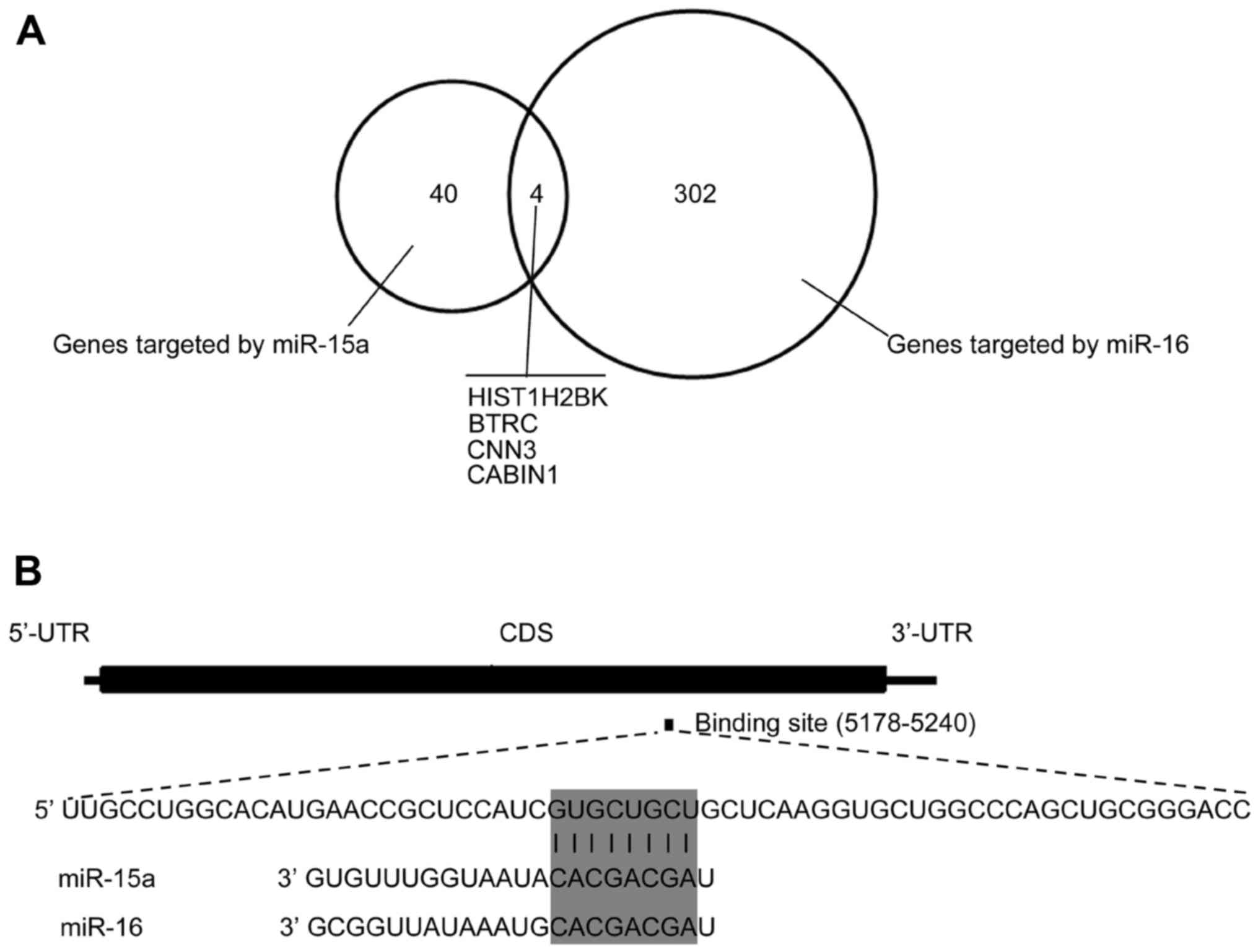
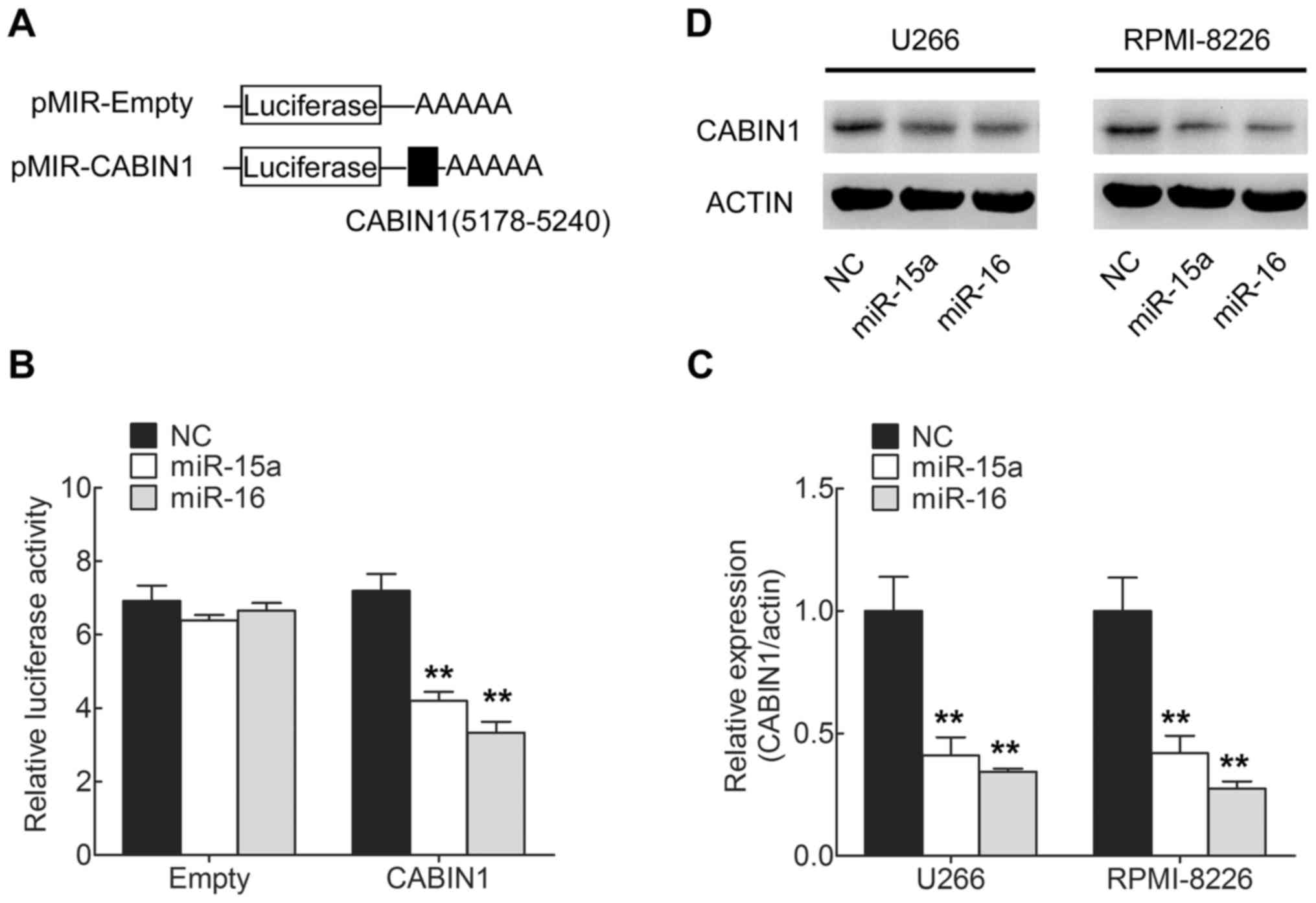
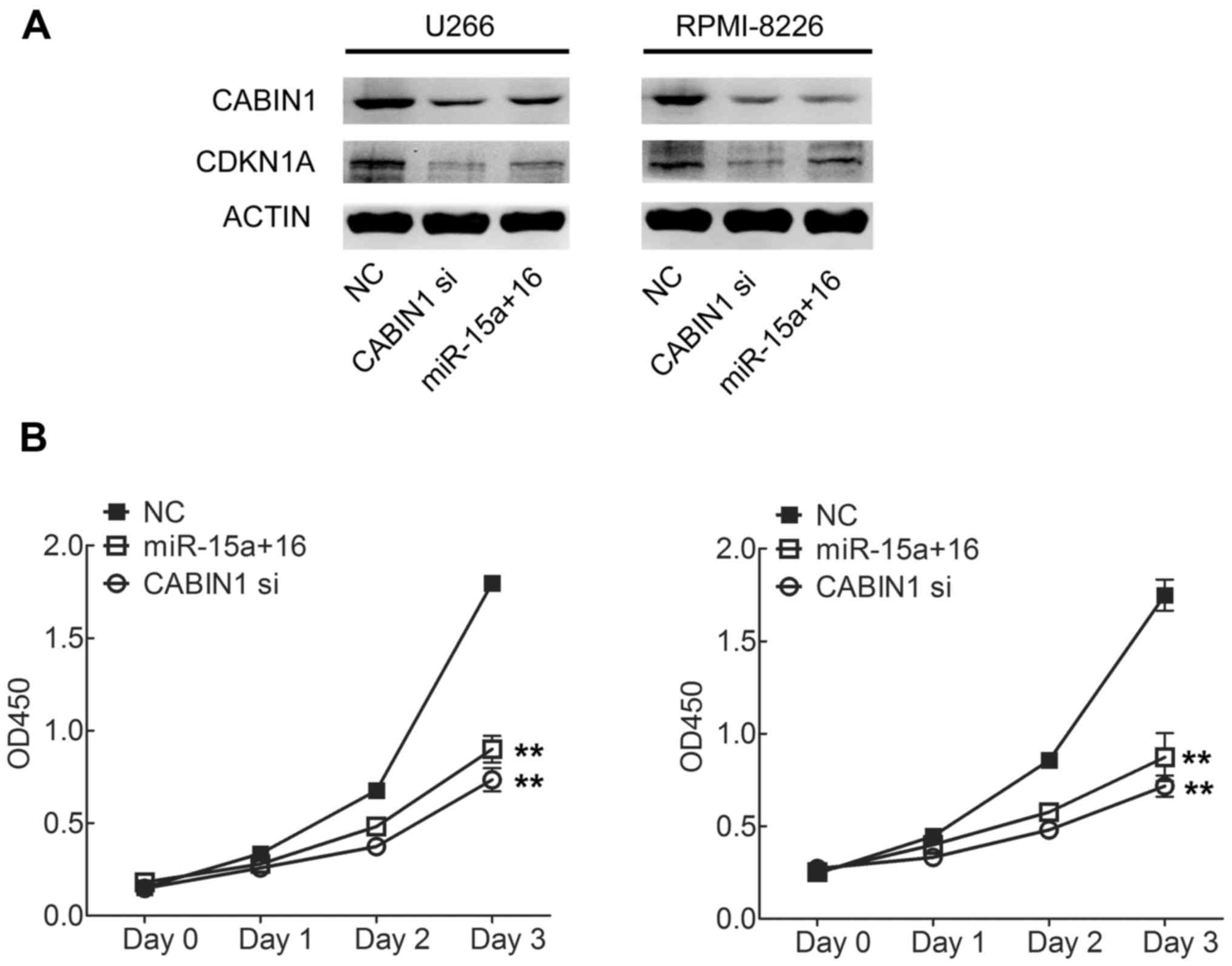
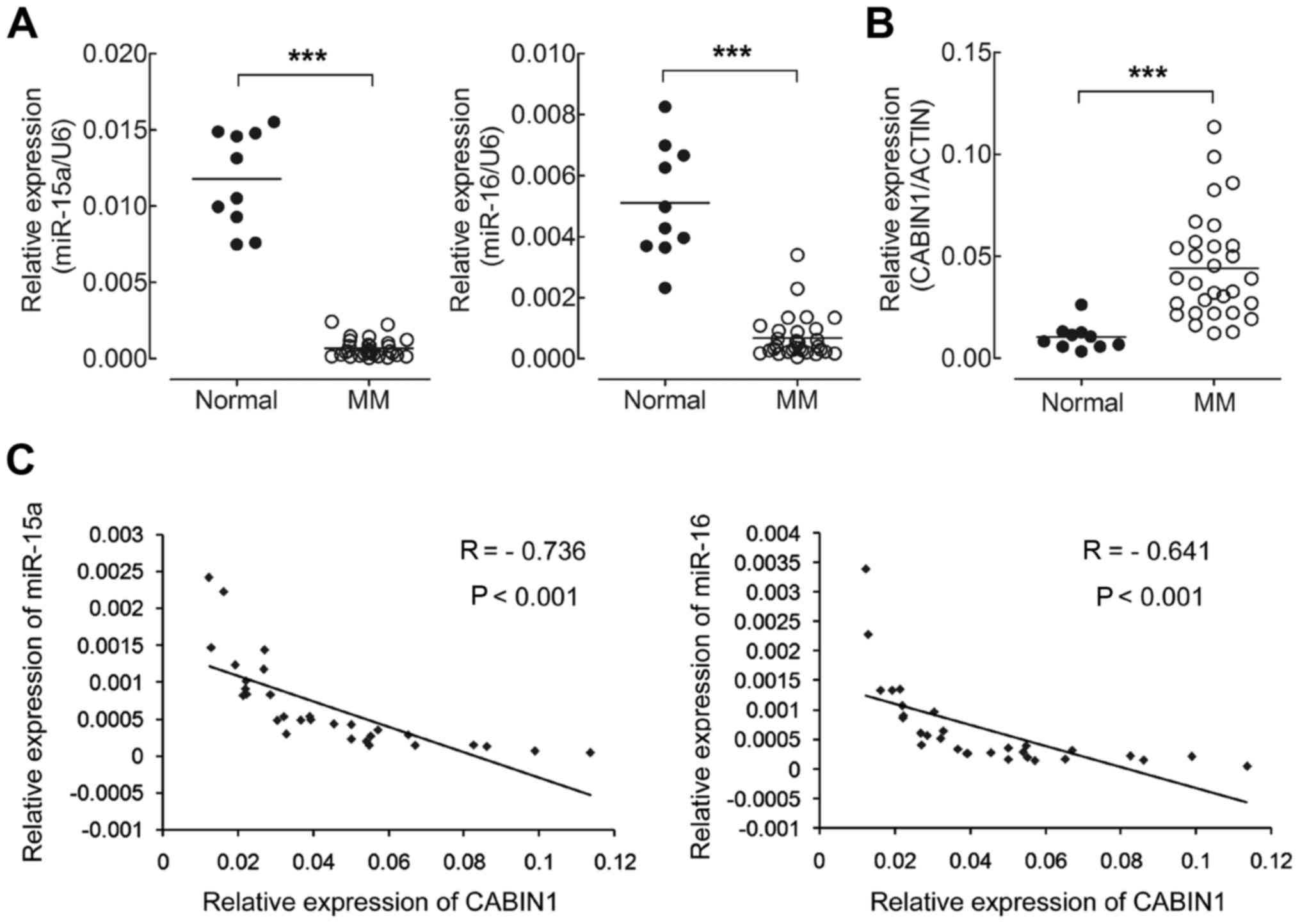
| miR-15a |
ENSG00000099991_ENST00000398319_CABIN1 |
| miR-16 |
ENSG00000099991_ENST00000398319_CABIN1 |
| miR-15a |
ENSG00000110429_ENST00000265651_FBXO3 |
| miR-16 |
ENSG00000110429_ENST00000265651_FBXO3 |
| miR-15a |
ENSG00000117519_ENST00000370206_CNN3 |
| miR-16 |
ENSG00000117519_ENST00000370206_CNN3 |
| miR-15a |
ENSG00000197903_ENST00000396891_HIST1H2BK |
| miR-16 |
ENSG00000197903_ENST00000396891_HIST1H2BK |
| miR-15a |
ENSG00000013275_ENST00000157812_PSMC4 |
| miR-15a |
ENSG00000015568_ENST00000016946_RGPD5 |
| miR-15a |
ENSG00000023909_ENST00000370238_GCLM |
| miR-15a |
ENSG00000025796_ENST00000369002_SEC63 |
| miR-15a |
ENSG00000057608_ENST00000380191_GDI2 |
| miR-15a |
ENSG00000069956_ENST00000261845_MAPK6 |
| miR-15a |
ENSG00000071553_ENST00000369762_ATP6AP1 |
| miR-15a |
ENSG00000073009_ENST00000369606_IKBKG |
| miR-15a |
ENSG00000110092_ENST00000227507_CCND1 |
| miR-15a |
ENSG00000110435_ENST00000227868_PDHX |
| miR-15a |
ENSG00000111641_ENST00000399466_NOP2 |
| miR-15a |
ENSG00000111642_ENST00000357008_CHD4 |
| miR-15a |
ENSG00000113312_ENST00000231238_TTC1 |
| miR-15a |
ENSG00000115758_ENST00000234111_ODC1 |
| miR-15a |
ENSG00000115966_ENST00000264110_ATF2 |
| miR-15a |
ENSG00000117215_ENST00000375105_PLA2G2D |
| miR-15a |
ENSG00000118971_ENST00000261254_CCND2 |
| miR-15a |
ENSG00000119777_ENST00000238788_TMEM214 |
| miR-15a |
ENSG00000121940_ENST00000415331_CLCC1 |
| miR-15a |
ENSG00000122786_ENST00000361901_CALD1 |
| miR-15a |
ENSG00000129515_ENST00000396526_SNX6 |
| miR-15a |
ENSG00000130294_ENST00000320389_KIF1A |
| miR-15a |
ENSG00000130726_ENST00000253024_TRIM28 |
| miR-15a |
ENSG00000131023_ENST00000253339_BX276089-1 |
| miR-15a |
ENSG00000132141_ENST00000314144_CCT6B |
| miR-15a |
ENSG00000133059_ENST00000367162_DSTYK |
| miR-15a |
ENSG00000135503_ENST00000257963_ACVR1B |
| miR-15a |
ENSG00000138073_ENST00000260643_PREB |
| miR-15a |
ENSG00000138381_ENST00000260952_ASNSD1 |
| miR-15a |
ENSG00000138386_ENST00000337386_NAB1 |
| miR-15a |
ENSG00000148688_ENST00000413330_RPP30 |
| miR-15a |
ENSG00000150907_ENST00000379561_FOXO1 |
| miR-15a |
