Previous studies have identified that the human
genome contains ~21,000 genes, and only <2% of them are
protein-coding genes (1,2). In the previous decades, studies of
protein-coding genes have led to improved understanding of their
participation in tumorigenesis and tumor characteristics,
consequentially establishing a number of protein prognostic markers
and therapeutic targets in numerous types of cancer (3–6).
Furthermore, larger numbers of non-coding RNAs (ncRNAs), including
microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), small
interfering RNAs (siRNAs), small nuclear RNAs (snRNAs), small
nucleolar RNAs (snoRNAs) and lncRNAs are expressed at lower levels
to fulfill regulatory functions to control complex
physiopathological processes in humans (7,8).
Therefore, it is important to characterize the functions of the
large majority of ncRNAs.
NcRNAs may be classified by their biological
functions: housekeeping ncRNAs and regulatory ncRNAs. Housekeeping
ncRNAs are usually expressed constitutively, including ribosomal
RNA (rRNAs), snRNAs, snoRNAs and transfer RNAs (tRNAs). Regulatory
ncRNAs, according to their length, comprise short regulatory
ncRNAs, including miRNAs, siRNAs and piRNAs and long regulatory
ncRNAs (8–10).
LncRNAs are a class of ncRNAs with >200
nucleotides in length. It is now recognized that lncRNAs function
as key regulatory players in a number of biological processes,
including embryonic development, cellular differentiation and
cancer (11). lncRNAs regulate their
target genes at transcriptional or post-transcriptional levels.
Previously, the dysregulation of lncRNAs has been closely
associated with carcinogenesis and cancer progression. Compared
with the protein-coding genes, lncRNAs exhibit more tissue- and
time-specific expression patterns, and their expressions are more
closely associated with their biological function and tumor status,
indicating enormous potential roles of lncRNAs as diagnostic and
prognostic biomarkers, and as therapeutic targets in cancer
(3,12–14). For
example, the variant genotypes of rs7763881 in the hepatocellular
carcinoma up-regulated long non-coding RNA gene may be responsible
for the decreased susceptibility to hepatitis B virus-associated
carcinogenesis in liver, suggesting that genetic variations in
lncRNAs are associated with cancer susceptibility (15). In addition, aberrant expression of
lncRNAs has been employed in cancer diagnosis and monitoring
(16). H19 is upregulated in the
plasma of patients with gastric cancer, and its expression enabled
the differentiation of early stage gastric cancer from healthy
controls (17). Subsequent studies
have indicated that the level of H19 may be used to monitor and
reflect the tumor dynamics in patients with gastric cancer
(18). Furthermore, lncRNA expression
profiles may also be used to identify clinically relevant cancer
subtypes that predict tumor biological behavior, therapeutic
responsiveness and clinical prognosis (19–23).
Gliomas represent 31% of all central nervous system
(CNS) tumors diagnosed in the United States, and 81% of all
malignant CNS tumor types with high morbidity and mortality
(2006–2010) (24). Despite the
treatment options of surgical resection followed by radiotherapy
and chemotherapy, the overall survival times of patients with
glioma, particularly patients with malignant glioma, were low. The
understanding of the genetic and molecular makeup of gliomas has
been advanced during the previous the decades. However, there
remains a lack of effective therapies for these tumors. Therefore,
an improved understanding of glioma pathogenesis is urgently
required. Previous studies suggest that lncRNAs have a close
association with glioma, but their roles and the underlying
mechanisms remain elusive. In the present review, the recent
progress on lncRNAs in the development of glioma was summarized,
and their possible functions and pathogenesis mechanisms in
regulating biological behaviors of glioma were discussed.
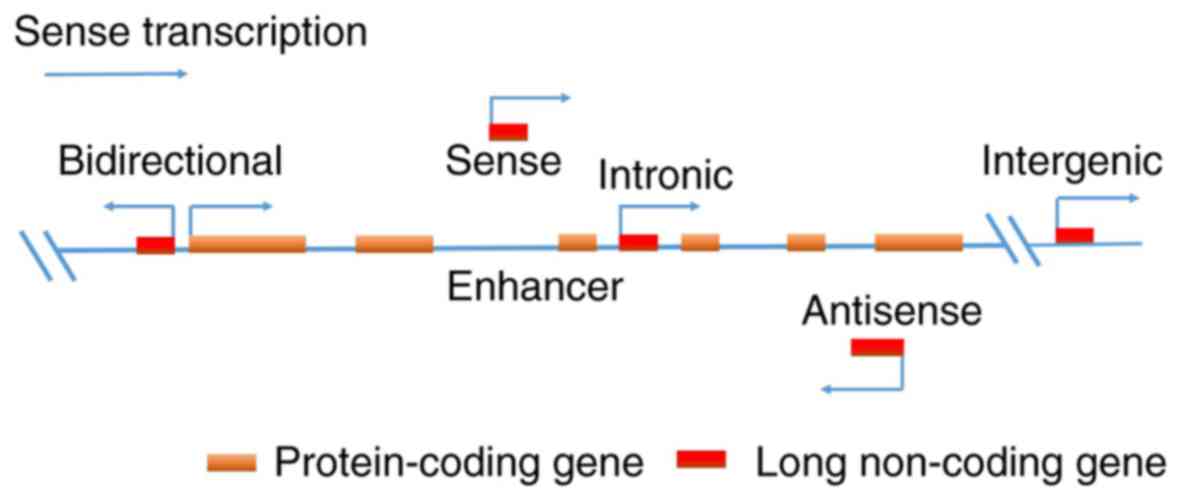
LncRNAs are a large and heterogeneous group of RNAs,
reflecting indirectly their enormous variety and structural
complexity. Based on its genomic location to protein-coding genes,
an lncRNA may be placed broadly into several categories: i)
bidirectional; ii) enhancer; iii) intergenic; iv) intronic; v)
sense and vi) anti-sense lncRNAs. The expression of bidirectional
lncRNAs is initiated within the vicinity (>1 kb) of a
neighboring coding transcript of the opposite strand. Enhancer
lncRNAs are located in the enhancer regions of the promoter of a
coding transcript. Intergenic lncRNAs are transcribed from regions
between two coding transcripts. Intronic lncRNAs are derived
entirely from within the introns of a coding transcript. Sense
lncRNAs overlap with a part of or the entire sense strand of a
transcript. Anti-sense lncRNAs are transcribed from the anti-sense
direction to the transcripts of a gene (Fig. 1) (25,26).
