Introduction
Hepatoblastoma (HB), derived from undifferentiated
hepatic progenitor cells (1), is a
highly malignant disease in the liver and the most common type of
hepatic malignancy in children (2).
Local and systemic metastasis are the critical causes for the poor
prognosis of this disease (3,4). Exploring the molecular mechanisms
underlying the metastatic process of HB is critical for identifying
novel biomarkers and therapeutic targets of HB.
Long non-coding RNAs (lncRNAs) are a group of
non-coding RNAs >200 nucleotides in length, with a limited
ability to code protein (5). However,
lncRNAs have been demonstrated to serve important roles in various
biological processes, including cell proliferation, differentiation
and motility (6). Emerging evidence
has demonstrated the aberrant expression of lncRNAs in various
types of human cancer (7,8). Deregulated lncRNAs are involved in the
growth, drug resistance and metastasis of human cancers (9). However, the pathophysiological functions
of lncRNAs in HB remain largely uncharacterized.
Nuclear paraspeckle assembly transcript 1 (NEAT1),
an lncRNA occurring in the nucleus that forms the core structural
component of the paraspeckle sub-organelles (10,11), was
revealed to be abnormally expressed in esophageal squamous cell
carcinoma (12), glioma (13), colorectal cancer (14) and prostate cancer (15). The abnormal expression of NEAT1 was
significantly associated with the prognosis of patients with cancer
(12). However, the expression
status, functional role and underlying mechanisms in HB remain
unknown.
In the present study, it was identified that the
expression of NEAT1 was significantly upregulated in human HB
tissues, and the HB tissues with metastasis had significantly
increased expression level of NEAT1 compared with those without
metastasis. Functionally, NEAT1 was revealed to promote the
metastatic ability and epithelial-mesenchymal transition (EMT) of
HB cells. Furthermore, the present study elucidated that NEAT1
exerts its functional effect in HB cells by inhibiting microRNA
(miR)-129-5p.
Materials and methods
Clinical samples and cell culture
A total of 32 human HB tissue samples and 15 normal
liver tissues were obtained from 32 patients with HB who underwent
surgical resection in the Department of Surgery, Children's
Hospital of Soochow University (Suzhou, China) between January 2005
and December 2010. The age range of HB patients was between 2 and 9
years old (mean age, 4.6 years), with 20 male and 12 female
patients. Clinical specimens were stored at −80°C. All samples were
obtained subsequent to obtaining written informed consent from
every patient. The protocols involving clinical samples were
approved by the Ethics Committee of Soochow University, in
accordance with the Declaration of Helsinki.
HepG2 HB cells, purchased from American Type Culture
Collection (ATCC; Manassas, VA, USA), were cultured in RPMI-1640
medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.), 100 U/ml penicillin and 100 U/ml
streptomycin. Cells were incubated at 37°C in a humidified
incubator containing 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the HB tissues and HepG2
cells using TRIzol (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. To investigate the level of NEAT1
in HB tissues and HepG2 cells, cDNA synthesis was performed using a
cDNA reverse transcription kit (Fermentas; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions, and
qPCR for 2 ng cDNA was performed with SYBR-Green I (Takara
Biotechnology Co., Ltd., Dalian, China). cDNA was denatured at 95°C
for 5 min and subjected to amplification (40 cycles of 98°C for 15
sec, 58°C for 30 sec and 72°C for 30 sec. The primers used were:
NEAT1 forward, 5′-CTTCCTCCCTTTAACTTATCCATTCAC-3′ and reverse,
5′-CTCTTCCTCCACCATTACCAACAATAC-3′; and β-actin forward,
5′-AAAGACCTGTACGCCAACAC-3′ and reverse,
5′-GTCATACTCCTGCTTGCTGAT-3′. β-actin was used as an endogenous
control. Relative gene expression was calculated using the
2−ΔΔCq method (16). Each
experiment was repeated at least 3 times.
To measure the relative miR-129-5p level in HepG2
cells, the TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.) was used according to the manufacturer's
instructions. qPCR was then performed, using TaqMan miR-129-5p and
U6 RNA assays (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Primers for miR-129-5p (cat. no.
CD201-0202) and U6 (cat. no. CD201-0145) were obtained from Tiangen
Biotech Co., Ltd. (Beijing, China). U6 was used as an endogenous
control.
Cell transfection
Small interfering RNAs (siRNAs) against NEAT1
(5′-UGGUAAUGGUGGAGGAAGAUU-3′, 5′-GUGAGAAGUUGCUUAGAAAUU-3′), a
scrambled negative control siRNA (cat. no. siN05815122147-1-5), a
miR-129-5p inhibitor (cat. no. miR20000242-1-5) and a corresponding
negative control vector (cat. no. NC-miR20000242-1-5) were
purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). A
NEAT1-expressing vector and a control vector were purchased from
Invitrogen (Thermo Fisher Scientific, Inc.). The aforementioned
vectors were transfected into HepG2 cells with Lipofectamine
2000® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions.