ENSG00000163249_ENST00000295414_CCNYL1 |
| miR-15a |
ENSG00000165494_ENST00000298281_PCF11 |
| miR-15a |
ENSG00000165672_ENST00000298510_PRDX3 |
| miR-15a |
ENSG00000166167_ENST00000370187_BTRC |
| miR-15a |
ENSG00000169710_ENST00000306749_FASN |
| miR-15a |
ENSG00000185658_ENST00000341322_BRWD1 |
| miR-15a |
ENSG00000185697_ENST00000402282_MYBL1 |
| miR-15a |
ENSG00000188647_ENST00000415701_PTAR1 |
| miR-16 |
ENSG00000005302_ENST00000337339_MSL3 |
| miR-16 |
ENSG00000005700_ENST00000306270_IBTK |
| miR-16 |
ENSG00000008438_ENST00000008938_PGLYRP1 |
| miR-16 |
ENSG00000008516_ENST00000336577_MMP25 |
| miR-16 |
ENSG00000008838_ENST00000394126_MED24 |
| miR-16 |
ENSG00000009954_ENST00000339594_BAZ1B |
| miR-16 |
ENSG00000010322_ENST00000345716_NISCH |
| miR-16 |
ENSG00000010404_ENST00000340855_IDS |
| miR-16 |
ENSG00000011028_ENST00000303375_MRC2 |
| miR-16 |
ENSG00000035403_ENST00000372755_VCL |
| miR-16 |
ENSG00000037474_ENST00000264670_NSUN2 |
| miR-16 |
ENSG00000050405_ENST00000341247_LIMA1 |
| miR-16 |
ENSG00000056277_ENST00000370978_ZNF280C |
| miR-16 |
ENSG00000063046_ENST00000262056_EIF4B |
| miR-16 |
ENSG00000064419_ENST00000265388_TNPO3 |
| miR-16 |
ENSG00000065150_ENST00000261574_IPO5 |
| miR-16 |
ENSG00000065526_ENST00000375759_SPEN |
| miR-16 |
ENSG00000065665_ENST00000379033_SEC61A2 |
| miR-16 |
ENSG00000066926_ENST00000262093_FECH |
| miR-16 |
ENSG00000067836_ENST00000322048_ROGDI |
| miR-16 |
ENSG00000069998_ENST00000336737_CECR5 |
| miR-16 |
ENSG00000070501_ENST00000265421_POLB |
| miR-16 |
ENSG00000070882_ENST00000313367_OSBPL3 |
| miR-16 |
ENSG00000072110_ENST00000193403_ACTN1 |
| miR-16 |
ENSG00000072135_ENST00000175756_PTPN18 |
| miR-16 |
ENSG00000072274_ENST00000360110_TFRC |
| miR-16 |
ENSG00000072803_ENST00000296933_FBXW11 |
| miR-16 |
ENSG00000074755_ENST00000381638_ZZEF1 |
| miR-16 |
ENSG00000075292_ENST00000409544_ZNF638 |
| miR-16 |
ENSG00000075624_ENST00000331789_ACTB |
| miR-16 |
ENSG00000076604_ENST00000262395_TRAF4 |
| miR-16 |
ENSG00000076826_ENST00000446248_
KIAA1543 |
| miR-16 |
ENSG00000083544_ENST00000411610_TDRD3 |
| miR-16 |
ENSG00000085872_ENST00000198939_CHERP |
| miR-16 |
ENSG00000086570_ENST00000261800_FAT2 |
| miR-16 |
ENSG00000086712_ENST00000380122_CXorf15 |
| miR-16 |
ENSG00000088387_ENST00000357329_DOCK9 |
