Introduction
Melatonin, a hormone secreted from the pineal gland
in the brain at night, mainly regulates the circadian rhythm
through seven transmembrane G-protein-coupled receptors, melatonin
receptor type 1A (MT1) and melatonin receptor type 1B (1–7). In
addition, melatonin is produced by other organs, including the
skin, bone marrow and lymphocytes (6,8). Breast
cancer is frequently identified in women, and demonstrates high
mortality once metastasized (9–11).
Patients with breast cancer present with low levels of melatonin,
which may be due to disruptions of the circadian rhythm (4,12–15). Melatonin treatment has been suggested
to halt breast cancer progression and improve quality of life in
patients with breast cancer (16,17).
Breast cancer is subcategorized by the expression
patterns of hormone receptors and human epidermal growth factor 2
(7,18). Triple-negative breast cancer is
aggressive and associated with poor prognosis (18). Triple-negative breast cancer cells are
also invasive, resulting in higher metastatic rates (18). However, therapeutic options are
lacking. Expression levels of MT1 are associated with survival
rates in African American and Caucasian women, suggesting that
melatonin may be a therapeutic option (19). However, the effect of melatonin on
triple-negative breast cancer cells remains largely unknown.
Kisspeptin (KiSS1), a gene product of KiSS1,
is known to regulate the onset of puberty and to suppress cancer
metastasis (20,21). KiSS1 expression levels are increased
in primary breast cancer lesions but reduced in metastatic lesions
(22–25), suggesting that KiSS1 may serve
pleiotropic functions during breast cancer development and
metastasis (20). While it was
revealed that melatonin regulates KiSS1 expression in the
hypothalamus in the brain (26,27), an
association between melatonin and KiSS1 in cancer has not been
identified. While 1 mM melatonin treatment inhibited metastatic
functions, including migration and invasion in triple-negative
MDA-MB-231 breast cancer cell lines, the pharmacological
concentration of melatonin also reduced the cell viability and
caused apoptosis (28–30). In addition, physiological
concentrations of melatonin only reduced the viabilities of less
invasive, non-triple-negative breast cancer cells (28,31,32), which
may be due to different MT1 expression levels in less or highly
invasive breast cancer cells (33).
Therefore, whether melatonin directly inhibits the invasion of
highly metastatic triple-negative breast cancer cells remains
unclear.
The GATA family of transcription factors is crucial
for determining cell fate, and is composed of six conserved members
that bind to the DNA sequence (A/T)GATA(A/G) (34–37). GATA
binding protein 3 (GATA3) has emerged as crucial for mammary
luminal cell fate (37,38). GATA3 expression patterns in breast
cancer appear to correlate with the expression patterns of estrogen
receptors and progesterone receptors (39,40).
Therefore, less invasive, hormone-positive breast cancer is likely
to express high levels of GATA3 (40,41).
Inversely, patients with highly invasive triple-negative breast
cancer may present with low levels of GATA3, which appears to be
maintained during metastasis (41,42).
The aim of the present study was to understand the
function of melatonin in triple-negative breast cancer cells. To
investigate the effect of melatonin on breast cancer cell
invasiveness, highly metastatic triple-negative breast cancer cells
were treated with melatonin at concentrations ranging from 1 nM to
10 µM. Melatonin at concentrations of 100 nM to 10 µM inhibited
triple-negative breast cancer cell migration and invasion with no
effect on cell proliferation. In addition, melatonin-induced KiSS1
expression prolonged its inhibition of metastatic abilities, as
confirmed by promoter assays and KiSS1 silencing in the
cells. It was also revealed that melatonin increased the expression
levels and activated transcriptional activity of GATA3 for KiSS1
expression. Therefore, the results of the present study suggested
that melatonin prolongs the inhibitory effect on breast cancer
metastasis by activating GATA3-mediated KiSS1 expression.
Materials and methods
Cell lines and reagents
MDA-MB-231, HCC-70 and 293T cells (American Type
Culture Collection, Manassas, VA, USA) were cultured in Dulbecco's
modified Eagle's medium supplemented with 5% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1%
penicillin/streptomycin solution (Gibco; Thermo Fisher Scientific,
Inc.). Melatonin was purchased from Sigma-Aldrich; Merck KGaA
(Darmstadt, Germany) and diluted in distilled water to a final
concentration of 50 ug/ml. KiSS1 was obtained from Phoenix
Pharmaceuticals, Inc. (Burlingame, CA, USA).
In silico promoter analyses, reporter
assays and chromatin immunoprecipitation assays
The human KiSS1 promoter region was analyzed in
silico using the AliBaba2 prediction tool (http://www.gene-regulation.com/pub/programs/alibaba2/index.html).