Transwell assay
The migration and invasion ability of HepG2 cells
was measured with a Transwell assay. A total of 2×103
HepG2 cells resuspended in serum-free medium were seeded into
Transwell inserts with 8-µm pores (EMD Millipore, Billerica, MA,
USA), whereas the lower chamber was filled with 600 µl medium
containing 10% FBS as an attractant. For the invasion assay, the
upper chamber was coated with a mixture of RPMI-1640 medium and
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) at a ratio of
7:1. Cells were then incubated in the previously described
conditions (37°C in a humidified incubator containing 5%
CO2). At 36 h after seeding, the migrated or invaded
HepG2 cells on the lower surface were stained with 0.1% crystal
violet for 5 min at room temperature, and cell numbers were counted
from 10 random fields of the lower surface of the filter using a
light microscope at ×20 magnification.
Western blot analysis
Radioimmunoprecipitation assay lysis buffer
(Beyotime Institute of Biotechnology, Shanghai, China) was used to
extract protein from HepG2 cells, and protein concentration was
measured using a bicinchoninic acid kit (Pierce; Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions. A
total of 40 µg of protein from each sample was separated on a
Novex® 10% Tris-glycine gel, then transferred to a
nitrocellulose membrane (both from Thermo Fisher Scientific, Inc.).
The membrane was blocked with 10% goat serum (BD Biosciences) at
room temperature for 30 min. The membranes were incubated with the
following primary antibodies overnight at 4°C: E-cadherin (1:1,000
dilution, cat. no. 3195; Cell Signaling Technologies, Inc.,
Danvers, MA, USA), vimentin (1:1,000 dilution, cat. no. sc-5565)
and GAPDH (1:1,500 dilution, cat. no. sc-32233; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). The membranes were then
incubated with anti-mouse (cat. no. 32430) or anti-rabbit (cat. no.
65-6120) secondary antibodies (1:10,000 dilution; Pierce; Thermo
Fisher Scientific, Inc.) at room temperature for 1 h, and
immunoreactive signals were detected using the Bio-Rad Gel imaging
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Quantifaction of immunoblots images were performed using Image Lab
software (version 6.0; Bio-Rad Laboratories, Inc.).
Prediction of NEAT1 targets
The prediction of the target miRNAs of NEAT1 was
performed using starBase (http://starbase.sysu.edu.cn/), a publicly available
database of miRNAs.
Statistical analysis
All quantitative data are expressed as the mean ±
standard deviation. GraphPad Prism 5 software (GraphPad Software,
Inc., La Jolla, CA, USA) was used to perform statistical analysis.
Pearson's χ2 test (for enumerative data) and Student's
t-test (for quantitative data) were performed in the present study.
P<0.05 was considered to indicate a statistically significant
difference.
Results
NEAT1 expression is significantly
upregulated in HB tissues
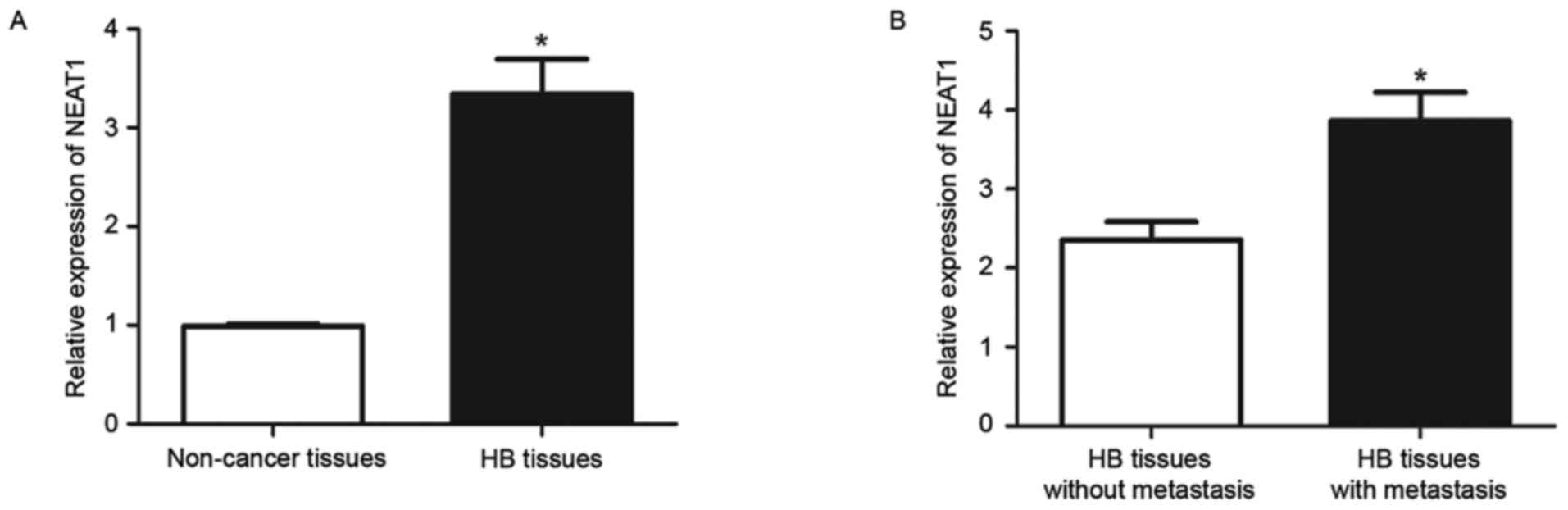
NEAT1 expression levels in 32 HB tissues and 15
non-tumor tissues were measured by RT-qPCR. The expression level of
NEAT1 in HB tissues was significantly increased compared with
non-tumor tissues (P<0.05; Fig.