| miR-16 |
ENSG00000088899_ENST00000329152_RP5-1187M17-10 |
| miR-16 |
ENSG00000089157_ENST00000228306_RPLP0 |
| miR-16 |
ENSG00000089693_ENST00000203630_MLF2 |
| miR-16 |
ENSG00000090376_ENST00000457197_IRAK3 |
| miR-16 |
ENSG00000090621_ENST00000372857_PABPC4 |
| miR-16 |
ENSG00000091164_ENST00000217515_TXNL1 |
| miR-16 |
ENSG00000093000_ENST00000347635_NUP50 |
| miR-16 |
ENSG00000093010_ENST00000361682_COMT |
| miR-16 |
ENSG00000095564_ENST00000265990_BTAF1 |
| miR-16 |
ENSG00000095739_ENST00000375533_BAMBI |
| miR-16 |
ENSG00000096968_ENST00000381652_JAK2 |
| miR-16 |
ENSG00000099203_ENST00000214869_TMED1 |
| miR-16 |
ENSG00000099991_ENST00000398319_CABIN1 |
| miR-16 |
ENSG00000100234_ENST00000266085_TIMP3 |
| miR-16 |
ENSG00000100241_ENST00000380817_SBF1 |
| miR-16 |
ENSG00000100304_ENST00000216129_TTLL12 |
| miR-16 |
ENSG00000100304_ENST00000216129_TTLL12 |
| miR-16 |
ENSG00000100697_ENST00000343455_DICER1 |
| miR-16 |
ENSG00000100764_ENST00000261303_PSMC1 |
| miR-16 |
ENSG00000101166_ENST00000355937_SLMO2 |
| miR-16 |
ENSG00000101190_ENST00000335351_TCFL5 |
| miR-16 |
ENSG00000101294_ENST00000340852_HM13 |
| miR-16 |
ENSG00000101452_ENST00000252011_DHX35 |
| miR-16 |
ENSG00000101782_ENST00000339486_RIOK3 |
| miR-16 |
ENSG00000102174_ENST00000379374_PHEX |
| miR-16 |
ENSG00000102931_ENST00000219204_ARL2BP |
| miR-16 |
ENSG00000103148_ENST00000399953_C16orf35 |
| miR-16 |
ENSG00000103222_ENST00000399410_ABCC1 |
| miR-16 |
ENSG00000103319_ENST00000263026_EEF2K |
| miR-16 |
ENSG00000103489_ENST00000261381_XYLT1 |
| miR-16 |
ENSG00000104331_ENST00000262644_IMPAD1 |
| miR-16 |
ENSG00000104341_ENST00000445593_LAPTM4B |
| miR-16 |
ENSG00000104824_ENST00000221419_HNRNPL |
| miR-16 |
ENSG00000104824_ENST00000221419_HNRNPL |
| miR-16 |
ENSG00000105173_ENST00000262643_CCNE1 |
| miR-16 |
ENSG00000105223_ENST00000409587_PLD3 |
| miR-16 |
ENSG00000105281_ENST00000306894_SLC1A5 |
| miR-16 |
ENSG00000105429_ENST00000334370_MEGF8 |
| miR-16 |
ENSG00000105784_ENST00000338056_RUNDC3B |
| miR-16 |
ENSG00000106009_ENST00000340611_C7orf27 |
| miR-16 |
ENSG00000106355_ENST00000450169_LSM5 |
| miR-16 |
ENSG00000106665_ENST00000361545_CLIP2 |
| miR-16 |
ENSG00000107937_ENST00000360803_GTPBP4 |
| miR-16 |
ENSG00000108510_ENST00000397786_MED13 |