Different sizes of KiSS1 promoter regions, constructed in pGL3
plasmids (Promega Corporation, Madison, WI, USA), were used in
luciferase assays. Each promoter region was amplified by PCR and
cloned into pGL3 plasmids. These plasmids were named KiSS1-luc
plasmids and the pGL3 backbone was the same as the KiSS1-luc. The
cells were transfected with KiSS1-luc plasmids using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instruction, and subjected to the luciferase assays
following the manufacturer's protocol (Dual-Luciferase Reporter
Assay system; Promega Corporation). Luciferase assays were
performed in triplicate, and three times independently. Chromatin
immunoprecipitation assays were performed according to the
manufacturer's protocol (ChIP Kit Magnetic One-Step; Abcam,
Cambridge, UK). The GATA3 antibody purchased from Abcam (cat no.
ab199428; dilution, 1:25), were used for the chromatin
immunoprecipitation assays. The GATA3 binding region was amplified
using a primer set as follows: Forward, 5′-CCAAAGTAAGTC-3′ and
reverse, 5′-CTTCCCTCCAGGG-3′.
RNA and protein analysis
For analyzing RNAs, 5×106 cells were
lysed with TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), and
cDNA was amplified with routine PCR procedures: For the RNA
analysis, the present study used the Reverse Transcription system
(Promega Corporation). Reverse transcription was performed using
oligo(dT)15 primers at 42°C, according to the
manufacturer's protocol. Samples were incubated for 10 min at 25°C,
reverse transcribed for 30 min at 42°C and then placed for 5 min at
95°C for the inactivation of the reverse transcription. Real-time
PCR was performed using SYBR-Green Real-Time PCR Master Mix (cat
no. 4309155; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Samples were incubated for 10 min at 95°C,
denatured for 15 sec at 95°C and annealed and extended for 1 min at
60°C, for 40 cycles. KiSS1 mRNA expression levels was
examined using the primer sequences as follows: Forward,
5′-GCCCACCATGAACTCACTG-3′ and reverse, 5′-CTGCCCCGCACCTGCG-3′.
β-actin mRNA was also amplified with primers, as follows:
Forward, 5′GGC TCC GGC ATG TGC AAG GC3′ and reverse, 5′CTG CCC CGC
ACC TGC G3′, as an internal control. PCRs were performed using a
LightCycler 480 Instrument II, and relative quantifications were
automatically conducted by the efficiency method (43,44) in
LightCycler 480 software 1.5 (both from Roche Diagnostics,
Indianapolis, IN, USA). For analyzing the protein levels,
1×106 cells were lysed using radioimmunoprecipitation
buffer for 30 min on ice, and centrifuged at 20,000 × g for 10 min
at 4°C. Subsequent to measuring the protein concentration with
Pierce BCA protein assay kit (Pierce; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol, 30 µg protein was
loaded and processed with 6–10% SDS-PAGE, and transferred to
polyvinylidene fluoride membranes. Subsequent to blocking with 5%
milk for 1 h at room temperature, the membrane was incubated with
KiSS1 (dilution, 1:250), GATA3 (dilution, 1:500) and β-actin
(dilution, 1:500) antibodies for an additional 1 h at room
temperature. β-actin was detected as an internal control. The
KiSS1, GATA3 and β-actin antibodies were obtained from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA). The membranes were then
incubated in secondary antibodies conjugated to horseradish
peroxidase (anti-rabbit, cat no. 7074; anti-mouse, cat no. 7076;
Cell Signaling Technology, Inc., Danvers, WI, USA) at 1:2,000 in
dilution for 1 h at room temperature. Band detection was performed
using LumiGLO chemiluminescent reagent and peroxidase (cat no.
7003; Cell Signaling Technology, Inc.). Western blot experiments
were replicated at least three times independently. For relative
quantifications, ImageJ software (version 1.50) was used (National
Institutes of Health, Bethesda, MA, USA).
Cell proliferation, migration and
invasion assays
Cells (1×105) were cultured in 96-well
plates, treated with different concentrations (0, 1, 10,
102, 103 and 104 nM) of melatonin
for 48 h, and then subjected to cell proliferation assays using the
CyQUANT Cell Proliferation Assay kit (Molecular Probes; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Experiments were performed in quadrate and independently repeated
in triplicate. For cell migration, 3×105 cells were
cultured in 6-well plates and scratched when the confluence reached
~80%. The cells were treated with 10 µM melatonin for 24 h at 37°C,
and then the number of migrated cells was counted. Experiments were
performed in triplicate. Non-treated cells were used as a control
in this migration assay. To examine invasiveness, cells were
cultured in the upper chambers of Matrigel-precoated Transwell
plates, and then treated with melatonin at 10 µM for 24 h at 37°C.