1A). The expression of NEAT1 was then compared between HB
tissues from patients with and without metastasis. The results of
RT-qPCR revealed that HB tissues with metastasis had significantly
increased levels of NEAT1 compared with those without metastasis
(P<0.05; Fig. 1B). These data
indicated that NEAT1 performs an oncogenic role in HB and possibly
contributes to the metastasis of HB.
NEAT1 promotes the migration and
invasion of HepG2 cells
Subsequent to confirming the aberrant expression of
NEAT1 in HB tissues, the functional effects of NEAT1 in HepG2 cells
were examined. Since HB tissues with metastasis had significantly
increased levels of NEAT1, it was investigated whether NEAT1 could
promote the migration and invasion of HepG2 cells. NEAT1-specific
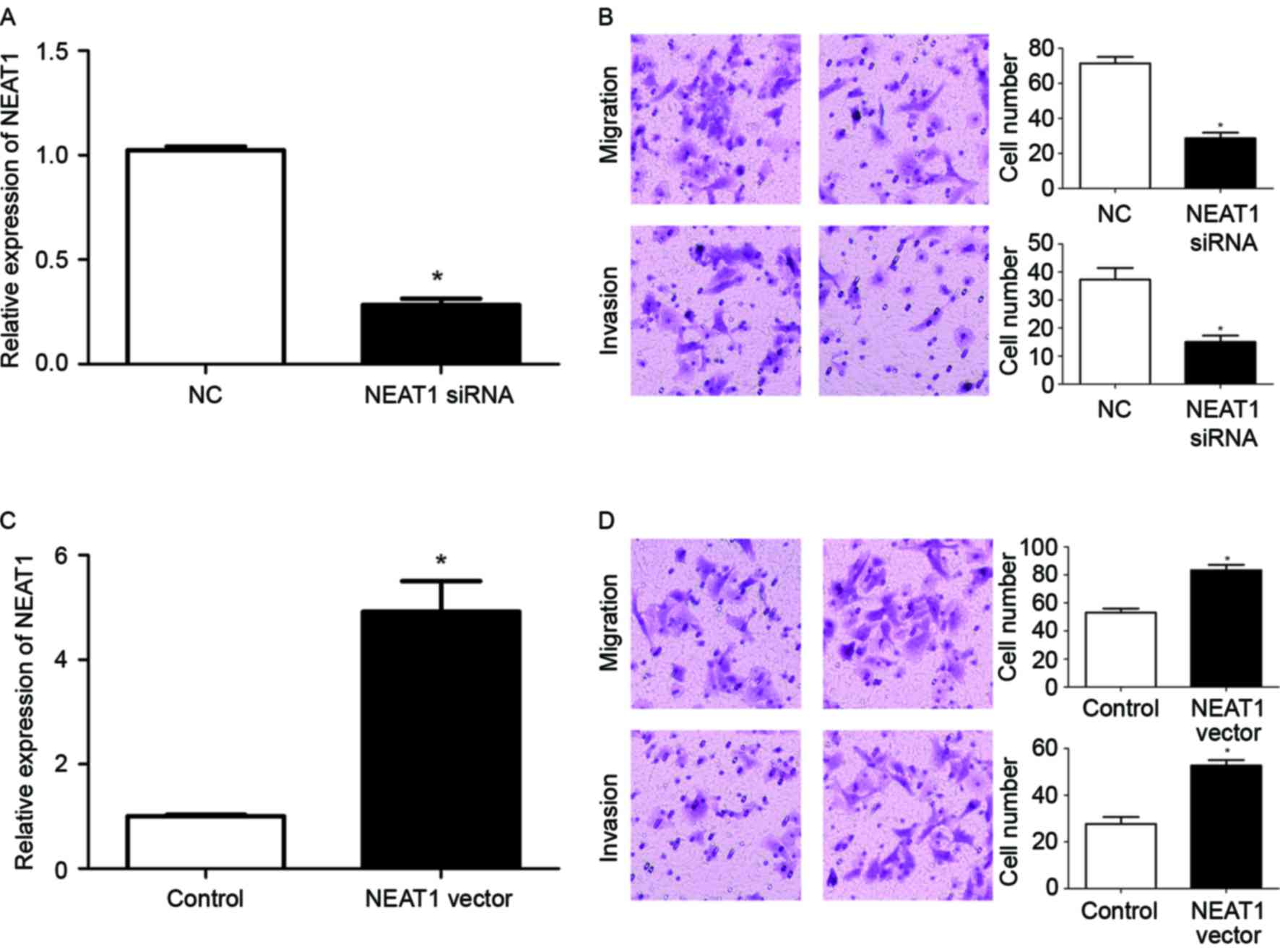
siRNA was used to inhibit the expression of NEAT1 in HepG2 cells.