The ways in which lncRNAs regulate gene expression
can also be grouped into three categories, which include
transcriptional and post-transcription regulation, and other
mechanisms. Transcriptional regulation is where lncRNAs regulate
gene expression through transcriptional interference and chromatin
remodeling (27). Post-transcription
regulation involves the regulation of RNA splicing by modulating
the functions of splicing factors or by directly binding to
pre-mRNA sequences. LncRNAs may also block translation through
interaction with translation factors or ribosomes (28–30). Other
mechanisms of gene expression by lncRNAs include protein
localization, telomere replication and RNA interference (31,32).
LncRNAs may also be classified into four archetypes
based on the molecular mechanisms of their functions: i) signal:
lncRNAs may serve as molecular signals for gene regulation; ii)
decoy: lncRNAs act as ‘molecular sinks’ that bind and sequestrate
protein targets but do not exert any additional functions; iii)
guide: lncRNAs interact with proteins and guide the localization of
ribonucleoprotein complexes to specific targets; iv) scaffold:
lncRNAs function as central platforms for multiple molecules to
form scaffolding complexes to regulate their functions (33).
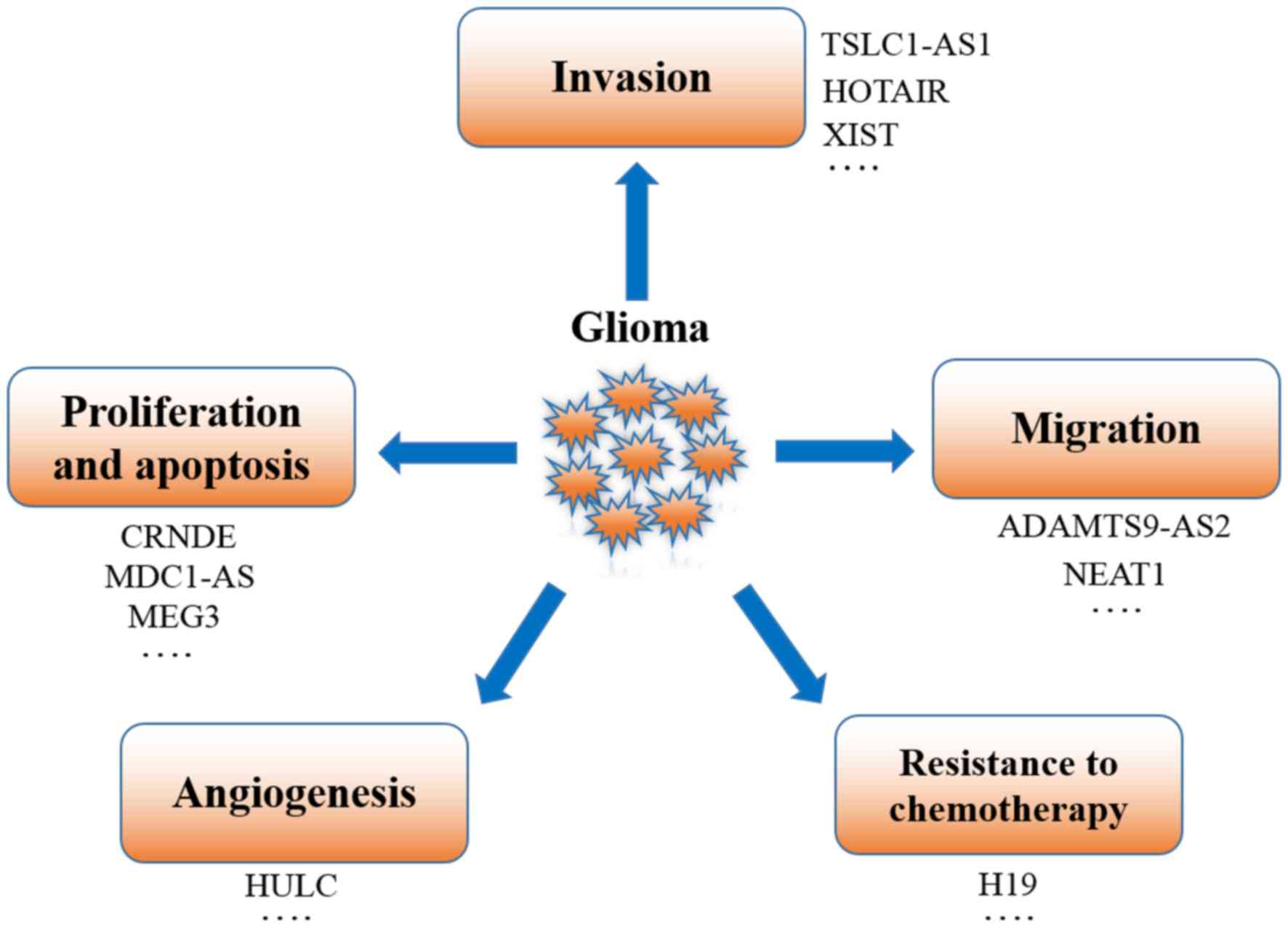
Previous studies have suggested that several lncRNAs
are involved in the development of gliomas and associated with
various biological behaviors of tumors, including proliferation,
migration, invasion and apoptosis. The lncRNAs that are associated
with gliomas are summarized in Table
I. In the following section, the potential roles of several
lncRNAs in the development of glioma, and their potential for
clinical applications for glioma treatment, are discussed.
lncRNA H19, a paternally imprinted gene residing
close to the telomeric region of chromosome 11p15.5, was first
identified as a tumor suppressor (34,35).