| miR-16 |
ENSG00000109775_ENST00000264689_UFSP2 |
| miR-16 |
ENSG00000109971_ENST00000227378_HSPA8 |
| miR-16 |
ENSG00000109971_ENST00000227378_HSPA8 |
| miR-16 |
ENSG00000110367_ENST00000264018_DDX6 |
| miR-16 |
ENSG00000110713_ENST00000324932_NUP98 |
| miR-16 |
ENSG00000111012_ENST00000228606_CYP27B1 |
| miR-16 |
ENSG00000112242_ENST00000346618_E2F3 |
| miR-16 |
ENSG00000112306_ENST00000230050_RPS12 |
| miR-16 |
ENSG00000112592_ENST00000392092_TBP |
| miR-16 |
ENSG00000112664_ENST00000358797_NUDT3 |
| miR-16 |
ENSG00000112701_ENST00000447266_SENP6 |
| miR-16 |
ENSG00000113569_ENST00000231498_NUP155 |
| miR-16 |
ENSG00000113732_ENST00000519374_ATP6V0E1 |
| miR-16 |
ENSG00000114030_ENST00000344337_KPNA1 |
| miR-16 |
ENSG00000114933_ENST00000403263_INO80D |
| miR-16 |
ENSG00000114982_ENST00000354204_KIAA1310 |
| miR-16 |
ENSG00000115307_ENST00000377526_AUP1 |
| miR-16 |
ENSG00000115616_ENST00000233969_SLC9A2 |
| miR-16 |
ENSG00000116396_ENST00000369787_KCNC4 |
| miR-16 |
ENSG00000116685_ENST00000376572_KIAA2013 |
| miR-16 |
ENSG00000116748_ENST00000369538_AMPD1 |
| miR-16 |
ENSG00000116786_ENST00000420314_PLEKHM2 |
| miR-16 |
ENSG00000117984_ENST00000236671_CTSD |
| miR-16 |
ENSG00000120158_ENST00000381750_RCL1 |
| miR-16 |
ENSG00000120438_ENST00000321394_TCP1 |
| miR-16 |
ENSG00000120833_ENST00000340600_SOCS2 |
| miR-16 |
ENSG00000122482_ENST00000370440_ZNF644 |
| miR-16 |
ENSG00000122566_ENST00000354667_HNRNPA2B1 |
| miR-16 |
ENSG00000123384_ENST00000243077_AC137834-1 |
| miR-16 |
ENSG00000123411_ENST00000262032_IKZF4 |
| miR-16 |
ENSG00000124529_ENST00000377364_HIST1H4B |
| miR-16 |
ENSG00000125166_ENST00000245206_GOT2 |
| miR-16 |
ENSG00000125166_ENST00000245206_GOT2 |
| miR-16 |
ENSG00000126016_ENST00000304758_AMOT |
| miR-16 |
ENSG00000126464_ENST00000418929_PRR12 |
| miR-16 |
ENSG00000126602_ENST00000246957_TRAP1 |
| miR-16 |
ENSG00000127152_ENST00000345514_BCL11B |
| miR-16 |
ENSG00000127616_ENST00000429416_SMARCA4 |
| miR-16 |
ENSG00000128714_ENST00000392539_HOXD13 |
| miR-16 |
ENSG00000129566_ENST00000262715_TEP1 |
| miR-16 |
ENSG00000130054_ENST00000252338_FAM155B |
| miR-16 |
ENSG00000130227_ENST00000252512_XPO7 |
| miR-16 |
ENSG00000130402_ENST00000252699_ACTN4 |
| miR-16 |
ENSG00000130640_ENST00000368563_TUBGCP2 |
| miR-16 |