Transfection of NEAT1 siRNA significantly reduced the expression
level of NEAT1 in HepG2 cells (P<0.05; Fig. 2A). Transwell assays were then used to
investigate the alteration of the migration and invasion of HepG2
cells subsequent to inhibiting the expression of NEAT1. Transwell
assays revealed that the downregulation of NEAT1 expression in
HepG2 cells significantly reduced the migration and invasion of
HepG2 cells (P<0.05; Fig. 2B). To
confirm the modulatory function of NEAT1 in the metastatic ability
of HepG2 cells, a NEAT1-expressing vector was used to overexpress
NEAT1 in HepG2 cells. Transfection of the NEAT1 vector
significantly increased the expression of NEAT1 in HepG2 cells
(P<0.05; Fig. 2C), and resulted in
a significantly increased extent of migration and invasion of HepG2
cells (P<0.05; Fig. 2D). Taken
together, these data indicated that NEAT1 may enhance the
metastatic ability of HepG2 cells.
NEAT1 contributes to the EMT of HepG2
cells
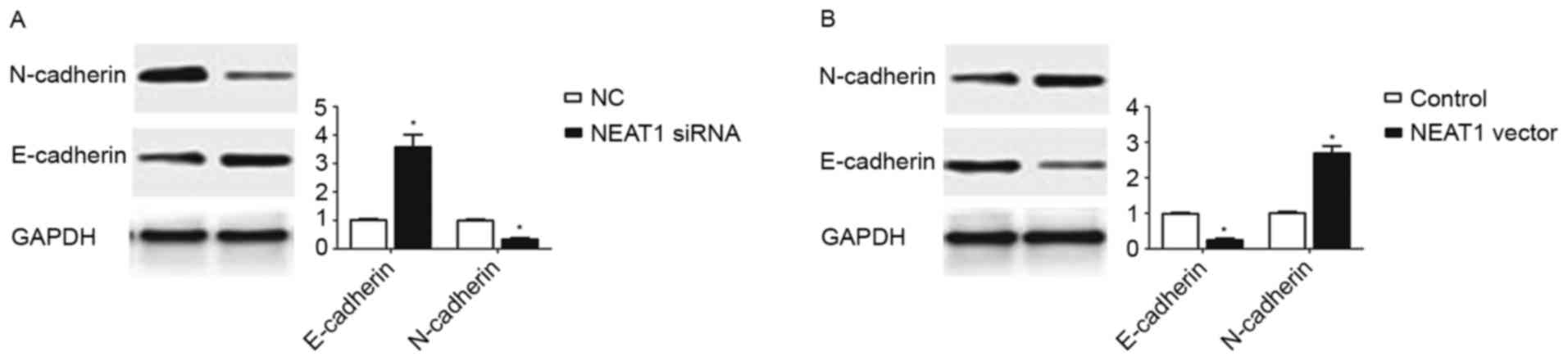
During the last two decades, EMT, which was
characterized as decreased expression of epithelial marker
(E-cadherin) and increased expression of mesenchymal marker
(N-cadherin), was recognized as an important mechanism in cancer
metastasis (17). Since the data of
the present study confirmed the enhancing effect of NEAT1 on the
metastatic ability of HepG2 cells, it was subsequently explored
whether NEAT1 expression could promote the EMT of HepG2 cells. The
results of western blotting revealed that inhibition of NEAT1 in
HepG2 cells resulted in significantly increased E-cadherin
expression (P<0.05; Fig. 3A) and
decreased N-cadherin expression (P<0.05; Fig. 3A). By contrast, overexpression of
NEAT1 in HepG2 cells led to the decreased expression of E-cadherin
(P<0.05; Fig. 3B) and the
increased expression of N-cadherin (P<0.05; Fig. 3B). These data indicated that the
expression of NEAT1 may promote EMT in HepG2 cells.