However, subsequent studies indicated that the function of H19 was
tissue and developmental stage specific. H19 is oncogenic in
thyroid cancer, hepatocellular and bladder carcinoma (36–38).
In gliomas, H19 contributes to tumorigenesis and
tumor progression via several mechanisms. Jiang et al
(39) identified that the increased
expression of H19 lncRNA promoted the invasion and angiogenesis of
glioblastoma cells in culture and increased the rate of growth of
xenograft tumors in mice. Jia et al (40) revealed that H19, as a molecular
sponge, promoted glioma-induced angiogenesis by downregulating
miRNA-29a. Chen et al (41)
demonstrated that H19 was upregulated in recurrent gliomas compared
with primary gliomas, suggesting that it was associated the
development of glioma. Shi et al (42) identified that H19 expression
associated with tumor grade, that H19 promoted glioma progression
via the H19-derived miR-675/CDH13 pathway, and that the suppression
of H19 expression inhibited the invasion of glioma cells.
Furthermore, H19 was expressed at high levels in the embryo and was
hypothesized to serve an important role in the maintenance of the
stemness in hematopoietic/embryonic stem cells (42–45). A
previous study has demonstrated that H19 is upregulated in
CD133+ glioblastoma cells compared with
CD133− tumor cells. The overexpression of H19 in CD133
tumor cells promoted tumor growth, indicating the importance of H19
in promoting stemness of glioblastoma cells (46). Li et al (47) reported that the knockdown of H19 was
able to significantly reduce the expression of stem cell markers. A
high expression of H19 was considered to transform normal
astrocytes into glioma stem cells, suggesting that H19 may have
role in contributing to the malignancy and stemness of glioblastoma
cells. In addition, aberrant expression of H19 was observed in
tumors from patients with temozolomide (TMZ) resistance, and
TMZ-resistant cell lines (48). The
silencing of H19 may regulate drug resistance genes, including
multidrug resistance protein 1, multidrug resistant associated
protein 1 and ATP-binding cassette subfamily G member 2; and may
promote apoptosis in sensitized tumor cells in drug-resistant
glioma (48). Furthermore, Jiang
et al (39) also demonstrated
that the stable overexpression of H19 in U87MG and U373MG cell
lines promoted tumor formation and induced tumor cell proliferation
and angiogenesis in an in vivo murine xenograft model.
HOTAIR, an lncRNA of >2,100-nucleotides in
length, is transcribed from the antisense of the HOXC gene locus in
chromosome 12 (60). It has been
demonstrated that the overexpression of HOTAIR is associated with
proliferation, invasion and chemoresistance of tumor cells.
Therefore, HOTAIR is considered to be a poor prognostic factor in
various types of cancer, including hepatocellular carcinoma,
gastric and lung cancer (61,62). HOTAIR has been investigated as an
important marker for molecular subtypes in glioma, which may serve
as a potential therapeutic target for classical and mesenchymal
gliomas (63). Zhou et al
(64) reported that the expression of
HOTAIR was associated with overall survival in patients with
glioblastoma. Additionally, cell cycle arrest and attenuation of
invasion in glioblastoma cells may be induced by HOTAIR depletion
and subsequent inhibition of Nemo-like kinase/β-catenin axis.
Similarly, Fang et al (65)
suggested that the inhibition of HOTAIR by superparamagnetic iron
oxide nanoparticles mediated siRNA transfection-induced programmed
cell death 4 expression, which suppressed the proliferation,
invasion and tumorigenicity of glioma stem cells. Recently,
accumulating evidence has suggested that the reciprocal association
between miRNA and lncRNA is actively involved in cancer
pathogenesis (66). Ke et al
(67) demonstrated that HOTAIR was
significantly upregulated in glioma tissues and cell lines compared
with normal controls. Furthermore, it was suggested that the
knockdown of HOTAIR may lead to the inhibition of FGF1 by
upregulating miR-326, which suppressed tumor growth in vitro
and in vivo. Yang et al (68) also confirmed that the survival time of
nude mice was extended in a short hairpin-HOTAIR group compared
with that of control groups. A recent study indicated that HOTAIR,
acting as an endogenous ‘sponge’, may bind with miR-141 to regulate
the epigenetic modification of the miRNA-induced repression of
spindle and kinetochore associated complex subunit 2 to promote the
proliferation and invasion of glioma cells (69). Additionally, Wang et al
(70) also demonstrated that
miR-148b-3p may inhibit glioma cell growth by directly
downregulating HOTAIR. These data suggest that the inhibition of
HOTAIR activity may potentially be used as a novel therapy for the
treatment of glioma.
CRNDE, which is transcribed from the strand opposite
to the adjacent iroquois homeobox 5 gene in chromosome 16, was
initially regarded as a pro-oncogenic lncRNA that is upregulated in
colorectal cancer (71). CRNDE serves
a vital role in the development of numerous organs including
breast, skin, and bronchial epithelium. Notably, an increased
expression of CRNDE has been identified in a variety of solid
tumors, including brain tumors (72).
Previous studies indicated that the expression of CRNDE was
markedly increased in primary and recurrent gliomas (73). Zhang et al (74) identified that CRNDE was significantly
overexpressed in glioma tissues, and that the expression level of
CRNDE was positively associated with the pathological grades of
glioma. Similarly, Zheng et al (75) demonstrated that CRNDE promoted
migration, invasion and proliferation, and inhibited apoptosis in
glioma cells through regulating the expression levels of the
miR-384/Piwi-like RNA-mediated gene silencing 4/signal transducer
and activator of transcription 3 axis. Consistent with these data,
Wang et al (76) demonstrated
that CRNDE was upregulated lncRNA in glioma compared with normal
tissues in their study, and that the overexpression of CRNDE
promoted proliferation and invasion in glioma through the
mechanistic target of rapamycin pathway in vitro and in
vivo. These results suggest that understanding the underlying
mechanisms whereby CRNDE functions in glioma may reveal a novel
therapeutic strategy for the treatment of glioma in future.
