Introduction
Pancreatic cancer is the fourth-leading cause of
cancer-associated mortality, with an overall 5-year survival rate
less than 5% (1). There are ~40,000
Americans annually that are diagnosed with pancreatic cancer, and
there is around an equivalent number of patients that succumb to
the disease (2). Pancreatic cancer
prognosis is so poor because the majority of patients have locally
advanced or distant metastatic disease at the time of presentation
(3). Although patient outcomes have
been improved by curative resection, chemotherapy and biological
therapies, the majority of patients succumb to disease within 5
years owing to the early recurrence and metastasis of the disease
(4). Therefore, a further
understanding the molecular mechanisms involved in pancreatic
cancer is required for improved diagnosis and treatment of the
disease.
Previous research demonstrated that multiple
signaling pathways are implicated in the pathogenesis of pancreatic
cancer, including the neurogenic locus notch homolog protein
(Notch), hedgehog, Wnt/β-catenin and transforming growth factor-β
(TGF-β) (5–7). The Notch receptor family includes
Notch1-4, which control pancreatic differentiation during
development and also participates in the initiation and progression
of pancreatic cancer (8). A previous
study indicates that Notch1 functions as a tumor suppressor in a
model of GTPase KRas (K-RAS)-induced pancreatic ductal
adenocarcinoma (PDAC) (9). Notch2 is
required for the progression of pancreatic intraepithelial
neoplasia and development of PDAC (10). Notch3 may be a prognostic biomarker in
PDAC. Moreover, several Notch receptors modify proteins; for
example, lunatic fringe (LFNG), which is a potential tumor
suppressor that regulates the activity of different Notch receptors
and can also serve important roles in pancreatic adenocarcinoma
(3). However, the function and
expression pattern of these Notch receptors in pancreatic cancer
remain poorly understood.
Dysregulation of the phosphoinositide-3 kinase
(PI3K)/AKT pathway is implicated in a number of human diseases,
including cancer, diabetes, cardiovascular disease and neurological
diseases (11). Activation is often
mediated by mutations occurring in the PIK3CA gene, which
encodes the p110α catalytic subunit of PI3Kα, a heterodimeric class
IA PI3K that is one of the most frequently mutated genes (16%) in
colorectal cancer (12). Certain
reports demonstrate that phosphoinositide-3 kinase catalytic
subunit-γ (PIK3CG) can also have important roles in sarcomagenesis
(13–15). A previous study suggested that PIK3CG
can be regulated by Notch signal transduction and is tightly
associated with stemness and migration of claudin-low breast cancer
cells (16). According to these
results, the level of PIK3CG expression seems to be tightly
associated with the metastasis and malignancy of different tumors.
Therefore, it is critical to investigate whether aberrant
expression of PIK3CG is associated with the initiation of
pancreatic cancer and its metastasis, advanced-stage diagnosis and
resistance to the majority of therapies (3).
The retinoblastoma gene (RB), a tumor
suppressor gene, is dysfunctional in several types of cancer
(17). One function of RB is
to prevent cell proliferation by inhibiting cell cycle progression
and recent studies have revealed a substantial role for RB and its
downstream effectors, particularly the E2F family of transcription
factors, in regulating various aspects of tumor progression,
angiogenesis and metastasis (18). In
pancreatic cancer, dysfunction of RB1 enables TGF-β to promote
cancer cell proliferation, and pancreatic carcinogenesis can be
accelerated by the deletion of RB (19,20). A
previous study reported that Notch1 exhibits oncogene-like
characteristics by inducing dephosphorylation of Rb in esophageal
squamous carcinoma cells (21).
However, the mechanism of the crosstalk between Notch, TGF-β and RB
requires further clarification.
In the present study, clinical pancreatic cancer
sections and the human pancreatic cancer MIA PaCa-2 cell line were
examined to assess the expression pattern of the Notch receptors,
P-Smad2, RB and PIK3CG and the crosstalk between these proteins in
pancreatic cancer cells to understand further the mechanisms
involved in pancreatic carcinogenesis.
Materials and methods
Tissue samples
A total of 20 patients (male, 16; female, 4) with
pancreatic cancer were included in this study. The median patient
age was 56 years (range, 23–79 years). All tumor samples were
obtained in December 2015 from the Department of Pathology, The
Second Xiangya Hospital of Central South University (Changsha,
China). Tumor samples were obtained during surgery and in routine
diagnostic biopsies. The samples were fixed in 10% formalin for 24
h at room temperature and paraffin-embedded. All research biopsies
were evaluated by pathologists specialized in pancreatic cancer
diagnostics and ensured adequate quantity of tumor tissues were
used for analysis. Adjacent tissue samples were located within 1 cm
of the tumor margin and were confirmed to be noncancerous by
pathological examination. Ethical approval for the present study
was provided by the Ethics Committee at The Second Xiangya Hospital
of Central South University (Hunan, China).