The cells in the upper chamber were removed with a swab, and cells
that had invaded through the Matrigel were stained with 0.4%
crystal violet and counted. Experiments were performed in
triplicate. Non-treated cells were used as a control. The migration
and invasion assays were observed using a Zeizz Axiovert inverted
microscope and images were analyzed using Zen software version 3.00
(Carl Zeizz, Oberkochen, Germany). A total of 4 fields were
randomly selected and the invaded cells were counted. Cells were
transfected with pcDNA (Invitrogen; Thermo Fisher Scientific,
Inc.), a pcDNA-GATA3 (human full length GATA3 sequence;
NM_001002295.1), control small interfering (si)RNA-A (cat no.
sc-37007) or KiSS1 siRNA (cat no. sc-37443) (both from Santa Cruz
Biotechnology, Inc.) for 24 h using Lipofectamine 2000®
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
protocol of the manufacturer, and then subjected to the cell
analyses including cell migration and invasion assays. In the
overexpression analysis, the pcDNA empty vector was used as the
control plasmid.
Statistical analysis
Statistical analysis was performed using unpaired
Student's t-tests or one-way analysis of variance with a post-hoc
Tukey's test was performed using SPSS version 22 (IBM Corp.,
Armonk, NY, USA). Results were expressed as the mean ± standard
deviation or mean ± standard error, and P<0.05 was considered to
indicate a statistically significant difference.
Results
Melatonin inhibits metastasis without
affecting cell proliferation
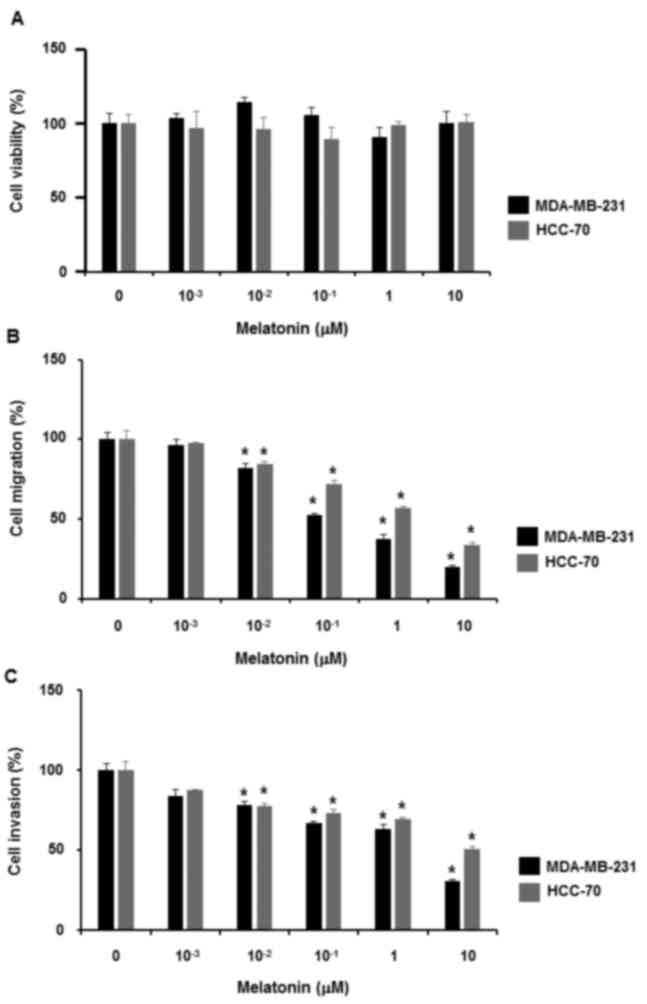
To examine the effect of melatonin on the
proliferation of triple-negative MDA-MB-231 and HCC-70 breast
cancer cells, the cells were treated with melatonin at different
concentrations (0, 1, 10, 102, 103 and
104 nM) for 48 h. Data from the cell proliferation
assays demonstrated that melatonin did not affect the proliferation
of triple-negative breast cancer cells (Fig. 1A).
Next, whether melatonin affects triple-negative
breast cancer cell migration and invasion was examined. When
triple-negative breast cancer cells were treated with melatonin at
different concentrations (0, 1, 10, 102, 103
and 104 nM) for 24 h, it was revealed that treatment
with 10 nM to 10 µM melatonin reduced cell migration (Fig. 1B). Likewise, 10 nM to 10 µM melatonin
treatment inhibited the invasiveness of triple-negative breast
cancer cells (Fig. 1C).