CASC2 has been identified as a tumor suppressor in
numerous types of solid tumors (77).
The role of CASC2 in glioma pathogenesis has been examined by
several studies. Wang et al (77) indicated that a low level of CASC2
expression was detected in gliomas. The findings of Wang et
al (78) suggested that CASC2
served as a tumor suppressor role via regulation of miR-21 in an
Ago2-dependent manner in gliomas. Furthermore, a study by Liao
et al (79) demonstrated that
the low expression of CASC2 was associated with malignant
characteristics and poor clinical prognosis in glioma. The
overexpression of CACS2 inhibits the proliferation of glioma cells
and amplifies TMZ-induced repression of cell proliferation through
the direct inhibition of miR-181a. However, whether CACS2 has the
same effect in vivo has not been reported in glioma.
Therefore, the role of CACS2 in a mouse model of glioma requires
additional investigation.
MEG3 is a maternal imprinting gene at the delta like
non-canonical notch ligand 1-MEG3 locus on chromosome 14q32.3 in
humans (80). A number of previous
studies demonstrated that MEG3 was expressed in a number of normal
tissues, with particularly marked expression in the brain, but
absent or low expression in multiple types of tumors, including
cervical carcinoma, breast adenocarcinoma, meningioma and glioma
(81,82). Wang et al (83) demonstrated that the expression of MEG3
was decreased in tumor tissues compared with adjacent non-tumor
tissues in gliomas. Furthermore, ectopic expression of MEG3
inhibited the growth of glioma cells by activation of the p53
signaling pathway. Liu et al (84) also suggested that MEG3 served an
important role in genotoxic stress-induced glioma cell death.
Similarly, Li et al (84)
indicated that a low level of MEG3 expression was observed in
glioma tissues. This low expression of MEG3 was due to DNA
methyltransferase 1 (DNMT1), which is mediated by hypermethylation
of the MEG3 promoter. Furthermore, the inhibition of DNMT1
repressed the growth and resulted in apoptosis of glioma cells in a
p53-dependent manner. Additionally, Zhang et al (85) also demonstrated that MEG3 markedly
reduced tumor volume and the expression of Ki-67 and proliferating
cell nuclear antigen in vivo.
With an improved understanding of their functions,
lncRNAs are becoming an important focus of study as a novel type of
cancer biomarker for diagnosis, treatment and prognostic
prediction. At present, certain lncRNAs have been examined for
their potential clinical applications. It has been demonstrated
that specific lncRNA profiles are associated with tumor subtypes,
mutational status and the survival time of patients with glioma,
based on the analysis of RNA-seq datasets from The Cancer Genome
Atlas (86). Gliomas with
oxalosuccinate decarboxylase (IDH1) mutations exhibited a unique
lncRNA gene expression signature that was different from that of
tumors exhibiting the wild-type IDH1 gene (87). As prognostic markers, the four lncRNAs
(AGAP2-AS1, TPT1-AS1, LINC01198 and MIR155HG) were suggested to
have prognostic value for patients with anaplastic gliomas
(88). Furthermore, lncRNAs have
demonstrated potential in applications for non-invasive detection
of cancer. Zhou et al (18)
identified that the level of H19 in the plasma of patients with
gastric cancer was significantly increased compared with healthy
controls. In addition, the plasma level of H19 was decreased
markedly in postoperative patients compared with that in
preoperative patients. The plasma level of H19 may be used as a
non-invasive method to evaluate glioma progression in the
future.
lncRNAs, due to their highly tissue-specific
expression in cancer phenotypes, are potential targets for cancer
therapy. Preclinical studies have demonstrated the therapeutic
efficacy of antisense oligonucleotides targeting cancer-associated
lncRNAs, including MALAT-1 and H19 (89,90). At
present, certain lncRNAs have been identified to be associated with
glioma therapy. Amit et al (91) revealed that a construct expressing the
diphtheria toxin A-fragment, under the control of H19 and
insulin-like growth factor 2 P4 promoters, demonstrated
anti-tumoral efficacy against glioblastoma in vitro and
in vivo. The BET bromodomain inhibitor, I-BET151 inhibited
the growth of glioblastoma cells in vitro and in vivo
by directly reducing HOTAIR expression (92). Similarly, Ke et al (67) also reported that HOTAIR knockdown
inhibited tumor growth. In addition, aberrant expression of growth
arrest specific transcript 5 (GAS5) lncRNA has also been associated
with chemoresistance in glioma. García-Claver et al
(93) revealed that the GAS5 lncRNA
was markedly upregulated following treatment with erlotinib (ERL)
in ERL-sensitive and -resistant glioma. The knockdown of GAS5
sensitized U87MG cells to ERL treatment. Similarly, in tumor
tissues and cell lines from patients with TMZ resistance, H19 was
significantly upregulated (48). The
silencing of H19 may decrease the half-maximal inhibitory
concentration values for TMZ, and increase the apoptotic rate of
glioma cells (48). In summary, these
results provide experimental basis for the use of lncRNAs as novel
therapeutic targets in gliomas. lncRNA-based therapeutics may
represent a novel direction for the treatment of glioma, although
studies concerning their safety, efficacy and more efficient
delivery systems are required.
LncRNAs may be regulators in the determination of
the development of particular organs, rather than simply
functioning as housekeeping genes. In contrast to other tissues,
the brain expresses high levels of numerous lncRNAs, which are
involved in neuro-development (94).