Cell culture
The pancreatic cancer MIA PaCa-2 cell line was
purchased from American Type Culture Collection (ATCC; Manassas,
VA, USA). The siRNA-Lfng and Notch3 intracellular domain
(N3IC)-overexpressing MIA PaCa-2 cell models were established as
previously described (3). The cells
were cultured according to ATCC protocols in Dulbecco's modified
Eagle's medium (DMEM) (Corning Incorporated, Corning, NY, USA)
supplemented with 10% fetal bovine serum (Atlanta Biologicals,
Inc., Flowery Branch, GA, USA) at 37°C with 100 U/ml penicillin and
100 µg/ml streptomycin maintained in a humidified environment
containing 5% CO2. For cell counting, MIA PaCa-2 cells
were respectively treated with 100, 50, 25, 12, 6, 3, 1.5, 0.8,
0.4, 0.2 and 0.1 µM PI3K inhibitor AS-605240 (catalog no. S1410;
Selleck Chemicals, Houston, TX, USA) at 37°C for 72 h. For wound
healing assays, MIA PaCa-2 cells were treated with AS-605240 at a
final concentration of 5 µM for 48 h.
Cell counting
For cell counting, MIA PaCa-2 cells were incubated
in 10% cell counting kit-8 solution (catalog no. 40203ES60;
Shanghai Yeasen Biotechnology Co., Ltd, Shanghai, China) diluted in
DMEM at 37°C for 3 h. The absorbance of each well was measured with
a microplate reader, set at an absorbance wavelength of 450 nm. All
experiments were performed in triplicate. The cell growth curve was
performing using GraphPad Prism 6.0 software (GraphPad Software,
Inc., La Jolla, CA, USA). Data are represented as the mean ±
standard error of three independent experiments.
Immunohistochemistry (IHC)
Formalin-fixed paraffin-embedded pancreatic tissues
were processed for histology and IHC as described previously
(22,23). For each sample, two adjacent tissue
sections (5-µm thickness) were mounted on one glass slide. Then,
the tissue sections were deparaffinized and rehydrated using xylene
and a graded ethanol series. Sections were then heated at 100°C in
a 10 mM citrate buffer solution (pH 6.0) for 30 min for antigen
retrieval. Endogenous peroxidase activity was quenched by immersing
the sections in 3% H2O2 for 10 min. Next, the
staining was performed using the ImmunoCruz™ ABC Staining system
(Santa Cruz Biotechnology, Inc., Dallas, TX, USA) according to the
manufacturer's protocol. The specimens for 1 h in 5% normal
blocking serum in PBS at room temperature. Following removal of
blocking reagent from the slides, the samples were incubated with
primary antibodies for overnight at 4°C. Following washing three
times with PBS for 5 mins each, incubation for 30 min at room
temperature with biotin-conjugated secondary antibody at
approximately 1 µg/ml was performed. Then the samples were washed
three times with PBS for 5 min each. Incubation for 30 mins at room
temperature with the ABC reagent was performed, followed by washing
three times with PBS for 5 min each. Subsequently, incubation in
peroxidase substrate and chromogen mixture until the desired stain
intensity developed was performed. Then sections were washed in
deionized H2O. When appropriate, dehydration with
alcohols and xylenes was performed as follows: Soak in 90% ethanol
twice for 3 mins each, 100% ethanol twice for 3 min each, then
xylene three times for 10 sec each and excess xylene was wiped off.
Immediately 1–2 drops of permanent mounting medium was added,
covered with a glass coverslip and observed by light microscopy. A
brown color was indicative of a positive result. Primary antibodies
used for immunostaining were as follows: Notch1 (catalog no. bTAN
20; 1:200;, Developmental Studies Hybridoma Bank (DSHB), University
of Iowa, IA, USA), Notch2 (catalog no. C651.6DbHN; DSHB; 1:200),
Notch3 (catalog no. 55114-1-AP; ProteinTech Group, Inc., Chicago,
IL, USA; 1:100), PI3K p110γ (catalog no. 5405; Cell Signaling
Technology, Inc., Danvers, MA, USA; 1:100), p-Smad2 (catalog no.
3101; Cell Signaling Technology, Inc; 1:100). Cytoplasmic and
nuclear staining was considered positive if >10% cells were
positively stained. Surrounding fibrous and inflammatory tissue was
not scored. The slides were graded independently by two observers
blinded to the clinical data.
Western blotting
For western blotting, the MIA PaCa-2 cells were
lysed in radioimmunoprecipitation buffer (Boston BioProducts Inc.,
Ashland, MA, USA) with protease inhibitor cocktail (Roche Applied
Science, Penzberg, Germany). Protein concentrations were determined
using the BCA protein assay kit (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Samples of cell lysate supernatant (50 µg
protein) were resolved by 8% SDS-PAGE and electrotransferred to
nitrocellulose membranes. The membranes were blocked in TBST buffer
with 5% w/v non-fat dry milk at room temperature for 1 h and then
probed at 4°C overnight with indicated antibodies with the
following dilutions: Akt (catalog no. 4691; Cell Signaling),
phospho-Akt (Ser473) (catalog no. 4060; Cell Signaling), phospho-RB
(catalog no. 8516; Cell Signaling), RB (catalog no. 9309; Cell
Signaling), PI3K p110γ (catalog no. 5405; Cell Signaling) and
β-actin (catalog no. sc-81178; Santa Cruz Biotechnology, Inc.), all
at 1:1,000 dilution. The secondary goat anti-rabbit IgG-HRP
(catalog no. sc-2004) and goat anti-mouse IgG-HRP (catalog no.
sc-2005) (both Santa Cruz Biotechnology, Inc.) were used at a
1:4,000 dilution at room temperature for 2 h. Detection was
performed using ECL Western Blotting Detection reagents (catalog
no. PI32106; Pierce; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). All antibodies were used in accordance with the
manufacturer's instructions.