ENSG00000130733_ENST00000393508_YIPF2 |
| miR-16 |
ENSG00000131269_ENST00000253577_ABCB7 |
| miR-16 |
ENSG00000131467_ENST00000441946_PSME3 |
| miR-16 |
ENSG00000132153_ENST00000443212_DHX30 |
| miR-16 |
ENSG00000132341_ENST00000448750_RAN |
| miR-16 |
ENSG00000132423_ENST00000254759_COQ3 |
| miR-16 |
ENSG00000132676_ENST00000368336_DAP3 |
| miR-16 |
ENSG00000132718_ENST00000368324_SYT11 |
| miR-16 |
ENSG00000132953_ENST00000255305_XPO4 |
| miR-16 |
ENSG00000133124_ENST00000372129_IRS4 |
| miR-16 |
ENSG00000133243_ENST00000255608_BTBD2 |
| miR-16 |
ENSG00000133433_ENST00000290765_GSTT2B |
| miR-16 |
ENSG00000133773_ENST00000256151_CCDC59 |
| miR-16 |
ENSG00000134049_ENST00000256433_IER3IP1 |
| miR-16 |
ENSG00000134817_ENST00000257254_APLNR |
| miR-16 |
ENSG00000134871_ENST00000360467_COL4A2 |
| miR-16 |
ENSG00000134970_ENST00000456936_TMED7 |
| miR-16 |
ENSG00000135269_ENST00000358204_TES |
| miR-16 |
ENSG00000135341_ENST00000369329_MAP3K7 |
| miR-16 |
ENSG00000135898_ENST00000392040_GPR55 |
| miR-16 |
ENSG00000136695_ENST00000393200_IL1F5 |
| miR-16 |
ENSG00000136827_ENST00000437532_TOR1A |
| miR-16 |
ENSG00000137713_ENST00000427203_PPP2R1B |
| miR-16 |
ENSG00000138041_ENST00000272313_SMEK2 |
| miR-16 |
ENSG00000138326_ENST00000435275_RPS24 |
| miR-16 |
ENSG00000138326_ENST00000435275_RPS24 |
| miR-16 |
ENSG00000138399_ENST00000453153_FASTKD1 |
| miR-16 |
ENSG00000138592_ENST00000433963_USP8 |
| miR-16 |
ENSG00000138802_ENST00000265175_SEC24B |
| miR-16 |
ENSG00000139117_ENST00000331366_CPNE8 |
| miR-16 |
ENSG00000140391_ENST00000267970_TSPAN3 |
| miR-16 |
ENSG00000140526_ENST00000355100_ABHD2 |
| miR-16 |
ENSG00000140829_ENST00000268482_DHX38 |
| miR-16 |
ENSG00000140955_ENST00000315906_ADAD2 |
| miR-16 |
ENSG00000141101_ENST00000268802_NOB1 |
| miR-16 |
ENSG00000141140_ENST00000378326_MYO19 |
| miR-16 |
ENSG00000141367_ENST00000269122_CLTC |
| miR-16 |
ENSG00000141367_ENST00000269122_CLTC |
| miR-16 |
ENSG00000141404_ENST00000334049_GNAL |
| miR-16 |
ENSG00000141720_ENST00000269554_PIP5K2B |
| miR-16 |
ENSG00000142235_ENST00000270238_LMTK3 |
| miR-16 |
ENSG00000142961_ENST00000371940_MOBKL2C |
| miR-16 |
ENSG00000144227_ENST00000272641_NXPH2 |
| miR-16 |
ENSG00000144659_ENST00000273158_SLC25A38 |
| miR-16 |
ENSG00000144852_ENST00000393716_NR1I2 |