Their deregulation may cause neurological disorders and brain
tumors (95). As aforementioned,
lncRNAs function as oncogenes or tumor suppressors by interacting
with DNA, mRNA, ncRNA and proteins, and regulate the proliferation,
migration, invasion, apoptosis, angiogenesis and stemness of glioma
cells (Fig. 2). Recent studies of
lncRNAs have highlighted their vital roles in the pathogenesis and
progression of glioma. Furthermore, the sensitivity and reliability
of RNA-based molecular technologies and tools to detect and target
lncRNAs in glioma have improved (96). However, it should be noted that the
current knowledge base regarding the biological roles of lncRNAs in
glioma is primarily concerned with the identification and
quantification of the expression levels of different lncRNAs and
associated molecules in tumor and normal tissues. To date, the
in vivo functions of a large proportion of the identified
lncRNAs remain unknown. Therefore, a short-term goal would be to
investigate the molecular, cellular and physiological functions of
lncRNAs and their roles in pathogenesis of glioma, which will
provide a foundation for developing novel medical therapies that
target lncRNAs. High throughput technologies and massively parallel
sequencing tools, in combination with bioinformatics methods, would
assist in identifying lncRNA candidate targets whose dysregulation
serves a pivotal role in the pathogenesis and progression of
glioma. Genetically engineered mouse models would also be an
indispensable tool to elucidate the functions and mechanisms of
lncRNA genes in glioma in vivo. In summary, the
identification of lncRNAs has revealed an additional facet of
glioma tumorigenesis. Understanding the precise molecular
mechanisms whereby lncRNAs function is important to advance
lncRNA-based diagnosis, prognosis and therapeutic interventions
against glioma.
The authors would like to thank Dr. Yunli Zhou
(Neuroendocrine Unit, Massachusetts General Hospital, Harvard
Medical School, Boston, MA, USA) for reviewing of the manuscript.
The present research is supported by the Medical Research Fund for
Young Scholars of the Sichuan Medical Association, Sichuan, China
(grant no., Q16076) and the Natural Science Foundation of Southwest
Medical University (grant no., 2016XNYD217).
|
1
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schmitt AM and Chang HY: Long Noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thomas J, Ohtsuka M, Pichler M and Ling H:
MicroRNAs: Clinical relevance in colorectal cancer. Int J Mol Sci.
16:28063–28076. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee JS: The mutational landscape of
hepatocellular carcinoma. Clin Mol Hepatol. 21:220–229. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet 15 Spec No. 1:R17–R29. 2006. View Article : Google Scholar
|
|
8
|
Han D, Wang M, Ma N, Xu Y, Jiang Y and Gao
X: Long noncoding RNAs: Novel players in colorectal cancer. Cancer
Lett. 361:13–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang J, Song YX and Wang ZN: Non-coding
RNAs in gastric cancer. Gene. 560:1–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li R, Zhu H and Luo Y: Understanding the
functions of long non-coding RNAs through their higher-order
structures. Int J Mol Sci. 17:E7022016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J,
Zhang Y, Chen J, Shen H and Hu Z: A genetic variant in long
non-coding RNA HULC contributes to risk of HBV-related
hepatocellular carcinoma in a Chinese population. PLoS One.
7:e351452012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hashad D, Elbanna A, Ibrahim A and Khedr
G: Evaluation of the role of circulating long non-coding RNA H19 as
a promising novel biomarker in plasma of patients with gastric
cancer. J Clin Lab Anal. 30:1100–1105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou X, Yin C, Dang Y, Ye F and Zhang G:
Identification of the long non-coding RNA H19 in plasma as a novel
biomarker for diagnosis of gastric cancer. Sci Rep. 5:115162015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Du Z, Fei T, Verhaak RG, Su Z, Zhang Y,
Brown M, Chen Y and Liu XS: Integrative genomic analyses reveal
clinically relevant long noncoding RNAs in human cancer. Nat Struct
Mol Biol. 20:908–913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou M, Zhao H, Xu W, Bao S, Cheng L and
Sun J: Discovery and validation of immune-associated long
non-coding RNA biomarkers associated with clinically molecular
subtype and prognosis in diffuse large B cell lymphoma. Mol Cancer.
16:162017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian
J, Zhang X and Fang JY: A long non-coding RNA signature to improve
prognosis prediction of colorectal cancer. Oncotarget. 5:2230–2242.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McCleland ML, Mesh K, Lorenzana E, Chopra
VS, Segal E, Watanabe C, Haley B, Mayba O, Yaylaoglu M, Gnad F and
Firestein R: CCAT1 is an enhancer-templated RNA that predicts BET
sensitivity in colorectal cancer. J Clin Invest. 126:639–652. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun J, Chen X, Wang Z, Guo M, Shi H, Wang
X, Cheng L and Zhou M: A potential prognostic long non-coding RNA
signature to predict metastasis-free survival of breast cancer
patients. Sci Rep. 5:165532015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the united states in 2006–2010. Neuro Oncol. 15
Suppl 2:ii1–ii56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Devaux Y, Zangrando J, Schroen B, Creemers
EE, Pedrazzini T, Chang CP, Dorn GW II, Thum T and Heymans S:
Cardiolinc network: Long noncoding RNAs in cardiac development and
ageing. Nat Rev Cardiol. 12:415–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takahashi H CPWgtnpfRtBBRC. 2014.
|
|
27
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno
M, Yoshida M and Nakagawa S: Competition between a noncoding exon
and introns: Gomafu contains tandem UACUAAC repeats and associates
with splicing factor-1. Genes Cells. 16:479–490. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rintala-Maki ND and Sutherland LC:
Identification and characterisation of a novel antisense non-coding
RNA from the RBM5 gene locus. Gene. 445:7–16. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin D, Pestova TV, Hellen CU and Tiedge H:
Translational control by a small RNA: Dendritic BC1 RNA targets the
eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol.