Expression level of miR-129-5p in
HepG2 cells is modulated by NEAT1
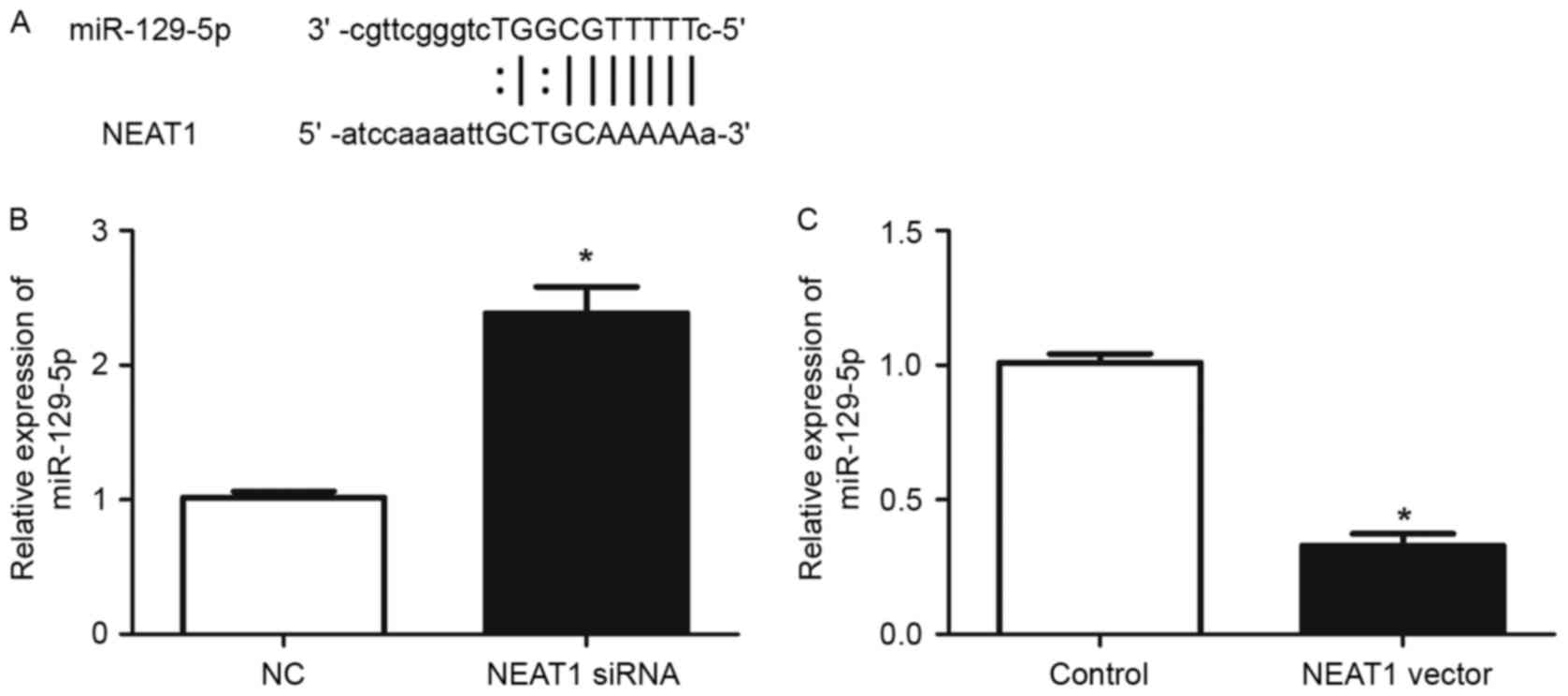
It has been demonstrated that lncRNAs contain
complementary sequences to miRNAs and may inhibit the expression
and activity of miRNAs (18). To
explore whether NEAT1 has similar mechanisms in HepG2 cells, the
prediction of microRNA target sites was performed with the starBase
database. Among numerous potential microRNA targets, the present
study focused on miR-129-5p, as it had previously been revealed to
serve an important role in modulating the metastasis and EMT of
human cancer (19–21). As demonstrated in Fig. 4A, NEAT1 contained a complementary
sequence against miR-129-5p. RT-qPCR was then used to confirm
whether NEAT1 affects the expression of miR-129-5p in HepG2 cells.
The results of RT-qPCR demonstrated that knockdown of NEAT1
significantly increased the expression of miR-129-5p (P<0.05;
Fig. 4B), while overexpression of
NEAT1 significantly reduced miR-129-5p level in HepG2 cells
(P<0.05; Fig. 4C). These results
demonstrated that NEAT1 may inhibit the expression of miR-129-5p in
HepG2 cells.