Melatonin induces KiSS1 expression via
GATA3
The results of the present study revealed that
melatonin inhibited the migration and invasion of triple-negative
breast cancer cells. This result was consistent with a previous
study (28). Nevertheless, the
mechanisms by which the inhibitory effect of melatonin is prolonged
were not investigated. It was assumed that melatonin may maintain
its inhibitory effect by inducing the production of anti-invasive
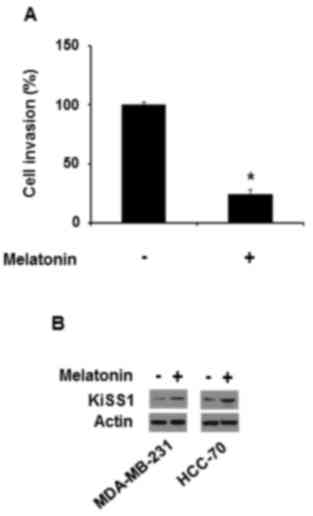
proteins. Therefore, the effect of conditional medium from the
cells treated with melatonin on the invasiveness of the breast
cancer cell lines was examined. Conditioned medium from MDA-MB-231
cells treated with 10 µM melatonin also repressed MDA-MB-231 cell
invasiveness (Fig. 2A), which
indicated that melatonin may prolong its inhibitory effect by
inducing the production of releasing factors that suppress
invasiveness.
KiSS1 is known to inhibit cancer cell migration and
invasion, resulting in a suppression of cancer metastases (20). Thus, whether melatonin affects KiSS1
expression in triple-negative breast cancer cells was examined.
Melatonin increased KiSS1 protein expression in MDA-MB-231 and
HCC-70 cells (Fig. 2B).
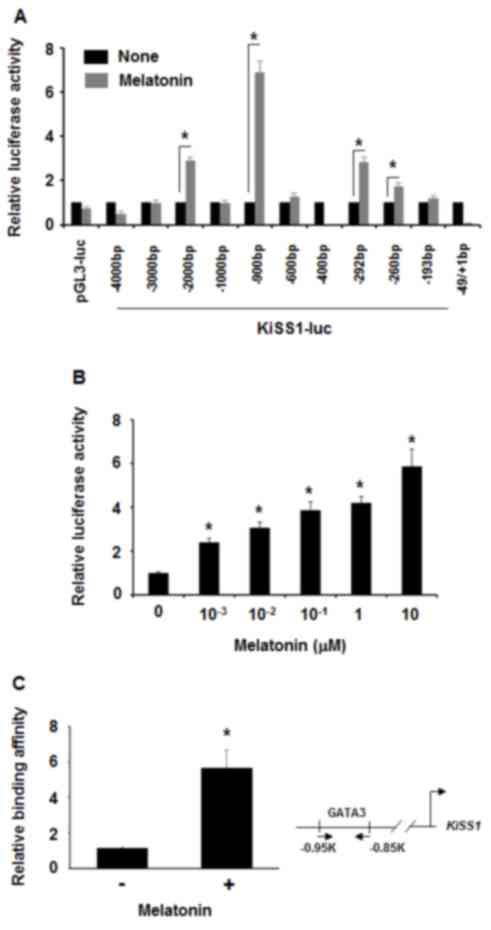
To examine whether melatonin affected KiSS1
expression at the transcriptional level, luciferase assays were
performed in 293T cells transfected with different KiSS1-luc
constructs. Treatment with 100 nM melatonin increased the
luciferase activity of −900 bp of the KiSS1 promoter region
(pKiSS1-900-luc) by ~7-fold, while not affecting the KiSS1 promoter
region of either −1 kb or −600 bp (Fig.
3A). Thus, it was hypothesized that melatonin may affect a
certain transcriptional factor that may bind to the KiSS1 promoter
region between −900 bp and −600 bp. When pKiSS1-900-luc was
transfected in MDA-MB-231 cells prior to treatment with different
concentrations of melatonin, it was revealed that luciferase
activities were increased by treatment with 1 nM to 1 µM melatonin
(Fig. 3B). When this region was
analyzed in silico, the GATA binding site was identified. A
previous study revealed that the zebra fish KiSS1 promoter contains
a GATA binding site (45). Thus, the
association between melatonin and GATA3 in transcriptional
regulation of the KiSS1 gene expression was also examined.
Consistently, in the chromatin immunoprecipitation assays,
melatonin induced GATA3 interaction with the KiSS1 promoter region
in MDA-MB-231 cells (Fig. 3C).
