenesis-related microRNAs in hepatocellular
carcinoma by expression profiling. Oncol Lett. 4:817–823.
2012.PubMed/NCBI
|
|
23
|
Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow
P, Chung AY, Jooi LL and Lee CG: Lethal-7 is down-regulated by the
hepatitis B virus × protein and targets signal transducer and
activator of transcription 3. J Hepatol. 53:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kong G, Zhang J, Zhang S, Shan C, Ye L and
Zhang X: Upregulated microRNA-29a by hepatitis B virus X protein
enhances hepatoma cell migration by targeting PTEN in cell culture
model. PLoS One. 6:e195182011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu G, Yu F, Xiao Z, Xu K, Xu J, Tang W,
Wang J and Song E: Hepatitis B virus X protein downregulates
expression of the miR-16 family in malignant hepatocytes in vitro.
Br J Cancer. 105:146–153. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng
X, Zhu Z, Jiao H, Lin J, Jiang K, et al: Hepatitis B virus X
protein represses miRNA-148a to enhance tumorigenesis. J Clin
Invest. 123:630–645. 2013.PubMed/NCBI
|
|
27
|
Liu FY, Zhou SJ, Deng YL, Zhang ZY, Zhang
EL, Wu ZB, Huang ZY and Chen XP: MiR-216b is involved in
pathogenesis and progression of hepatocellular carcinoma through
HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis.
6:e16702015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hassan MM, Spitz MR, Thomas MB, Curley SA,
Patt YZ, Vauthey JN, Glover KY, Kaseb A, Lozano RD, El-Deeb AS, et
al: The association of family history of liver cancer with
hepatocellular carcinoma: A case-control study in the United
States. J Hepatol. 50:334–341. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang ZZ, Liu X, Wang DQ, Teng MK, Niu LW,
Huang AL and Liang Z: Hepatitis B virus and hepatocellular
carcinoma at the miRNA level. World J Gastroenterol. 17:3353–3358.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X,
Zhou X and Gan J: A serum microRNA panel as potential biomarkers
for hepatocellular carcinoma related with hepatitis B virus. PLoS
One. 9:e1079862014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan
J, Wu J and Li M: MicroRNA-374a activates Wnt/β-catenin signaling
to promote breast cancer metastasis. J Clin Invest. 123:566–579.
2013.PubMed/NCBI
|
|
32
|
Della Vittoria, Scarpati G, Falcetta F,
Carlomagno C, Ubezio P, Marchini S, De Stefano A, Singh VK,
D'Incalci M, De Placido S and Pepe S: A specific miRNA signature
correlates with complete pathological response to neoadjuvant
chemoradiotherapy in locally advanced rectal cancer. Int J Radiat
Oncol Biol Phys. 83:1113–1119. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang H, Qi S, Zhang T, Wang A, Liu R, Guo
J, Wang Y and Xu Y: miR-188-5p inhibits tumour growth and
metastasis in prostate cancer by repressing LAPTM4B expression.
Oncotarget. 6:6092–6104. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao
Y, Tang J, Chen X, Dai J, Wei Q, et al: Serum microRNA profiling
and breast cancer risk: The use of miR-484/191 as endogenous
controls. Carcinogenesis. 33:828–834. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Macconi D, Tomasoni S, Romagnani P,
Trionfini P, Sangalli F, Mazzinghi B, Rizzo P, Lazzeri E, Abbate M,
Remuzzi G and Benigni A: MicroRNA-324-3p promotes renal fibrosis
and is a target of ACE inhibition. J Am Soc Nephrol. 23:1496–1505.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dharap A, Pokrzywa C, Murali S, Pandi G
and Vemuganti R: MicroRNA miR-324-3p induces promoter-mediated
expression of RelA gene. PLoS One. 8:e794672013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zearo S, Kim E, Zhu Y, Zhao JT, Sidhu SB,
Robinson BG and Soon PSh: MicroRNA-484 is more highly expressed in
serum of early breast cancer patients compared to healthy
volunteers. BMC Cancer. 14:2002014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang K, Long B, Jiao JQ, Wang JX, Liu JP,
Li Q and Li PF: miR-484 regulates mitochondrial network through
targeting Fis1. Nat Commun. 3:7812012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ,
Wang TY, Li HC and Wu XN: Differential expression of miRNAs in
esophageal cancer tissue. Oncol Lett. 5:1639–1642. 2013.PubMed/NCBI
|
|
40
|
Niu G, Li B, Sun J and Sun L: miR-454 is
down-regulated in osteosarcomas and suppresses cell proliferation
and invasion by directly targeting c-Met. Cell Prolif. 48:348–355.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu L, Nie J, Chen L, Dong G, Du X, Wu X,
Tang Y and Han W: The oncogenic role of microRNA-130a/301a/454 in
human colorectal cancer via targeting Smad4 expression. PLoS One.
8:e555322013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu L, Gong X, Sun L, Yao H, Lu B and Zhu
L: miR-454 functions as an oncogene by inhibiting CHD5 in
hepatocellular carcinoma. Oncotarget. 6:39225–39234.
2015.PubMed/NCBI
|
|
43
|
Zhao M, Luo R, Liu Y, Gao L, Fu Z, Fu Q,
Luo X, Chen Y, Deng X, Liang Z, et al: miR-3188 regulates
nasopharyngeal carcinoma proliferation and chemosensitivity through
a FOXO1-modulated positive feedback loop with
mTOR-p-PI3K/AKT-c-JUN. Nat Commun. 7:113092016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Deng M, Tang H, Zhou Y, Zhou M, Xiong W,
Zheng Y, Ye Q, Zeng X, Liao Q, Guo X, et al: miR-216b suppresses
tumor growth and invasion by targeting KRAS in nasopharyngeal
carcinoma. J Cell Sci. 124:2997–3005. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ye H, Zhang C, Wang BJ, Tan XH, Zhang WP,
Teng Y and Yang X: Synergistic function of Kras mutation and HBx in
initiation and progression of hepatocellular carcinoma in mice.
Oncogene. 33:5133–5138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim SY, Lee YH and Bae YS: MiR-186,
miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular
senescence by targeting α subunit of protein kinase CKII in human
colorectal cancer cells. Biochem Biophys Res Commun. 429:173–179.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ali S, Banerjee S, Logna F, Bao B, Philip
PA, Korc M and Sarkar FH: Inactivation of Ink4a/Arf leads to
deregulated expression of miRNAs in K-Ras transgenic mouse model of
pancreatic cancer. J Cell Physiol. 227:3373–3380. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Anson M, Crain-Denoyelle AM, Baud V,
Chereau F, Gougelet A, Terris B, Yamagoe S, Colnot S, Viguier M,
Perret C and Couty JP: Oncogenic β-catenin triggers an inflammatory
response that determines the aggressiveness of hepatocellular
carcinoma in mice. J Clin Invest. 122:586–599. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cha MY, Kim CM, Park YM and Ryu WS:
Hepatitis B virus X protein is essential for the activation of
Wnt/beta-catenin signaling in hepatoma cells. Hepatology.
39:1683–1693. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fang F, Chang RM, Yu L, Lei X, Xiao S,
Yang H and Yang LY: MicroRNA-188-5p suppresses tumor cell
proliferation and metastasis by directly targeting FGF5 in
hepatocellular carcinoma. J Hepatol. 63:874–885. 2015. View Article : Google Scholar : PubMed/NCBI
|