NEAT1 exerts its functional effects on
the metastasis and EMT of HepG2 cells through modulating
miR-129-5p
To confirm that NEAT1 exerts its functional effect
in HepG2 cells by modulating miR-129-5p, HepG2 cells transfected
with NEAT1 siRNA were then transfected with an miR-129-5p
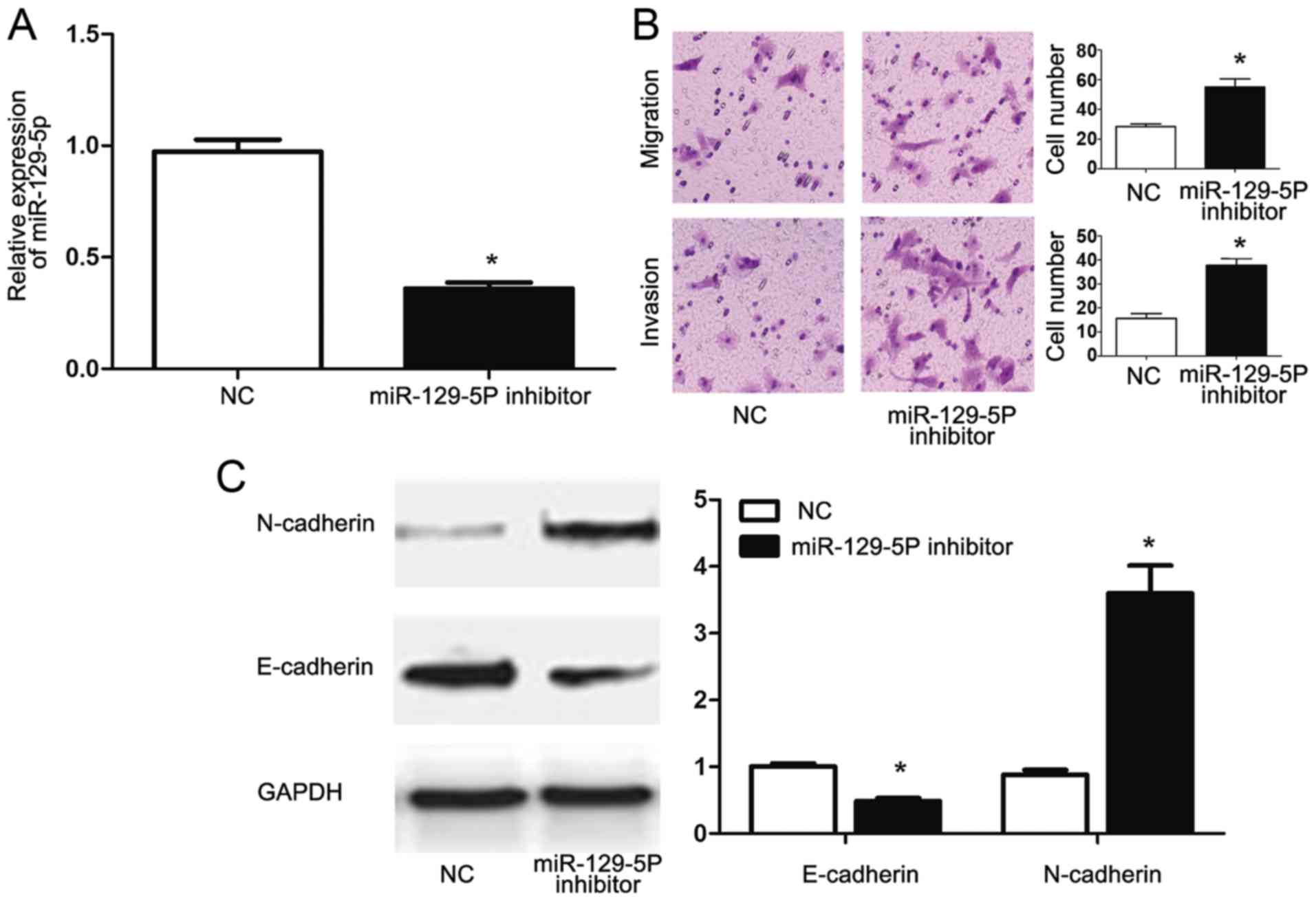
inhibitor. Transfection with the miR-129-5p inhibitor significantly
reduced the miR-129-5p expression of HepG2 cells (P<0.05;
Fig. 5A). Functionally, the
inhibition of miR-129-5p in HepG2 cells transfected with NEAT1
siRNA reversed the inhibitory effects of NEAT1 siRNA on the
migration and invasion of HepG2 cells. Furthermore, inhibition of
miR-129-5p decreased E-cadherin (P<0.05; Fig. 5B) and increased N-cadherin (P<0.05;
Fig. 5B) expression in HepG2 cells
transfected with NEAT1 siRNA, indicating that miR-129-5p inhibition
reversed the inhibitory effects of NEAT1-knockdown on EMT of HepG2
cells. These data indicated that NEAT1 may not only modulate the
expression of miR-129-5p, but also exert its functional effects on
metastasis and EMT of HepG2 cells through modulating miR-129-5p
expression.