Therefore, these data indicated that melatonin activated KiSS1
expression at a transcriptional level.
Melatonin inhibits breast cancer cell
invasiveness via GATA3-dependent KiSS1 expression
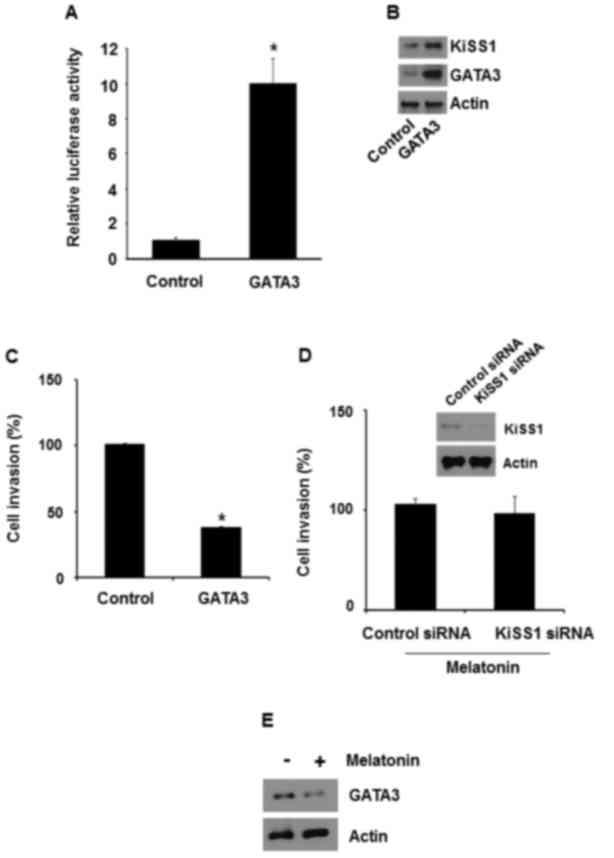
GATA3 overexpression has been revealed to inhibit
metastasis of MDA-MB-231 cells. Therefore, whether GATA3-induced
KiSS1 expression is required to inhibit the invasiveness of breast
cancer cells was investigated. In MDA-MB-231 cells, GATA3
overexpression increased the luciferase activity of pKiSS1-900-luc
(Fig. 4A). Accordingly, it was
revealed that GATA3 overexpression in MDA-MB-231 cells increased
KiSS1 and GATA3 expression (Fig. 4B).
Thus, the present study also examined whether GATA3 regulated
MDA-MB-231 cell invasion. When GATA3 was overexpressed in
MDA-MB-231 cells, the number of invading cells was reduced by ~62%
(Fig. 4C).
Whether melatonin-induced KiSS1 expression is
required for the inhibition of the invasiveness was then examined.
When KiSS1 was silenced in MDA-MB-231 cells with KiSS1 siRNA,
melatonin failed to inhibit the invasiveness (Fig. 4D). In addition, melatonin increased
GATA3 protein levels when MDA-MB-231 cells were treated with
melatonin (Fig. 4E). Therefore, these
results suggested that melatonin repressed the invasiveness via
GATA3-mediated KiSS1 expression.
Discussion
The present study revealed how melatonin prolongs
its inhibitory effect on the invasiveness of triple-negative breast
cancer cells. Melatonin induced GATA3-mediated KiSS1 expression in
triple-negative breast cancer cells. Consequently, KiSS1 maintained
a melatonin-induced inhibitory effect.
MT1 deficiency in metastatic breast cancer cells may
explain why melatonin fails to inhibit proliferation (33). However, previous studies have
demonstrated that melatonin inhibited the metastatic abilities of
MDA-MB-231 breast cancer cells by inhibiting either Rho-associated
protein kinase 1 or p38 mitogen-activated protein kinases (31,32,46).
Likewise, the results of the present study revealed that melatonin
inhibited the invasiveness of triple-negative breast cancer cells
with no effect on proliferation. In addition, melatonin was
revealed to maintain its inhibitory effect by inducing KiSS1
expression. Likewise, KiSS1 silencing prevented melatonin-induced
inhibition of invasiveness. KiSS1 is known to inhibit the
metastatic abilities of cancer cells including the invasiveness
(20). Thus, KiSS1 appears to prolong
the inhibitory effect of melatonin on the invasiveness of breast
cancer cells via GATA3 transcriptional activation.
The present study demonstrated that melatonin
promoted KiSS1 expression in triple-negative breast cancer cells.