28:3008–3019. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Raveh E, Matouk IJ, Gilon M and Hochberg
A: The H19 Long non-coding RNA in cancer initiation, progression
and metastasis-a proposed unifying theory. Mol Cancer. 14:1842015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hao Y, Crenshaw T, Moulton T, Newcomb E
and Tycko B: Tumour-suppressor activity of H19 RNA. Nature.
365:764–767. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu L, Yang J, Zhu X, Li D, Lv Z and Zhang
X: Long noncoding RNA H19 competitively binds miR-17-5p to regulate
YES1 expression in thyroid cancer. FEBS J. 283:2326–2339. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang
J, Dong N, He J, Sun Q, Lv G, et al: Long noncoding RNA H19
indicates a poor prognosis of colorectal cancer and promotes tumor
growth by recruiting and binding to eIF4A3. Oncotarget.
7:22159–22173. 2016.PubMed/NCBI
|
|
38
|
Matouk IJ, Raveh E, Abu-lail R, Mezan S,
Gilon M, Gershtain E, Birman T, Gallula J, Schneider T, Barkali M,
et al: Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys
Acta. 1843:1414–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang X, Yan Y, Hu M, Chen X, Wang Y, Dai
Y, Wu D, Wang Y, Zhuang Z and Xia H: Increased level of H19 long
noncoding RNA promotes invasion, angiogenesis, and stemness of
glioblastoma cells. J Neurosurg. 124:129–136. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jia P, Cai H, Liu X, Chen J, Ma J, Wang P,
Liu Y, Zheng J and Xue Y: Long non-coding RNA H19 regulates glioma
angiogenesis and the biological behavior of glioma-associated
endothelial cells by inhibiting microRNA-29a. Cancer Lett.
381:359–369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Y, Wu JJ, Lin XB, Bao Y, Chen ZH,
Zhang CR, Cai Z, Zhou JY, Ding MH, Wu XJ, et al: Differential
lncRNA expression profiles in recurrent gliomas compared with
primary gliomas identified by microarray analysis. Int J Clin Exp
Med. 8:5033–5043. 2015.PubMed/NCBI
|
|
42
|
Shi Y, Wang Y, Luan W, Wang P, Tao T,
Zhang J, Qian J, Liu N and You Y: Long non-coding RNA H19 promotes
glioma cell invasion by deriving miR-675. PLoS One. 9:e862952014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Poirier F, Chan C, Timmons P, Robertson
EJ, Evans MJ and Rigby PW: The murine H19 gene is activated during
embryonic stem cell differentiation in vitro and at the time of
implantation in the developing embryo. Development. 113:1105–1114.
1991.PubMed/NCBI
|
|
44
|
Venkatraman A, He XC, Thorvaldsen JL,
Sugimura R, Perry JM, Tao F, Zhao M, Christenson MK, Sanchez R, Yu
JY, et al: Maternal imprinting at the H19-Igf2 locus maintains
adult haematopoietic stem cell quiescence. Nature. 500:345–349.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yin Y, Wang H, Liu K, Wang F, Ye X, Liu M,
Xiang R, Liu N and Liu L: Knockdown of H19 enhances differentiation
capacity to epidermis of parthenogenetic embryonic stem cells. Curr
Mol Med. 14:737–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jiang X, Yan Y, Hu M, Chen X, Wang Y, Dai
Y, Wu D, Wang Y, Zhuang Z and Xia H: Increased level of H19 long
noncoding RNA promotes invasion, angiogenesis, and stemness of
glioblastoma cells. J Neurosurg. 2016:129–136. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li W, Jiang P, Sun X, Xu S, Ma X and Zhan
R: Suppressing H19 modulates tumorigenicity and stemness in U251
and U87MG glioma cells. Cell Mol Neurobiol. 36:1219–1227. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jiang P, Wang P, Sun X, Yuan Z, Zhan R, Ma
X and Li W: Knockdown of long noncoding RNA H19 sensitizes human
glioma cells to temozolomide therapy. Onco Targets Ther.
9:3501–3509. 2016.PubMed/NCBI
|
|
49
|
Wang J, Pan Y, Wu J, Zhang C, Huang Y,
Zhao R, Cheng G, Liu J, Qin C, Shao P, et al: The association
between abnormal long Noncoding RNA MALAT-1 expression and cancer
lymph node metastasis: A meta-analysis. Biomed Res Int.
2016:18234822016.PubMed/NCBI
|
|
50
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhou Y, Xu X, Lv H, Wen Q, Li J, Tan L, Li
J and Sheng X: The Long Noncoding RNA MALAT-1 is highly expressed
in ovarian cancer and induces cell growth and migration. PLoS One.
11:e01552502016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Han T, Jiao F, Hu H, Yuan C, Wang L, Jin
ZL, Song WF and Wang LW: EZH2 promotes cell migration and invasion
but not alters cell proliferation by suppressing E-cadherin, partly
through association with MALAT-1 in pancreatic cancer. Oncotarget.