Wound-healing experiment
Collective cell migration was measured using
wound-healing assay. The cells were seeded in a 6-well plate at a
density of 5×105 cells per well. After incubation at
37°C for 12 h, a portion of the monolayer was scratched with a
1,000 µl pipette tip, and examined for resealing of the ‘wounded’
monolayer after 48 h using a light microscope.
Statistical analysis
Data from all experiments are presented as the mean
± standard deviation of at least three independent experiments
performed in duplicate for each construct. Statistical analysis was
performed using the two-tailed Student's t-test. The software used
for statistical analysis was GraphPad Prism 6.0 software (GraphPad
Software, Inc., La Jolla, CA, USA). P<0.001 was considered to
indicate a statistically significant difference.
Results
Changes in Notch receptor expression
in in pancreatic cancer tissues compared with tumor-adjacent
tissues
The expression levels of all Notch receptors has
been previously found to be upregulated in the
KrasLSL-G12D/+;Pdx1-Cre pancreatic mouse model (3). However, the present study observed that
the expression pattern of Notch receptors in human pancreatic
tissue samples is different from the expression pattern in mouse
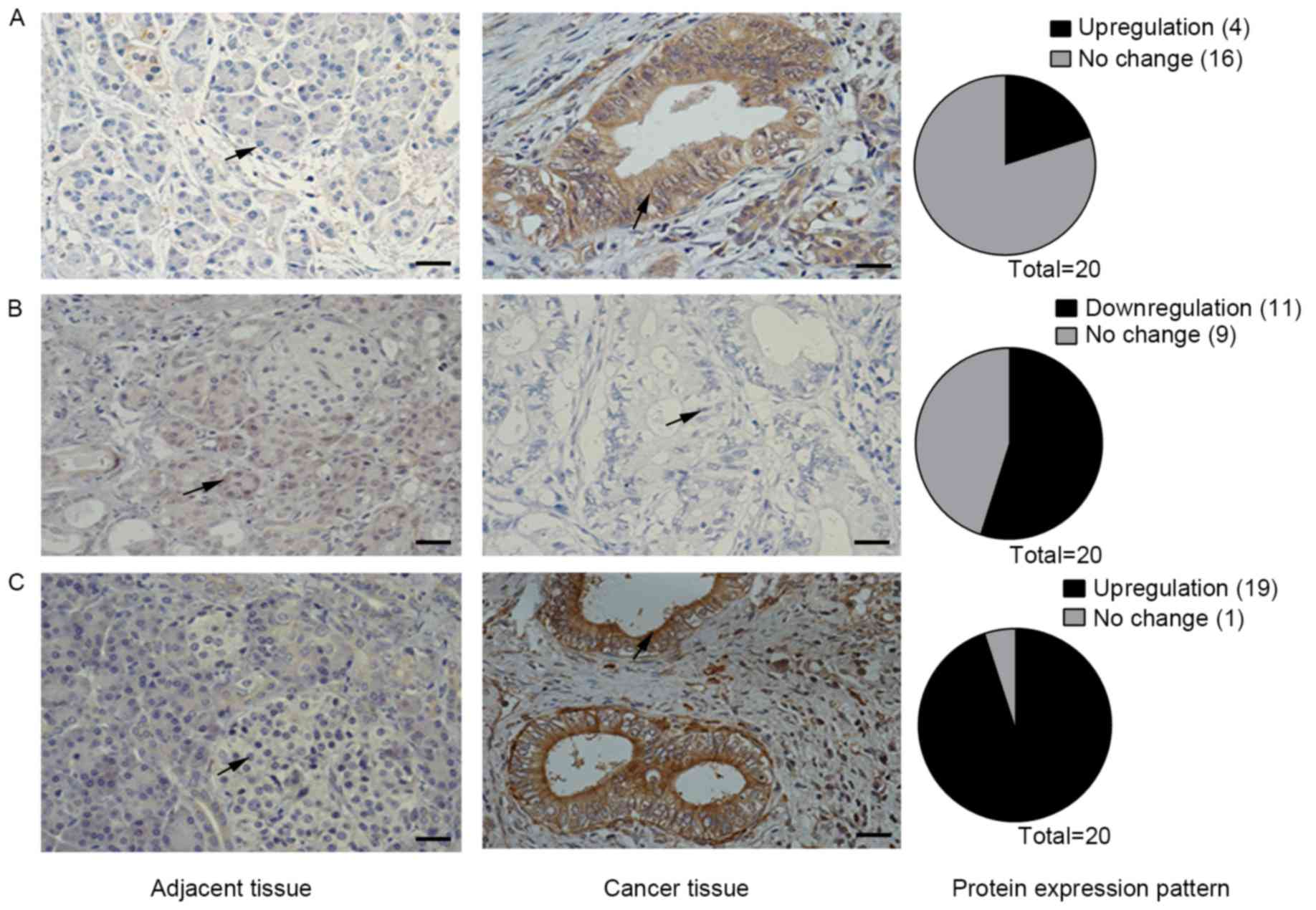
tumor tissues (Fig. 1). Using IHC
analysis of samples from 20 patients, Notch1 staining was found to
be positive in 4/20 (20%) pancreatic cancer tissues, whereas the
adjacent tissues were all negative for staining (Fig. 1A). Staining for Notch3 was positive in
19/20 pancreatic cancer tissues (95%) and negative in all adjacent
tissues (Fig. 1C). However, Notch2
staining results were negative in 11/20 (55%) pancreatic cancer
tissues, but positive in all adjacent tissues (Fig. 1B). Therefore, the Notch2 receptor may
serve a different role to Notch1 and Notch3 in human pancreatic
cancer.