| miR-16 |
ENSG00000145495_ENST00000274140_MARCH6 |
| miR-16 |
ENSG00000145495_ENST00000274140_MARCH6 |
| miR-16 |
ENSG00000146833_ENST00000349062_TRIM4 |
| miR-16 |
ENSG00000147255_ENST00000370904_IGSF1 |
| miR-16 |
ENSG00000147403_ENST00000424325_RPL10 |
| miR-16 |
ENSG00000147408_ENST00000454498_CSGALNACT1 |
| miR-16 |
ENSG00000147533_ENST00000520817_GOLGA7 |
| miR-16 |
ENSG00000149273_ENST00000278572_RPS3 |
| miR-16 |
ENSG00000150764_ENST00000440460_DIXDC1 |
| miR-16 |
ENSG00000151612_ENST00000379448_ZNF827 |
| miR-16 |
ENSG00000152234_ENST00000282050_ATP5A1 |
| miR-16 |
ENSG00000152270_ENST00000282096_PDE3B |
| miR-16 |
ENSG00000152291_ENST00000377386_TGOLN2 |
| miR-16 |
ENSG00000154124_ENST00000284274_FAM105B |
| miR-16 |
ENSG00000154760_ENST00000285013_SLFN13 |
| miR-16 |
ENSG00000155393_ENST00000299192_HEATR3 |
| miR-16 |
ENSG00000156110_ENST00000372734_ADK |
| miR-16 |
ENSG00000156261_ENST00000286788_CCT8 |
| miR-16 |
ENSG00000156304_ENST00000286835_SFRS15 |
| miR-16 |
ENSG00000156486_ENST00000521839_KCNS2 |
| miR-16 |
ENSG00000156508_ENST00000316292_EEF1A1 |
| miR-16 |
ENSG00000159202_ENST00000360943_UBE2Z |
| miR-16 |
ENSG00000159348_ENST00000367249_CYB5R1 |
| miR-16 |
ENSG00000160209_ENST00000291565_PDXK |
| miR-16 |
ENSG00000160767_ENST00000361361_FAM189B |
| miR-16 |
ENSG00000160991_ENST00000356387_ORAI2 |
| miR-16 |
ENSG00000161542_ENST00000446526_PRPSAP1 |
| miR-16 |
ENSG00000162300_ENST00000294258_ZFPL1 |
| miR-16 |
ENSG00000163013_ENST00000521871_FBXO41 |
| miR-16 |
ENSG00000163113_ENST00000369135_OTUD7B |
| miR-16 |
ENSG00000163527_ENST00000295770_STT3B |
| miR-16 |
ENSG00000163946_ENST00000431842_C3orf63 |
| miR-16 |
ENSG00000164076_ENST00000477224_CAMKV |
| miR-16 |
ENSG00000164300_ENST00000507668_SERINC5 |
| miR-16 |
ENSG00000164818_ENST00000297440_HEATR2 |
| miR-16 |
ENSG00000164889_ENST00000485713_SLC4A2 |
| miR-16 |
ENSG00000165629_ENST00000356708_ATP5C1 |
| miR-16 |
ENSG00000165995_ENST00000377315_CACNB2 |
| miR-16 |
ENSG00000165995_ENST00000396576_CACNB2 |
| miR-16 |
ENSG00000166105_ENST00000431683_GLB1L3 |
| miR-16 |
ENSG00000166167_ENST00000370187_BTRC |
| miR-16 |
ENSG00000166681_ENST00000372645_NGFRAP1 |
| miR-16 |
ENSG00000166833_ENST00000396085_NAV2 |
| miR-16 |
ENSG00000167397_ENST00000394975_VKORC1 |