7:11194–11207. 2016.PubMed/NCBI
|
|
53
|
Tano K, Mizuno R, Okada T, Rakwal R,
Shibato J, Masuo Y, Ijiri K and Akimitsu N: MALAT-1 enhances cell
motility of lung adenocarcinoma cells by influencing the expression
of motility-related genes. FEBS Lett. 584:4575–4580. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu S, Song L, Zeng S and Zhang L:
MALAT1-miR-124-RBG2 axis is involved in growth and invasion of
HR-HPV-positive cervical cancer cells. Tumour Biol. 37:633–640.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P
and Wu JW: Long noncoding RNA MALAT1 associates with the malignant
status and poor prognosis in glioma. Tumour Biol. 36:3355–3359.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xiang J, Guo S, Jiang S, Xu Y, Li J, Li L
and Xiang J: Silencing of Long Non-coding RNA MALAT1 promotes
apoptosis of glioma cells. J Korean Med Sci. 31:688–694. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Vassallo I, Zinn P, Lai M, Rajakannu P,
Hamou MF and Hegi ME: WIF1 re-expression in glioblastoma inhibits
migration through attenuation of non-canonical WNT signaling by
downregulating the lncRNA MALAT1. Oncogene. 35:12–21. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ma J, Wang P, Yao Y, Liu Y, Li Z, Liu X,
Li Z, Zhao X, Xi Z, Teng H, et al: Knockdown of long non-coding RNA
MALAT1 increases the blood-tumor barrier permeability by
up-regulating miR-140. Biochim Biophys Acta. 1859:324–338. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X,
Sun T, Xie X, Zhou Y and Du Z: Tumor-suppressive function of long
noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and
inactivation of ERK/MAPK signaling. Cell Death Dis. 7:e21232016.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cai B, Song XQ, Cai JP and Zhang S:
HOTAIR: A cancer-related long non-coding RNA. Neoplasma.
61:379–391. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xue X, Yang YA, Zhang A, Fong KW, Kim J,
Song B, Li S, Zhao JC and Yu J: LncRNA HOTAIR enhances ER signaling
and confers tamoxifen resistance in breast cancer. Oncogene.
35:2746–55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang
H, Liang WC, Wang SS, Ko CH, Waye MM, et al: Hotair mediates
hepatocarcinogenesis through suppressing miRNA-218 expression and
activating P14 and P16 signaling. J Hepatol. 63:886–895. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang JX, Han L, Bao ZS, Wang YY, Chen LY,
Yan W, Yu SZ, Pu PY, Liu N, You YP, et al: HOTAIR, a cell
cycle-associated long noncoding RNA and a strong predictor of
survival, is preferentially expressed in classical and mesenchymal
glioma. Neuro Oncol. 15:1595–1603. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhou X, Ren Y, Zhang J, Zhang C, Zhang K,
Han L, Kong L, Wei J, Chen L, Yang J, et al: HOTAIR is a
therapeutic target in glioblastoma. Oncotarget. 6:8353–8365.
2015.PubMed/NCBI
|
|
65
|
Fang K, Liu P, Dong S, Guo Y, Cui X, Zhu
X, Li X, Jiang L, Liu T and Wu Y: Magnetofection based on
superparamagnetic iron oxide nanoparticle-mediated low lncRNA
HOTAIR expression decreases the proliferation and invasion of
glioma stem cells. Int J Oncol. 49:509–518. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma
J, Li Z, Liu XB, Li ZQ, Wang ZH and Xue YX: Knockdown of long
non-coding RNA HOTAIR inhibits malignant biological behaviors of
human glioma cells via modulation of miR-326. Oncotarget.
6:21934–21949. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yang B, Wei ZY, Wang BQ, Yang HC, Wang JY
and Bu XY: Down-regulation of the long noncoding RNA-HOX transcript
antisense intergenic RNA inhibits the occurrence and progression of
glioma. J Cell Biochem. 119:2278–2287. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bian EB, Ma CC, He XJ, Wang C, Zong G,
Wang HL and Zhao B: Epigenetic modification of miR-141 regulates
SKA2 by an endogenous ‘sponge’ HOTAIR in glioma. Oncotarget.
7:30610–30625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wang G, Li Z, Tian N, Han L, Fu Y, Guo Z
and Tian Y: miR-148b-3p inhibits malignant biological behaviors of
human glioma cells induced by high HOTAIR expression. Oncol Lett.
12:879–886. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Graham LD, Pedersen SK, Brown GS, Ho T,
Kassir Z, Moynihan AT, Vizgoft EK, Dunne R, Pimlott L, Young GP, et
al: Colorectal neoplasia differentially expressed (CRNDE), a novel
gene with elevated expression in colorectal adenomas and
adenocarcinomas. Genes Cancer. 2:829–840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ellis BC, Molloy PL and Graham LD: CRNDE:
A long non-coding RNA involved in cancer, neurobiology, and
development. Front Genet. 3:2702012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Kiang K, Zhang XQ and Leung GK: Long
non-coding RNAs: The key players in glioma pathogenesis. Cancers
(Basel). 7:1406–1424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C
and Liu Y: CRNDE promotes malignant progression of glioma by
attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 24:1199–1215.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang Y, Wang Y, Li J, Zhang Y, Yin H and
Han B: CRNDE, a long-noncoding RNA, promotes glioma cell growth and
invasion through mTOR signaling. Cancer Lett. 367:122–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Palmieri G, Paliogiannis P, Sini MC, Manca
A, Palomba G, Doneddu V, Tanda F, Pascale MR and Cossu A: Long
non-coding RNA CASC2 in human cancer. Crit Rev Oncol Hematol.
111:31–38. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J
and Xue YX: Long non-coding RNA CASC2 suppresses malignancy in
human gliomas by miR-21. Cell Signal. 27:275–282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Liao Y, Shen L, Zhao H, Liu Q, Fu J, Guo
Y, Peng R and Cheng L: LncRNA CASC2 interacts with miR-181a to
modulate glioma growth and resistance to TMZ through PTEN pathway.
J Cell Biochem. 118:1889–1899. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang X, Zhou Y, Mehta KR, Danila DC,
Scolavino S, Johnson SR and Klibanski A: A pituitary-derived MEG3
isoform functions as a growth suppressor in tumor cells. J Clin
Endocrinol Metab. 88:5119–5126. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Balci T, Yilmaz Susluer S, Kayabasi C,
Ozmen Yelken B, Biray Avci C and Gunduz C: Analysis of dysregulated
long non-coding RNA expressions in glioblastoma cells. Gene.