PIK3CG and TGF-β signal play important
roles in pancreatic carcinogenesis
Notch signaling antagonizes TGF-β signaling in the
mouse pancreas and in PDAC cells through downregulation of Tgfb and
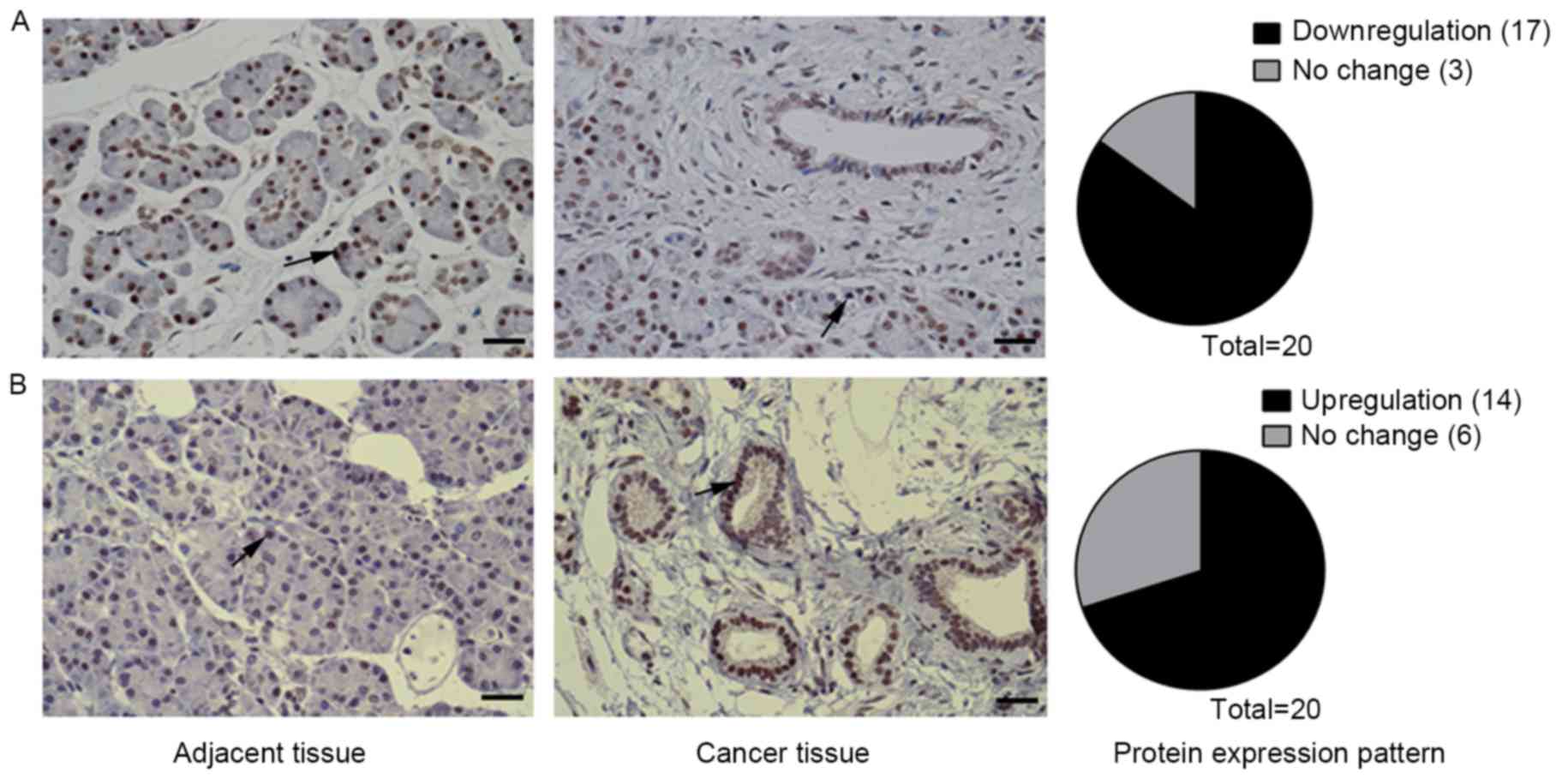
Tgfbr gene expression (3). The IHC
results of the present study revealed that nuclear staining of
p-smad2 was weaker or the p-smad2 expression decreased in
pancreatic cancer tissues compared with the adjacent tissues in
17/20 (85%) patients (Fig. 2A), which
was consistent with results in mice found in a previous study
(3). Staining for PIK3CG was found to
be stronger in pancreatic cancer tissue compared with adjacent
tissues in 14/20 (70%) patients. To the best of our knowledge, the
results of the present study revealed that PIK3CG expression is
upregulated in human pancreatic cancer for the first time (Fig. 2B) and that PIK3CG may serve an
important role in human pancreatic cancer.