As a disruption of light cycles increases the risk of breast cancer
(13,47,48), it is
plausible that melatonin and KiSS1 may exhibit similar expression
patterns in breast cancer. Melatonin has been revealed to regulate
circadian rhythms by inhibiting KiSS1 expression in the brain of
rodents (27). However, melatonin
induces KiSS1 expression in fish, and vice versa (49,50).
Therefore, the mechanisms that result in the different effects of
melatonin on KiSS1 expression in different experimental conditions,
including animal models, remain unknown. The present study
demonstrated that melatonin activated GATA3-mediated KiSS1
expression in triple-negative breast cancer cells. Previous in
situ hybridization assays have revealed GATA3 expression in the
hypothalamus of C57B6 mouse brains (51). Therefore, GATA3 may regulate
melatonin-induced KiSS1 expression differently in the brain.
Nevertheless, it remains to be determined how melatonin
differentially regulates KiSS1 expression via GATA3 in different
cells and/or tissues.
In conclusion, the present study suggested that
melatonin prolongs its anti-metastatic effect by activating
GATA3-mediated KiSS1 expression. While the effect of melatonin on
breast cancer has been examined, to the best of our knowledge the
present study is the first to reveal the effect of melatonin on
triple-negative breast cancer cells.
Acknowledgements
The present study was supported by Korea National
University of Transportation in 2015, and partly by Basic Science
Research Program through the National Research Foundation of Korea
funded by the Ministry of Science, ICT and Future Planning (grant
no. NRF-2014R1A1A1035831).
References
|
1
|
Brainard GC, Hanifin JP, Greeson JM, Byrne
B, Glickman G, Gerner E and Rollag MD: Action spectrum for
melatonin regulation in humans: Evidence for a novel circadian
photoreceptor. J Neurosci. 21:6405–6412. 2001.PubMed/NCBI
|
|
2
|
Lerner AB, Case JD and Takahashi Y:
Isolation of melatonin and 5-methoxyindole-3-acetic acid from
bovine pineal glands. J Biol Chem. 235:1992–1997. 1960.PubMed/NCBI
|
|
3
|
Stehle JH, von Gall C and Korf HW:
Melatonin: A clock-output, a clock-input. J Neuroendocrinol.
15:383–389. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hastings MH, Reddy AB and Maywood ES: A
clockwork web: Circadian timing in brain and periphery, in health
and disease. Nat Rev Neurosci. 4:649–661. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Slominski RM, Reiter RJ,
Schlabritz-Loutsevitch N, Ostrom RS and Slominski AT: Melatonin
membrane receptors in peripheral tissues: Distribution and
functions. Mol Cell Endocrinol. 351:152–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Claustrat B, Brun J and Chazot G: The
basic physiology and pathophysiology of melatonin. Sleep Med Rev.
9:11–24. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dubocovich ML, Delagrange P, Krause DN,
Sugden D, Cardinali DP and Olcese J: International union of basic
and clinical pharmacology. LXXV. Nomenclature, classification, and
pharmacology of G protein-coupled melatonin receptors. Pharmacol
Rev. 62:343–380. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Acuña-Castroviejo D, Escames G, Venegas C,
Díaz-Casado ME, Lima-Cabello E, López LC, Rosales-Corral S, Tan DX
and Reiter RJ: Extrapineal melatonin: Sources, regulation, and
potential functions. Cell Mol Life Sci. 71:2997–3025. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Burstein HJ, Polyak K, Wong JS, Lester SC
and Kaelin CM: Ductal carcinoma in situ of the breast. N Engl J
Med. 350:1430–1441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pagani O, Senkus E, Wood W, Colleoni M,
Cufer T, Kyriakides S, Costa A, Winer EP and Cardoso F; ESO-MBC
Task Force, : International guidelines for management of metastatic
breast cancer: Can metastatic breast cancer be cured? J Natl Cancer
Inst. 102:456–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Virnig BA, Tuttle TM, Shamliyan T and Kane
RL: Ductal carcinoma in situ of the breast: A systematic review of
incidence, treatment, and outcomes. J Natl Cancer Inst.
102:170–178. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rabstein S, Harth V, Justenhoven C, Pesch
B, Plöttner S, Heinze E, Lotz A, Baisch C, Schiffermann M, Brauch
H, et al: Polymorphisms in circadian genes, night work and breast
cancer: Results from the GENICA study. Chronobiol Int.
31:1115–1122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Van Dycke KC, Rodenburg W, van Oostrom CT,
van Kerkhof LW, Pennings JL, Roenneberg T, van Steeg H and van der
Horst GT: Chronically alternating light cycles increase breast
cancer risk in mice. Curr Biol. 25:1932–1937. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Basler M, Jetter A, Fink D, Seifert B,
Kullak-Ublick GA and Trojan A: Urinary excretion of melatonin and
association with breast cancer: Meta-analysis and review of the
literature. Breast Care (Basel). 9:182–187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schernhammer ES and Hankinson SE: Urinary
melatonin levels and breast cancer risk. J Natl Cancer Inst.