590:120–122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wang P, Ren Z and Sun P: Overexpression of
the long non-coding RNA MEG3 impairs in vitro glioma cell
proliferation. J Cell Biochem. 113:1868–1874. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Liu Q, Sun S, Yu W, Jiang J, Zhuo F, Qiu
G, Xu S and Jiang X: Altered expression of long non-coding RNAs
during genotoxic stress-induced cell death in human glioma cells. J
Neurooncol. 122:283–292. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang L, Liang X and Li Y: Long non-coding
RNA MEG3 inhibits cell growth of gliomas by targeting miR-93 and
inactivating PI3K/AKT pathway. Oncol Rep. 38:2408–2416. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Reon BJ, Anaya J, Zhang Y, Mandell J,
Purow B, Abounader R and Dutta A: Expression of lncRNAs in
low-grade gliomas and glioblastoma multiforme: An In silico
analysis. PLoS Med. 13:e10021922016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang XQ, Kiang KM, Wang YC, Pu JK, Ho A,
Cheng SY, Lee D, Zhang PD, Chen JJ, Lui WM, et al: IDH1
mutation-associated long non-coding RNA expression profile changes
in glioma. J Neurooncol. 125:253–263. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang W, Yang F, Zhang L, Chen J, Zhao Z,
Wang H, Wu F, Liang T, Yan X, Li J, et al: LncRNA profile study
reveals four-lncRNA signature associated with the prognosis of
patients with anaplastic gliomas. Oncotarget. 7:77225–77236.
2016.PubMed/NCBI
|
|
89
|
Evans JR, Feng FY and Chinnaiyan AM: The
bright side of dark matter: lncRNAs in cancer. J Clin Invest.
126:2775–2782. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Arun G, Diermeier S, Akerman M, Chang KC,
Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, et
al: Differentiation of mammary tumors and reduction in metastasis
upon Malat1 lncRNA loss. Genes Dev. 30:34–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Amit D, Matouk IJ, Lavon I, Birman T,
Galula J, Abu-Lail R, Schneider T, Siegal T, Hochberg A and Fellig
Y: Transcriptional targeting of glioblastoma by diphtheria toxin-A
driven by both H19 and IGF2-P4 promoters. Int J Clin Exp Med.
5:124–135. 2012.PubMed/NCBI
|
|
92
|
Pastori C, Kapranov P, Penas C, Peschansky
V, Volmar CH, Sarkaria JN, Bregy A, Komotar R, St Laurent G, Ayad
NG and Wahlestedt C: The Bromodomain protein BRD4 controls HOTAIR,
a long noncoding RNA essential for glioblastoma proliferation. Proc
Natl Acad Sci USA. 112:pp. 8326–8331. 2015; View Article : Google Scholar : PubMed/NCBI
|
|
93
|
García-Claver A, Lorente M, Mur P,
Campos-Martín Y, Mollejo M, Velasco G and Meléndez B: Gene
expression changes associated with erlotinib response in glioma
cell lines. Eur J Cancer. 49:1641–1653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Shi C, Zhang L and Qin C: Long non-coding
RNAs in brain development, synaptic biology, and Alzheimer's
disease. Brain Res Bull. 132:160–169. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Subhramanyam CS and Hu Q: Non-coding rna
in brain development and disorder. Curr Med Chem. 24:1983–1997.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Hassan A, Mosley J, Singh S and Zinn PO: A
comprehensive review of genomics and noncoding RNA in gliomas. Top
Magn Reson Imaging. 26:3–14. 2017.PubMed/NCBI
|
|
97
|
Hu L, Lv QL, Chen SH, Sun B, Qu Q, Cheng
L, Guo Y, Zhou HH and Fan L: Up-Regulation of long non-coding RNA
AB073614 predicts a poor prognosis in patients with glioma. Int J
Environ Res Public Health. 13:4332016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zhu Y, Zhang X, Qi L, Cai Y, Yang P, Xuan
G and Jiang Y: HULC long noncoding RNA silencing suppresses
angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling
pathway in human gliomas. Oncotarget. 7:14429–14440.
2016.PubMed/NCBI
|
|
99
|
He C, Jiang B, Ma J and Li Q: Aberrant
NEAT1 expression is associated with clinical outcome in high grade
glioma patients. APMIS. 124:169–74. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhen L, Yun-Hui L, Hong-Yu D, Jun M and
Yi-Long Y: Long noncoding RNA NEAT1 promotes glioma pathogenesis by
regulating miR-449b-5p/c-Met axis. Tumour Biol. 37:673–683. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Guo H, Wu L, Yang Q, Ye M and Zhu X:
Functional linc-POU3F3 is overexpressed and contributes to
tumorigenesis in glioma. Gene. 554:114–119. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Liu H, Lv Z and Guo E: Knockdown of long
noncoding RNA SPRY4-IT1 suppresses glioma cell proliferation,
metastasis and epithelial-mesenchymal transition. Int J Clin Exp
Pathol. 8:9140–9146. 2015.PubMed/NCBI
|
|
103
|
Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J,
Chen L, Xi Z, Teng H, Wang Z, et al: Knockdown of long non-coding
RNA XIST exerts tumor-suppressive functions in human glioblastoma
stem cells by up-regulating miR-152. Cancer Lett. 359:75–86. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Yao J, Zhou B, Zhang J, Geng P, Liu K, Zhu
Y and Zhu W: A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated
by DNMT1 and inhibits migration of glioma cells. Tumour Biol.
35:7935–7944. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Li J, Bian EB, He XJ, Ma CC, Zong G, Wang
HL and Zhao B: Epigenetic repression of long non-coding RNA MEG3
mediated by DNMT1 represses the p53 pathway in gliomas. Int J
Oncol. 48:723–733. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Qin X, Yao J, Geng P, Fu X, Xue J and
Zhang Z: LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma.
Int J Clin Exp Pathol. 7:3065–3072. 2014.PubMed/NCBI
|