Notch signaling pathway modulates RB
and p-AKT expression in pancreatic cancer cells
In pancreatic cancer, RB1 dysfunction enabled TGF-β
signaling, which was modulated by Notch signaling to promote cancer
cell proliferation (19). The present
authors previously found that PIK3CG gene expression was modulated
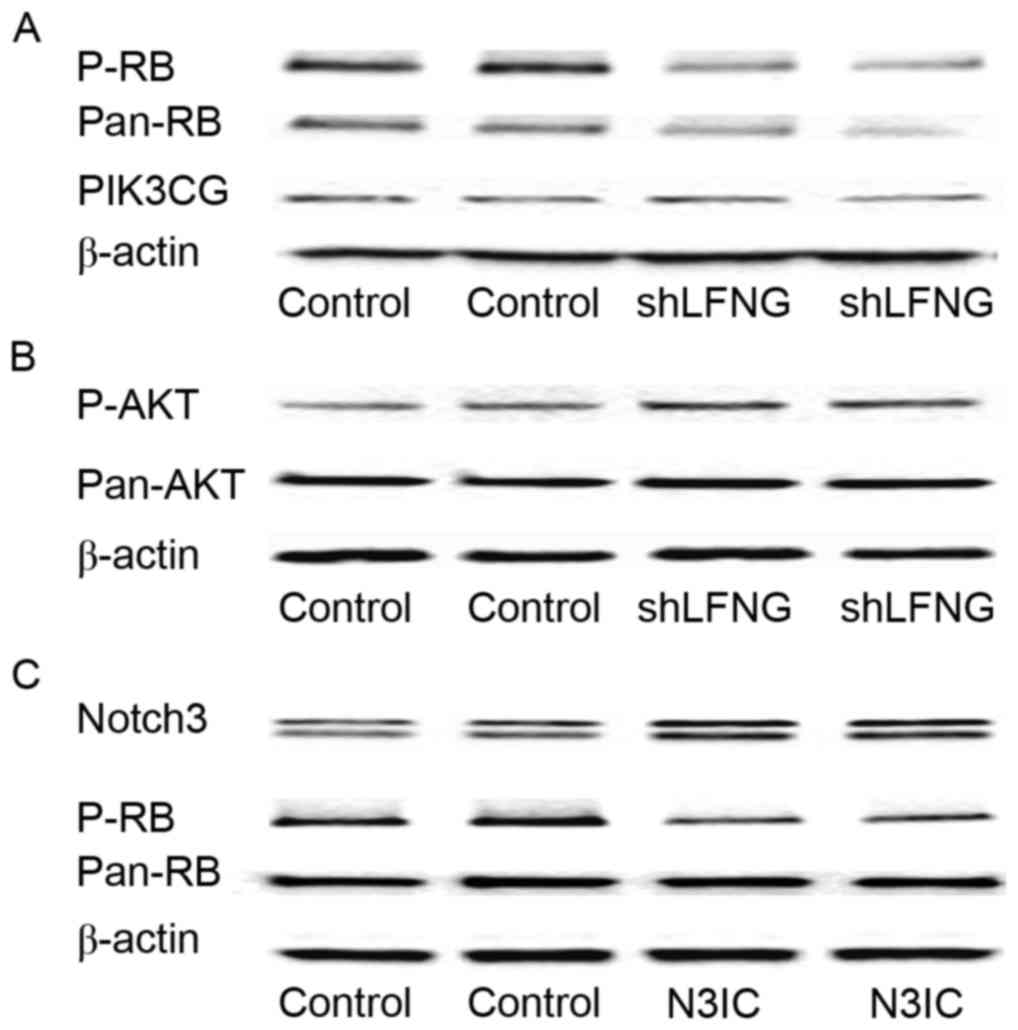
by Notch signaling in claudin-low breast cancer (16). Using the siRNA-Lfng MIA PaCa-2 cell
model, where Notch signaling is activated (3), it was observed by western blotting that
the protein expression level of p-RB and pan-RB was downregulated
compared with the control, the PIK3CG expression level remained the
same and p-AKT expression was upregulated (Fig. 3A and B). These results indicated that
RB1 and p-AKT, but not PIK3CG, were the downstream targets of the
Notch signaling pathway in the siRNA-Lfng MIA PaCa-2 cell model,
suggesting that P-AKT may be regulated by Notch signaling through
pathways other than PIK3CG/AKT. Western blot analysis was also
performed to assess the level of RB expression, using another MIA
PaCa-2 cell model in which Notch3IC was over-expressed. The results
confirmed that RB expression was inhibited when Notch signaling was
activated in MIA PaCa-2 cells (Fig.