Discussion
The metastasis of HB is the major reason for the
poor prognosis of patients with HB (1–4). There are
currently few effective treatments available for patients with
metastasis. lncRNAs have been identified to serve an important role
in cancer development and progression (12,15).
Exploring the expression status and biological functions of lncRNAs
in HB may contribute to the identification of novel biomarkers and
therapeutic targets for patients with HB.
Among a number of cancer-associated lncRNAs, NEAT1
has been identified to be an oncogenic lncRNA; it may be
overexpressed in prostate cancer (22), colorectal cancer (20), glioma (13) and esophageal squamous cell carcinoma
(12), and its overexpression was
associated with poor prognosis of the patients with cancer
(12). In the present study, it was
revealed that NEAT1 was overexpressed in HB tissues. Notably, tumor
tissues from patients with metastasis were found to have
significantly increased expression levels of NEAT1 compared with
patients without metastasis, indicating its oncogenic function and
potential ability to promote metastasis in HB.
Previous studies have demonstrated that NEAT1 has
various biological functions in various types of human cancer. For
example, a previous study on prostate cancer revealed that NEAT1
may promote the growth of prostate cancer by affecting the
epigenetic status of the target gene promoter (15). An additional study on lung cancer
demonstrated that NEAT1 was associated with the apoptosis and cell
cycle progression modulated by miR-449a (23). NEAT1 has also been reported to serve a
role in the regulation of the pro-tumorigenic hypoxia phenotype in
breast cancer (24). Therefore, these
previous studies demonstrated that the biological role of NEAT1 in
human cancers varies between cancer types. In the present study,
the results of Transwell assays demonstrated that the knockdown of
NEAT1 inhibits the migration and invasion of HB cells, while
overexpression of NEAT1 promotes the migration and invasion of HB
cells. In addition, the results of western blotting revealed that
NEAT1 may promote the EMT of HepG2 cells. Therefore, the results of
the present study demonstrated that NEAT1 promoted the metastatic
ability and EMT of HB cells.
lncRNAs may act as a ‘sponge’ for miRNAs and
regulate the expression and function of miRNAs (22,25,26), thus
affecting the development and progression of various types of human
cancer. Therefore, it was investigated whether NEAT1 may exert its
functional effects on HB through similar mechanisms. Using
starBase, NEAT1 was revealed to contain a complementary sequence of
miR-129-5p, indicating that NEAT1 may regulate the expression of
miR-129-5p. RT-qPCR was then used to confirm whether NEAT1
regulates the expression of miR-129-5p in HepG2 cells. The results
of RT-qPCR revealed that the knockdown of NEAT1 increased the level
of miR-129-5p, while overexpression of NEAT1 decreased the level of
miR-129-5p in HepG2 cells. These results demonstrated that NEAT1
may inhibit the expression of miR-129-5p in HepG2 cells.
Since previous studies have confirmed that
miR-129-5p regulates the metastasis and EMT of cancer cells
(19–21), the present study investigated whether
NEAT1 exerted its functional effects on HepG2 cells through
modulating the expression of miR-129-5p. An miR-129-5p inhibitor
was used to inhibit the expression level of miR-129-5p in HepG2
cells transfected with NEAT1 siRNA. The results of Transwell assays
revealed that inhibiting the expression of miR-129-5p partially
reversed the inhibitory effects of NEAT1 knockdown on cell
migration and invasion. Furthermore, the western blotting results
demonstrated that inhibiting miR-129-5p may abrogate the inhibitory
effects of NEAT1 knockdown on EMT. These results indicated that
NEAT1 may not only inhibit the expression of NEAT1 in HepG2 cells,
but also exert its functional effects on HepG2 cells by modulating
the expression of miR-129-5p.
In conclusion, to the best of our knowledge, the
present study demonstrated for the first time that the expression
of NEAT1 is significantly elevated in HB tissues compared with
non-cancerous liver tissue. HB tissue from patients with metastasis
exhibited a significantly increased level of NEAT1 expression
relative to HB tissue from patients that did not exhibit
metastasis. NEAT1 may promote the EMT and metastatic behaviors of
HB cells. Furthermore, NEAT1 may inhibit miR-129-5p, and the
functional effects of NEAT1 in HB may be mediated by modulating
miR-129-5p expression.