| miR-16 |
ENSG00000167552_ENST00000301071_TUBA1A |
| miR-16 |
ENSG00000167721_ENST00000301364_TSR1 |
| miR-16 |
ENSG00000167842_ENST00000381165_MIS12 |
| miR-16 |
ENSG00000168137_ENST00000402198_SETD5 |
| miR-16 |
ENSG00000168268_ENST00000422318_NT5DC2 |
| miR-16 |
ENSG00000168488_ENST00000336783_ATXN2L |
| miR-16 |
ENSG00000168758_ENST00000305476_SEMA4C |
| miR-16 |
ENSG00000169180_ENST00000304658_XPO6 |
| miR-16 |
ENSG00000169251_ENST00000351193_NMD3 |
| miR-16 |
ENSG00000169813_ENST00000443950_HNRNPF |
| miR-16 |
ENSG00000170004_ENST00000380358_CHD3 |
| miR-16 |
ENSG00000170006_ENST00000304385_TMEM154 |
| miR-16 |
ENSG00000170500_ENST00000393437_LONRF2 |
| miR-16 |
ENSG00000170759_ENST00000302418_KIF5B |
| miR-16 |
ENSG00000170776_ENST00000394518_AKAP13 |
| miR-16 |
ENSG00000171150_ENST00000306503_SOCS5 |
| miR-16 |
ENSG00000171385_ENST00000369697_KCND3 |
| miR-16 |
ENSG00000171490_ENST00000396503_RSL1D1 |
| miR-16 |
ENSG00000171570_ENST00000406058_EGLN2 |
| miR-16 |
ENSG00000172354_ENST00000393926_GNB2 |
| miR-16 |
ENSG00000173068_ENST00000380672_BNC2 |
| miR-16 |
ENSG00000173638_ENST00000311124_SLC19A1 |
| miR-16 |
ENSG00000173786_ENST00000393892_CNP |
| miR-16 |
ENSG00000173825_ENST00000309880_TIGD3 |
| miR-16 |
ENSG00000173875_ENST00000343325_ZNF791 |
| miR-16 |
ENSG00000173915_ENST00000337003_USMG5 |
| miR-16 |
ENSG00000174231_ENST00000304992_PRPF8 |
| miR-16 |
ENSG00000174292_ENST00000311668_TNK1 |
| miR-16 |
ENSG00000174405_ENST00000356922_LIG4 |
| miR-16 |
ENSG00000174595_ENST00000310992_KLF14 |
| miR-16 |
ENSG00000175414_ENST00000310389_ARL10 |
| miR-16 |
ENSG00000175467_ENST00000312397_SART1 |
| miR-16 |
ENSG00000177169_ENST00000321867_ULK1 |
| miR-16 |
ENSG00000179915_ENST00000406316_NRXN1 |
| miR-16 |
ENSG00000180596_ENST00000396984_HIST1H2BC |
| miR-16 |
ENSG00000180758_ENST00000377411_GPR157 |
| miR-16 |
ENSG00000181031_ENST00000323434_RPH3AL |
| miR-16 |
ENSG00000181192_ENST00000263035_DHTKD1 |
| miR-16 |
ENSG00000181222_ENST00000322644_POLR2A |
| miR-16 |
ENSG00000181418_ENST00000421952_DDN |
| miR-16 |
ENSG00000181472_ENST00000325144_ZBTB2 |
| miR-16 |
ENSG00000182149_ENST00000329908_KIAA0174 |
| miR-16 |
ENSG00000182158_ENST00000330387_CREB3L2 |
| miR-16 |
ENSG00000182447_ENST00000327928_OTOL1 |