3C).
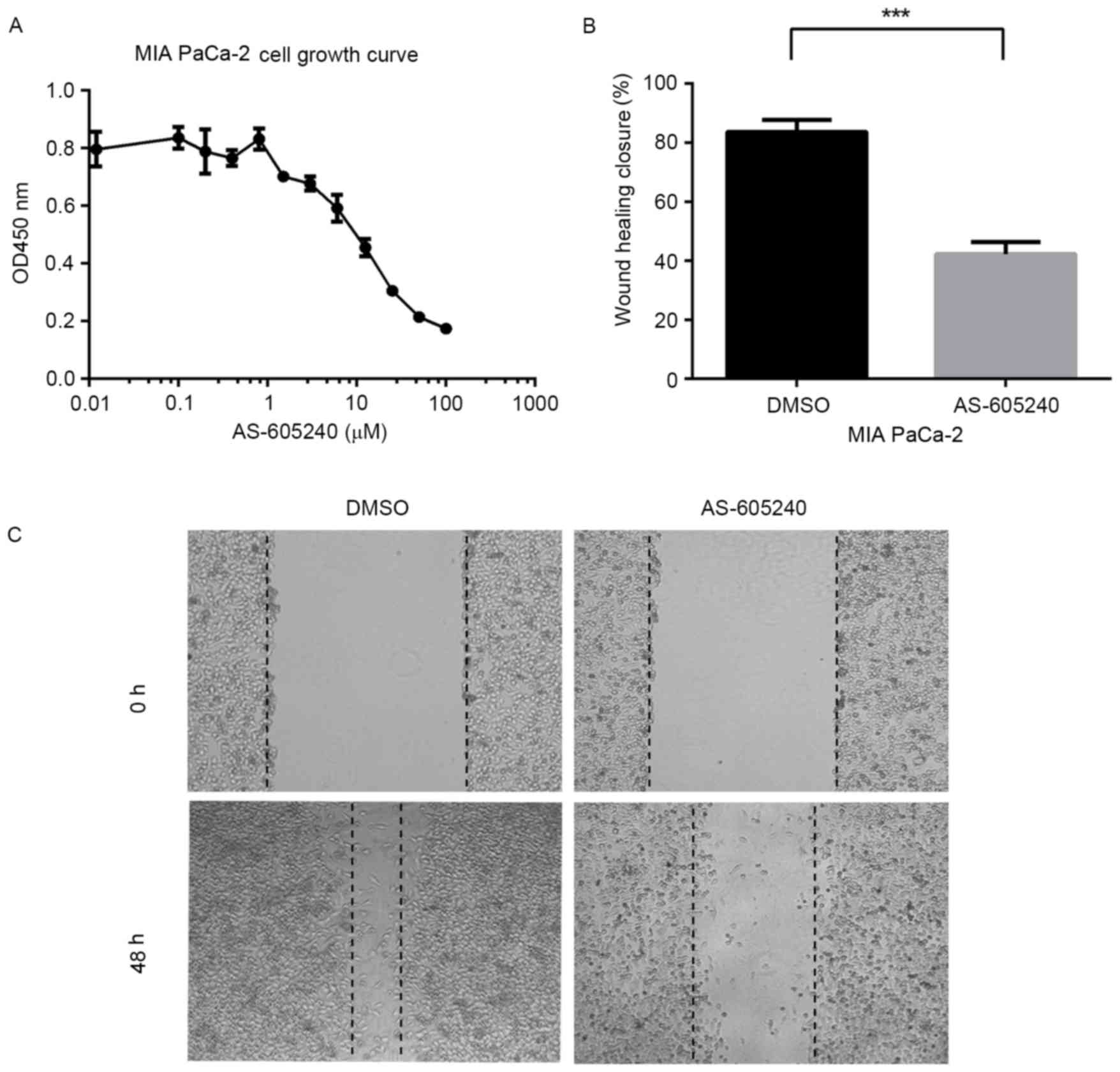
Inhibition of PIK3CG blocks the growth
and migration of MIA PaCa-2 cells
PIK3CG has been tightly associated with claudin-low
breast cancer migration and the potential targets of multiple types
of cancer, including sarcoma and gastric cancer (24). The results of the present study
demonstrated that PIK3CG was a key gene involved in the growth and
migration of pancreatic cancer cells. The growth of MIA PaCa-2
cells was markedly reduced upon treatment with the PIK3CG-specific
inhibitor AS-605240 (Fig. 4A). The
migration of MIA PaCa-2 cells was significantly blocked when the
cells were incubated with 5 µM AS-605240 for 48 h, as assessed
using wound healing experiments (Fig. 4B
and C).
Discussion
Recently, increasing evidence has demonstrated that
the Notch signaling pathway has important roles in the initiation,
progression and metastasis of a variety of human malignancies,
including pancreatic cancer (25,26).
However, the mechanism by which this happens remains poorly
understood. The present study focused on the current understanding
of the molecular pathogenesis of pancreatic cancer, particularly on
the Notch signaling pathway and its novel molecular targets,
including RB and PIK3CG. The current study may aid the prevention
and treatment of pancreatic cancer.