Acknowledgements
The present study was supported by the Natural
Science Foundation of China (grant no. 81502496), the Science
Foundation of Jiangsu Province Health Department (grant nos.
Q201503 and Q201304), Planning Project of Science and Technology
Development of Suzhou (grant no. SYS201763) and the Jiangsu
Government Scholarship for Overseas Studies.
References
|
1
|
Weinberg AG and Finegold MJ: Primary
hepatic tumors of childhood. Hum Pathol. 14:512–537. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ishak KG and Glunz PR: Hepatoblastoma and
hepatocarcinoma in infancy and childhood. Report of 47 cases.
Cancer. 20:396–422. 1967. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Czauderna P, Otte JB, Roebuck DJ, von
Schweinitz D and Plaschkes J: Surgical treatment of hepatoblastoma
in children. Pediatr Radiol. 36:187–191. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haas JE, Muczynski KA, Krailo M, Ablin A,
Land V, Vietti TJ and Hammond GD: Histopathology and prognosis in
childhood hepatoblastoma and hepatocarcinoma. Cancer. 64:1082–1095.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takahashi K, Yan I, Haga H and Patel T:
Long noncoding RNA in liver diseases. Hepatology. 60:744–753. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang H, Chen Z, Wang X, Huang Z, He Z and
Chen Y: Long non-coding RNA: A new player in cancer. J Hematol
Oncol. 6:372013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol Cell. 33:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Souquere S, Beauclair G, Harper F, Fox A
and Pierron G: Highly ordered spatial organization of the
structural long noncoding NEAT1 RNAs within paraspeckle nuclear
bodies. Mol Biol Cell. 21:4020–4027. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Kong J, Ma Z, Gao S and Feng X: Up
regulation of the long non-coding RNA NEAT1 promotes esophageal
squamous cell carcinoma cell progression and correlates with poor
prognosis. Am J Cancer Res. 5:2808–2815. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhen L, Yun-Hui L, Hong-Yu D, Jun M and
Yi-Long Y: Long noncoding RNA NEAT1 promotes glioma pathogenesis by
regulating miR-449b-5p/c-Met axis. Tumour Biol. 37:673–683. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Li Y, Chen W, He F, Tan Z, Zheng J,
Wang W, Zhao Q and Li J: NEAT expression is associated with tumor
recurrence and unfavorable prognosis in colorectal cancer.
Oncotarget. 6:27641–27650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chakravarty D, Sboner A, Nair SS,
Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K,
Kossai M, et al: The oestrogen receptor alpha-regulated lncRNA
NEAT1 is a critical modulator of prostate cancer. Nat Commun.
5:53832014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jalali S, Bhartiya D, Lalwani MK,
Sivasubbu S and Scaria V: Systematic transcriptome wide analysis of
lncRNA-miRNA interactions. PLoS One. 8:e538232013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li M, Tian L, Wang L, Yao H, Zhang J, Lu
J, Sun Y, Gao X, Xiao H and Liu M: Down-regulation of miR-129-5p
inhibits growth and induces apoptosis in laryngeal squamous cell
carcinoma by targeting APC. PLoS One. 8:e778292013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Hei Y, Shu Q, Dong J, Gao Y, Fu H,
Zheng X and Yang G: VCP/p97, down-regulated by microRNA-129-5p,
could regulate the progression of hepatocellular carcinoma. PLoS
One. 7:e358002012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duan L, Hao X, Liu Z, Zhang Y and Zhang G:
miR-129-5p is down-regulated and involved in the growth, apoptosis
and migration of medullary thyroid carcinoma cells through
targeting RET. FEBS Lett. 588:1644–1651. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
You J, Zhang Y, Liu B, Li Y, Fang N, Zu L,
Li X and Zhou Q: MicroRNA-449a inhibits cell growth in lung cancer
and regulates long noncoding RNA nuclear enriched abundant
transcript. Indian J Cancer. 51 Suppl 3:e77–e81. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Choudhry H, Albukhari A, Morotti M, Haider
S, Moralli D, Smythies J, Schödel J, Green CM, Camps C, Buffa F, et
al: Tumor hypoxia induces nuclear paraspeckle formation through
HIF-2α dependent transcriptional activation of NEAT1 leading to
cancer cell survival. Oncogene. 34:4482–4490. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer
A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J
and Califano A: An extensive microRNA-mediated network of RNA-RNA
interactions regulates established oncogenic pathways in
glioblastoma. Cell. 147:370–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Deng L, Yang SB, Xu FF and Zhang JH: Long
noncoding RNA CCAT1 promotes hepatocellular carcinoma progression
by functioning as let-7 sponge. J Exp Clin Cancer Res. 34:182015.
View Article : Google Scholar : PubMed/NCBI
|