97:1084–1087. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Innominato PF, Lim AS, Palesh O, Clemons
M, Trudeau M, Eisen A, Wang C, Kiss A, Pritchard KI and Bjarnason
GA: The effect of melatonin on sleep and quality of life in
patients with advanced breast cancer. Support Care Cancer.
24:1097–1105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hill SM, Belancio VP, Dauchy RT, Xiang S,
Brimer S, Mao L, Hauch A, Lundberg PW, Summers W, Yuan L, et al:
Melatonin: An inhibitor of breast cancer. Endocr Relat Cancer.
22:R183–R204. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kumar P and Aggarwal R: An overview of
triple-negative breast cancer. Arch Gynecol Obstet. 293:247–269.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Oprea-Ilies G, Haus E, Sackett-Lundeen L,
Liu Y, McLendon L, Busch R, Adams A and Cohen C: Expression of
melatonin receptors in triple negative breast cancer (TNBC) in
African American and Caucasian women: Relation to survival. Breast
Cancer Res Treat. 137:677–687. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cho SG, Li D, Tan K, Siwko SK and Liu M:
KiSS1 and its G-protein-coupled receptor GPR54 in cancer
development and metastasis. Cancer Metastasis Rev. 31:585–591.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tena-Sempere M: Roles of kisspeptins in
the control of hypothalamic-gonadotropic function: Focus on sexual
differentiation and puberty onset. Endocr Dev. 17:52–62. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Papaoiconomou E, Lymperi M, Petraki C,
Philippou A, Msaouel P, Michalopoulou F, Kafiri G, Vassilakos G,
Zografos G and Koutsilieris M: Kiss-1/GPR54 protein expression in
breast cancer. Anticancer Res. 34:1401–1407. 2014.PubMed/NCBI
|
|
23
|
Marot D, Bieche I, Aumas C, Esselin S,
Bouquet C, Vacher S, Lazennec G, Perricaudet M, Kuttenn F, Lidereau
R and de Roux N: High tumoral levels of Kiss1 and G-protein-coupled
receptor 54 expression are correlated with poor prognosis of
estrogen receptor-positive breast tumors. Endocr Relat Cancer.
14:691–702. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kostadima L, Pentheroudakis G and Pavlidis
N: The missing kiss of life: Transcriptional activity of the
metastasis suppressor gene KiSS1 in early breast cancer. Anticancer
Res. 27:2499–2504. 2007.PubMed/NCBI
|
|
25
|
Stark AM, Tongers K, Maass N, Mehdorn HM
and Held-Feindt J: Reduced metastasis-suppressor gene
mRNA-expression in breast cancer brain metastases. J Cancer Res
Clin Oncol. 131:191–198. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Revel FG, Saboureau M, Masson-Pévet M,
Pévet P, Mikkelsen JD and Simonneaux V: Kisspeptin mediates the
photoperiodic control of reproduction in hamsters. Curr Biol.
16:1730–1735. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gingerich S, Wang X, Lee PK, Dhillon SS,
Chalmers JA, Koletar MM and Belsham DD: The generation of an array
of clonal, immortalized cell models from the rat hypothalamus:
Analysis of melatonin effects on kisspeptin and
gonadotropin-inhibitory hormone neurons. Neuroscience.