Pancreatic cancer is a malignant cancer that
frequently metastasizes prior to diagnosis, meaning there is a
pressing requirement to understand the mechanism by which it
initiates and progresses. Data from a previous study demonstrated
that deletion of Lfng in the KrasLSL-G12D/+;Pdx1-Cre
mouse model caused increased activation of Notch3 throughout the
initiation and progression of PDAC (3). In the present study, the expression
pattern of Notch1-3 receptors was first assessed using human
clinical samples to confirm the function of the Notch signaling
pathway in pancreatic cancer. The results of the present study
demonstrated that Notch3 expression was upregulated in the majority
of tumor tissues assessed, and Notch1 staining was observed to be
positive in a number of pancreatic cancer tissues by IHC staining.
The degree of Notch1 and Notch3 staining was consistent with
previous data from mouse models and clinical specimens (27). The current study found that Notch2
expression was downregulated in tumor tissues, meaning that Notch
receptors have different functions in the process of pancreatic
carcinogenesis and that use of a γ-secretase inhibitor to block the
Notch signaling pathway for the treatment of pancreatic cancer
should be used cautiously.
A previous study by the present authors demonstrated
that TGF-β, which is regulated by Notch signaling, is tightly
associated with the metastasis of pancreatic cancer (3). The present study revealed that the gene
expression level of p-Smad2 and PIK3CG was altered in pancreatic
cancer tissues compared with the adjacent tissues. By
over-activating the Notch signaling pathway in pancreatic cancer
MIA PaCa-2 cells, the expression of RB was downregulated and P-AKT
was upregulated, whereas PIK3CG expression did not change. These
results demonstrated that Notch signaling and PIK3CG may have
important roles in the initiation and progression of pancreatic
cancer. Notch signaling can be an upstream regulator of RB but not
PIK3CG in the pancreas, although PIK3CG could be a target for Notch
signaling in triple-negative breast cancer. In pancreatic cancer,
PIK3CG may function independent of the Notch signaling pathway and
the mechanism involved in the process is worthy of being further
clarified. It is very promising that both cell growth and migration
rate decreased when MIA PaCa-2 cells were treated with the PIK3CG
inhibitor AS-605240. These data reveal that PIK3CG may be a
potential target for the treatment of malignant pancreatic cancer.
For future studies, the present authors will investigate the
function of AS-605240 using more pancreatic cell lines and
KrasLSL−G12D/+; Pdx1-Cre mouse models.
In conclusion, the present study revealed the
expression pattern of the Notch receptors, RB and PIK3CG in
pancreatic cancer and adjacent tissue samples, although more
pancreatic cancer tissue samples are required to confirm the
experimental data of the study further. The Notch signaling pathway
and PIK3CG are involved in pancreatic carcinogenesis. Cell model
results revealed that Notch signal pathway is able to modulate RB
expression but not PIK3CG, which is tightly associated with cell
proliferation and migration in pancreatic cancer cells. The use of
more pancreatic cell lines to confirm the present results is
required, as is investigating the different transcription
factor-binding patterns of the PIK3CG promoter between pancreatic
cancer and claudin-low breast cancer cells. Finally, the finding of
PIK3CG function in pancreas cancer provides new insights into the
functions of this gene, which may be a novel target gene for the
treatment of pancreatic cancer.
Acknowledgements
The present study was supported by the Scientific
Research Foundation for the Returned Overseas Chinese Scholars, the
State Education Ministry and the Fund of the National Forestry
Bureau, China.
References
|
1
|
Chen Huang, Jiawei Du and Keping Xie:
FOXM1 and its oncogenic signaling in pancreatic cancer
pathogenesis. Biochim Biophys Acta. 1845:104–116. 2014.PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang S, Chung WC and Xu K: Lunatic Fringe
is a potent tumor suppressor in Kras-initiated pancreatic cancer.
Oncogene. 35:2485–2495. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boyle J, Czito B, Willett C and Palta M:
Adjuvant radiation therapy for pancreatic cancer: a review of the
old and the new. J Gastrointest Oncol. 6:436–444. 2015.PubMed/NCBI
|
|
5
|
Kunnimalaiyaan S, Trevino J, Tsai S,
Gamblin TC and Kunnimalaiyaan M: Xanthohumol-mediated suppression
of Notch1 signaling is associated with antitumor activity in human
pancreatic cancer cells. Mol Cancer Ther. 14:1395–1403. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marechal R, Bachet JB, Calomme A, Demetter
P, Delpero JR, Svrcek M, Cros J, Bardier-Dupas A, Puleo F, Monges
G, et al: Sonic hedgehog and Gli1 expression predict outcome in
resected pancreatic adenocarcinoma. Clin Cancer Res. 21:1215–1224.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang S, Zhu L, Tang H, Zhang M, Chen Z,
Fei J, Han B and Zou GM: Ape1 regulates WNT/β-catenin signaling
through its redox functional domain in pancreatic cancer cells. Int
J Oncol. 47:610–620. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Court H, Amoyel M, Hackman M, Lee KE, Xu
R, Miller G, Bar-Sagi D, Bach EA, Bergo MO and Philips MR:
Isoprenylcysteine carboxylmethyltransferase deficiency exacerbates
KRAS-driven pancreatic neoplasia via Notch suppression. J Clin
Invest. 123:4681–4694. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hanlon L, Avila JL, Demarest RM, Troutman
S, Allen M, Ratti F, Rustgi AK, Stanger BZ, Radtke F, Adsay V, et
al: Notch1 functions as a tumor suppressor in a model of
K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res.