| miR-16 |
ENSG00000182611_ENST00000333151_HIST1H2AJ |
| miR-16 |
ENSG00000183638_ENST00000382483_RP1L1 |
| miR-16 |
ENSG00000184009_ENST00000331925_ACTG1 |
| miR-16 |
ENSG00000184436_ENST00000215742_THAP7 |
| miR-16 |
ENSG00000184634_ENST00000374080_MED12 |
| miR-16 |
ENSG00000184675_ENST00000330258_FAM123B |
| miR-16 |
ENSG00000184779_ENST00000330339_RPS17 |
| miR-16 |
ENSG00000184867_ENST00000328766_ARMCX2 |
| miR-16 |
ENSG00000185176_ENST00000407834_AQP12B |
| miR-16 |
ENSG00000185818_ENST00000331662_NAT8L |
| miR-16 |
ENSG00000185915_ENST00000379499_KLHL34 |
| miR-16 |
ENSG00000186765_ENST00000334850_FSCN2 |
| miR-16 |
ENSG00000187239_ENST00000446176_FNBP1 |
| miR-16 |
ENSG00000187498_ENST00000375820_COL4A1 |
| miR-16 |
ENSG00000187583_ENST00000379410_PLEKHN1 |
| miR-16 |
ENSG00000187837_ENST00000343677_HIST1H1C |
| miR-16 |
ENSG00000187837_ENST00000343677_HIST1H1C |
| miR-16 |
ENSG00000188486_ENST00000375167_H2AFX |
| miR-16 |
ENSG00000188493_ENST00000378313_C19orf54 |
| miR-16 |
ENSG00000189091_ENST00000302516_SF3B3 |
| miR-16 |
ENSG00000189292_ENST00000403610_FAM150B |
| miR-16 |
ENSG00000189334_ENST00000368702_S100A14 |
| miR-16 |
ENSG00000196090_ENST00000373198_PTPRT |
| miR-16 |
ENSG00000196235_ENST00000378524_SUPT5H |
| miR-16 |
ENSG00000196365_ENST00000360614_LONP1 |
| miR-16 |
ENSG00000196604_ENST00000357462_A26C1B |
| miR-16 |
ENSG00000196924_ENST00000369863_FLNA |
| miR-16 |
ENSG00000196981_ENST00000330689_WDR5B |
| miR-16 |
ENSG00000197157_ENST00000354725_SND1 |
| miR-16 |
ENSG00000197353_ENST00000359228_LYPD2 |
| miR-16 |
ENSG00000197386_ENST00000355072_HTT |
| miR-16 |
ENSG00000197858_ENST00000355091_GPAA1 |
| miR-16 |
ENSG00000198586_ENST00000431350_TLK1 |
| miR-16 |
ENSG00000198804_ENST00000361624_MT-CO1 |
| miR-16 |
ENSG00000205581_ENST00000380749_HMGN1 |
| miR-16 |
ENSG00000205707_ENST00000381356_LYRM5 |
| miR-16 |
ENSG00000215041_ENST00000399464_NEURL4 |
| miR-16 |
ENSG00000215114_ENST00000399598_UBXN2B |
| miR-16 |
ENSG00000231555_ENST00000445736_HSPA1B |
| miR-16 |
ENSG00000232804_ENST00000450744_HSPA1B |
| miR-16 |
ENSG00000235067_ENST00000419792_TUBB |
| miR-16 |
ENSG00000237405_ENST00000447921_AGER |
| miR-16 |
ENSG00000244476_ENST00000472091_RP4-761I2-4 |
| miR-16 |
ENSG00000253953_ENST00000519479_PCDHGB4 |