162:1134–1140. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Borin TF, Arbab AS, Gelaleti GB, Ferreira
LC, Moschetta MG, Jardim-Perassi BV, Iskander AS, Varma NR, Shankar
A, Coimbra VB, et al: Melatonin decreases breast cancer metastasis
by modulating Rho-associated kinase protein-1 expression. J Pineal
Res. 60:3–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jardim-Perassi BV, Lourenco MR, Doho GM,
Grígolo IH, Gelaleti GB, Ferreira LC, Borin TF, Moschetta MG and de
Campos Zuccari DA Pires: Melatonin regulates angiogenic factors
under hypoxia in breast cancer cell lines. Anticancer Agents Med
Chem. 16:347–358. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jardim-Perassi BV, Arbab AS, Ferreira LC,
Borin TF, Varma NR, Iskander AS, Shankar A, Ali MM and de Campos
Zuccari DA: Effect of melatonin on tumor growth and angiogenesis in
xenograft model of breast cancer. PLoS One. 9:e853112014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mao L, Yuan L, Slakey LM, Jones FE, Burow
ME and Hill SM: Inhibition of breast cancer cell invasion by
melatonin is mediated through regulation of the p38
mitogen-activated protein kinase signaling pathway. Breast Cancer
Res. 12:R1072010. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ortiz-López L, Morales-Mulia S,
Ramirez-Rodriguez G and Benitez-King G: ROCK-regulated cytoskeletal
dynamics participate in the inhibitory effect of melatonin on
cancer cell migration. J Pineal Res. 46:15–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mao L, Yuan L, Xiang S, Zeringue SB,
Dauchy RT, Blask DE, Hauch A and Hill SM: Molecular deficiency
(ies) in MT1 melatonin signaling pathway underlies the
melatonin-unresponsive phenotype in MDA-MB-231 human breast cancer
cells. J Pineal Res. 56:246–253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bresnick EH, Lee HY, Fujiwara T, Johnson
KD and Keles S: GATA switches as developmental drivers. J Biol
Chem. 285:31087–31093. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Morceau F, Schnekenburger M, Dicato M and
Diederich M: GATA-1: Friends, brothers, and coworkers. Ann NY Acad
Sci. 1030:537–554. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zheng R and Blobel GA: GATA transcription
factors and cancer. Genes Cancer. 1:1178–1188. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Naylor MJ and Ormandy CJ: Gata-3 and
mammary cell fate. Breast Cancer Res. 9:3022007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kouros-Mehr H, Kim JW, Bechis SK and Werb
Z: GATA-3 and the regulation of the mammary luminal cell fate. Curr
Opin Cell Biol. 20:164–170. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Oh DS, Troester MA, Usary J, Hu Z, He X,
Fan C, Wu J, Carey LA and Perou CM: Estrogen-regulated genes
predict survival in hormone receptor-positive breast cancers. J
Clin Oncol. 24:1656–1664. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Engelsen IB, Stefansson IM, Akslen LA and
Salvesen HB: GATA3 expression in estrogen receptor alpha-negative
endometrial carcinomas identifies aggressive tumors with high
proliferation and poor patient survival. Am J Obstet Gynecol.
199:543.e1–e7. 2008. View Article : Google Scholar
|
|
41
|
Demir H, Turna H, Can G and Ilvan S:
Clinicopathologic and prognostic evaluation of invasive breast
carcinoma molecular subtypes and GATA3 expression. J BUON.
15:774–782. 2010.PubMed/NCBI
|
|
42
|
Cimino-Mathews A, Subhawong AP, Illei PB,
Sharma R, Halushka MK, Vang R, Fetting JH, Park BH and Argani P:
GATA3 expression in breast carcinoma: Utility in triple-negative,
sarcomatoid, and metastatic carcinomas. Hum Pathol. 44:1341–1349.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nocillado JN, Mechaly AS and Elizur A: In
silico analysis of the regulatory region of the Yellowtail Kingfish
and Zebrafish Kiss and Kiss receptor genes. Fish Physiol Biochem.
39:59–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Borin TF, Arbab AS, Gelaleti GB, Ferreira
LC, Moschetta MG, Jardim-Perassi BV, Iskander AS, Varma NR, Shankar
A, Coimbra VB, et al: Melatonin decreases breast cancer metastasis
by modulating Rho-associated kinase protein-1 expression. J Pineal
Res. 60:3–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Papantoniou K, Pozo OJ, Espinosa A, Marcos
J, Castaño-Vinyals G, Basagaña X, Pagès E Juanola, Mirabent J,
Martín J, Faro P Such, et al: Increased and mistimed sex hormone
production in night shift workers. Cancer Epidemiol Biomarkers
Prev. 24:854–863. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Stevens RG, Brainard GC, Blask DE, Lockley
SW and Motta ME: Breast cancer and circadian disruption from
electric lighting in the modern world. CA Cancer J Clin.
64:207–218. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kim NN, Choi YU, Park HS and Choi CY:
Kisspeptin regulates the somatic growth-related factors of the
cinnamon clownfish Amphiprion melanopus. Comp Biochem Physiol A Mol
Integr Physiol. 179:17–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Alvarado MV, Carrillo M and Felip A:
Melatonin-induced changes in kiss/gnrh gene expression patterns in
the brain of male sea bass during spermatogenesis. Comp Biochem
Physiol A Mol Integr Physiol. 185:69–79. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao GY, Li ZY, Zou HL, Hu ZL, Song NN,
Zheng MH, Su CJ and Ding YQ: Expression of the transcription factor
GATA3 in the postnatal mouse central nervous system. Neurosci Res.
61:420–428. 2008. View Article : Google Scholar : PubMed/NCBI
|