70:4280–4286. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mazur PK, Einwachter H, Lee M, Sipos B,
Nakhai H, Rad R, Zimber-Strobl U, Strobl LJ, Radtke F, Klöppel G,
et al: Notch2 is required for progression of pancreatic
intraepithelial neoplasia and development of pancreatic ductal
adenocarcinoma. Proc Natl Acad Sci USA. 107:pp. 13438–13443. 2010;
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao HF, Wang J and Tony To SS: The
phosphatidylinositol 3-kinase/Akt and c-Jun N-terminal kinase
signaling in cancer: Alliance or contradiction? (Review). Int J
Oncol. 47:429–436. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Manceau G, Marisa L, Boige V, Duval A,
Gaub MP, Milano G, Selves J, Olschwang S, Jooste V, le Legrain M,
et al: PIK3CA mutations predict recurrence in localized
microsatellite stable colon cancer. Cancer Med. 4:371–382. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martin D, Galisteo R, Molinolo AA, Wetzker
R, Hirsch E and Gutkind JS: PI3Kγ mediates kaposi's
sarcoma-associated herpesvirus vGPCR-induced sarcomagenesis. Cancer
Cell. 19:805–813. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Wang K, Jiang YZ, Chang XW, Dai CF
and Zheng J: 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) inhibits
human ovarian cancer cell proliferation. Cell Oncol (Dordr).
37:429–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Forbes SA, Bindal N, Bamford S, Cole C,
Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al:
COSMIC: Mining complete cancer genomes in the catalogue of somatic
mutations in cancer. Nucleic Acids Res. 39((Database Issue):
D945–D950. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang S, Chung WC, Wu G, Egan SE, Miele L
and Xu K: Manic fringe promotes a claudin-low breast cancer
phenotype through notch-mediated PIK3CG induction. Cancer Res.
75:1936–1943. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Murphree AL and Benedict WF:
Retinoblastoma: Clues to human oncogenesis. Science. 223:1028–1033.
1984. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schaal C, Pillai S and Chellappan SP: The
Rb-E2F transcriptional regulatory pathway in tumor angiogenesis and
metastasis. Adv Cancer Res. 121:147–182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gore AJ, Deitz SL, Palam LR, Craven KE and
Korc M: Pancreatic cancer-associated retinoblastoma 1 dysfunction
enables TGF-γ to promote proliferation. J Clin Invest. 124:338–352.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Carriere C, Gore AJ, Norris AM, Gunn JR,
Young AL, Longnecker DS and Korc M: Deletion of Rb accelerates
pancreatic carcinogenesis by oncogenic Kras and impairs senescence
in premalignant lesions. Gastroenterology. 141:1091–1101. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kagawa S, Natsuizaka M, Whelan KA,
Facompre N, Naganuma S, Ohashi S, Kinugasa H, Egloff AM, Basu D,
Gimotty PA, et al: Cellular senescence checkpoint function
determines differential Notch1-dependent oncogenic and
tumor-suppressor activities. Oncogene. 34:2347–2359. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Zhao YJ, Zou QY, Zhang K, Wu YM,
Zhou C, Wang K and Zheng J: Preeclampsia does not alter vascular
growth and expression of CD31 and vascular endothelial cadherin in
human placentas. J Histochem Cytochem. 63:22–31. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao YJ, Zou QY, Li Y, Li HH, Wu YM, Li
XF, Wang K and Zheng J: Expression of G-protein subunit γ-14 is
increased in human placentas from preeclamptic pregnancies. J
Histochem Cytochem. 62:347–354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao Z, Song Y, Piao D, Liu T and Zhao L:
Identification of genes and long non-coding RNAs associated with
the pathogenesis of gastric cancer. Oncol Rep. 34:1301–1310. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bailey JM, Alsina J, Rasheed ZA,
McAllister FM, Fu YY, Plentz R, Zhang H, Pasricha PJ, Bardeesy N,
Matsui W, et al: DCLK1 marks a morphologically distinct
subpopulation of cells with stem cell properties in preinvasive
pancreatic cancer. Gastroenterology. 146:245–1256. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miyamoto Y, Maitra A, Ghosh B, Zechner U,
Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM,
Wolfe MS, et al: Notch mediates TGF alpha-induced changes in
epithelial differentiation during pancreatic tumorigenesis. Cancer
Cell. 3:565–576. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mann CD, Bastianpillai C, Neal CP, Masood
MM, Jones DJ, Teichert F, Singh R, Karpova E, Berry DP and Manson
MM: Notch3 and HEY-1 as prognostic biomarkers in pancreatic
adenocarcinoma. PLoS One. 7:e511192012. View Article : Google Scholar : PubMed/NCBI
|