Introduction
Cancer is a genetic disease characterized by genomic
abnormalities that alter the transcriptome and influence the
pathways that control proliferation and survival (1). The application of next-generation
sequencing technology and single-cell sequencing in oncology has
provided evidence that cellular heterogeneity is common in cancer
(2). In the majority of tumor
studies, key information may be disregarded owing to the
tissue-centric nature of research. Malignant solid tumor tissues
consist mainly of tumor cells, but also contain tumor-associated
stromal, immune and vascular cells. Although non-tumor cells
constitute a relatively small proportion of the cancer tissue,
their role as potent tumor promoters has been previously indicated
(3). A previous study, using a
network approach to identify the functional gene modules in cancer
cells, verified the presence of immune, stromal and vascular gene
modules in cancer tissues (4).
The majority of genomic and transcriptomic studies
into cancer do not explicitly consider genetic heterogeneity, and
the generated inferences usually refer to mixed cell populations
(5). However, the experimental
isolation of single cells from tissues is expensive and may affect
cell physiology. Additionally, the single-cell sequencing of a
large cohort is unrealistic. An efficient solution to this
limitation may be the de-convolution of genomic data from
heterogeneous samples. Publicly available transcriptome databases
can provide resources that allow for this type of analysis. To
date, only one method, referred to as ‘Estimation of STromal and
Immune cells in MAlignant Tumors using Expression data’ (ESTIMATE)
has been described that can be used to score the stromal and immune
fraction in transcriptomic data of cancer tissue (5,6). However,
to the best of our knowledge, the association of the proportion of
immune and stromal fraction with patient survival has not been
thoroughly investigated.
Several studies have examined the
microenvironment-associated transcriptional tumor profile using
transcriptomic data. Calon et al (7) identified transforming growth factor-β
(TGF-β) response signatures in tumor-associated stromal cells that
could predict disease relapse in colorectal cancer (CRC). Cheng
et al (8) used principle
component analysis and clustering methods to identify a signature
of stromal activation that was associated with late recurrence in
breast cancer. Teschendorff et al (9) described an immune response gene
expression module associated with a good prognostic subtype in
estrogen receptor negative breast cancer. Finak et al
(10) used laser capture
microdissection (LCM) to compare the gene expression profiles of
tumor stroma from primary breast tumors and derived signatures that
were strongly associated with the clinical outcome by clustering.
Isella et al (11) used Gene
Set Enrichment Analysis (GSEA) and examined the gene signatures of
subtypes for expression in stromal cell subpopulations vs. CRC
cells. Wu et al (12)
identified a stromal gene super-module associated with gastric
cancer patient survival using gene co-expression network analysis.
Furthermore, extensive experimental research has indicated the role
served by stromal and immune cells in breast cancer (8,10,13), CRC (7,11,14), lymphoma (15) and drug resistance (16,17).
Transcriptome-based subtyping of cancer identifies
different subtypes by clustering; however, non-tumor components are
usually ignored (18). The ESTIMATE
algorithm scores stromal and immune cells that form the major
non-tumor components of tumor samples. In the present study, the
scoring of stromal and immune cells in healthy and cancerous
tissues, as well as in disease prognosis and drug resistance was
investigated. The scores were associated with the
clinicopathological characteristics of various cancer types and
chemotherapeutic drug resistance. The results of the present study
indicated that ESTIMATE could be used as a metric for patient
prognosis assessment.
Materials and methods
Microarray datasets of healthy and
disease tissue
The normal tissue dataset GSE45878 and cancer tissue
dataset GSE2109 were obtained from the Gene Expression Omnibus
(GEO) database (www.ncbi.nlm.nih.gov/geo/). A validation RNA-Seq
dataset E-MTAB-2836 from 32 different normal tissues was downloaded
from EBI ArrayExpress database (www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2836/)
(19).
ESTIMATE algorithm
Stromal and immune scores were calculated by the
ESTIMATE package in R (version 2.15.3) (20). ESTIMATE algorithm exploits the unique
properties of the transcriptional profiles of cancer samples to
infer tumor cellularity and identify the infiltrating normal cells
(6). Five rounds of gene filtering
identified two distinct gene signatures: i) A ‘stromal signature’
that indicates the stroma, and ii) an ‘immune signature’ that
represents the infiltration of immune cells in tumor tissue.
ESTIMATE outputs stromal, immune and ESTIMATE scores by performing
single-sample GSEA. For a given sample, gene expression values were
rank-normalized and rank-ordered. The empirical cumulative
distribution functions of the signature genes and the remaining
genes were calculated. A value of statistical significance was
calculated by integrating the difference between the empirical
cumulative distribution function, which is similar to the one used
in GSEA, but based on absolute expression rather than differential
expression (6).
Survival analysis
The breast cancer (GSE31448), CRC (GSE17538,
GSE41258, GSE39396), Ewing's sarcoma (GSE17679), glioma (GSE16011),
hepatocellular carcinoma (GSE20140), leukemia (GSE12417), lung
cancer (GSE3141), lymphoma (GSE10846), melanoma (GSE65904) and
ovarian cancer (GSE32062) datasets, and the respective clinical
information were obtained from the GEO repository.
For metastasis and relapse analysis, the sarcoma
(GSE21050), breast cancer (GSE1456), hepatocellular carcinoma
(GSE10140), gastric cancer (GSE26253) and prostate cancer
(GSE46691) datasets were obtained from the GEO database. The Cancer
Genome Atlas (TCGA) expression dataset was obtained from Firebrowse
at Broad Institute of the Massachusetts Institute of Technology
& Harvard (firebrowse.org/).
Statistical analysis
The ESTIMATE scores for each dataset were calculated
and patients were divided into two equal groups of high or low
ESTIMATE score by median split. The ESTIMATE scores were normalized
prior to Cox proportional hazards multivariate analysis. Overall
survival time curves were plotted using the Kaplan-Meier method.
Distributions of overall survival were compared using the log-rank
test. Metastasis-free survival curves were plotted similarly for
the samples that metastasis information was available. Furthermore,
TCGA datasets' scores were associated with clinical information
using Cox proportional hazards multivariate analysis. All analyses
were conducted using SPSS software (version 17.0; SPSS, Inc.,
Chicago, IL, USA). The concordance index was used to indicate the
probability that a patient with decreased survival time is
associated with a high value of a predictor. It was estimated using
the rms R package (21). ESTIMATE
score differences between two groups were assessed using unpaired
two-tailed t-tests in Microsoft Excel. P<0.05 was considered to
indicate a statistically significant difference. For multiple
comparisons, Bonferroni corrections were applied following analysis
of variance, and P<0.05/number of tests were used as
significance threshold. The correlation between lymphoblastoid cell
line immune score and 5-FU treatment response was calculation by
Pearson's correlation analysis in SPSS.
Results
Stromal and immune scores in healthy
and disease tissues
To investigate the difference in ESTIMATE scores
between healthy and malignant tissues, two public microarray
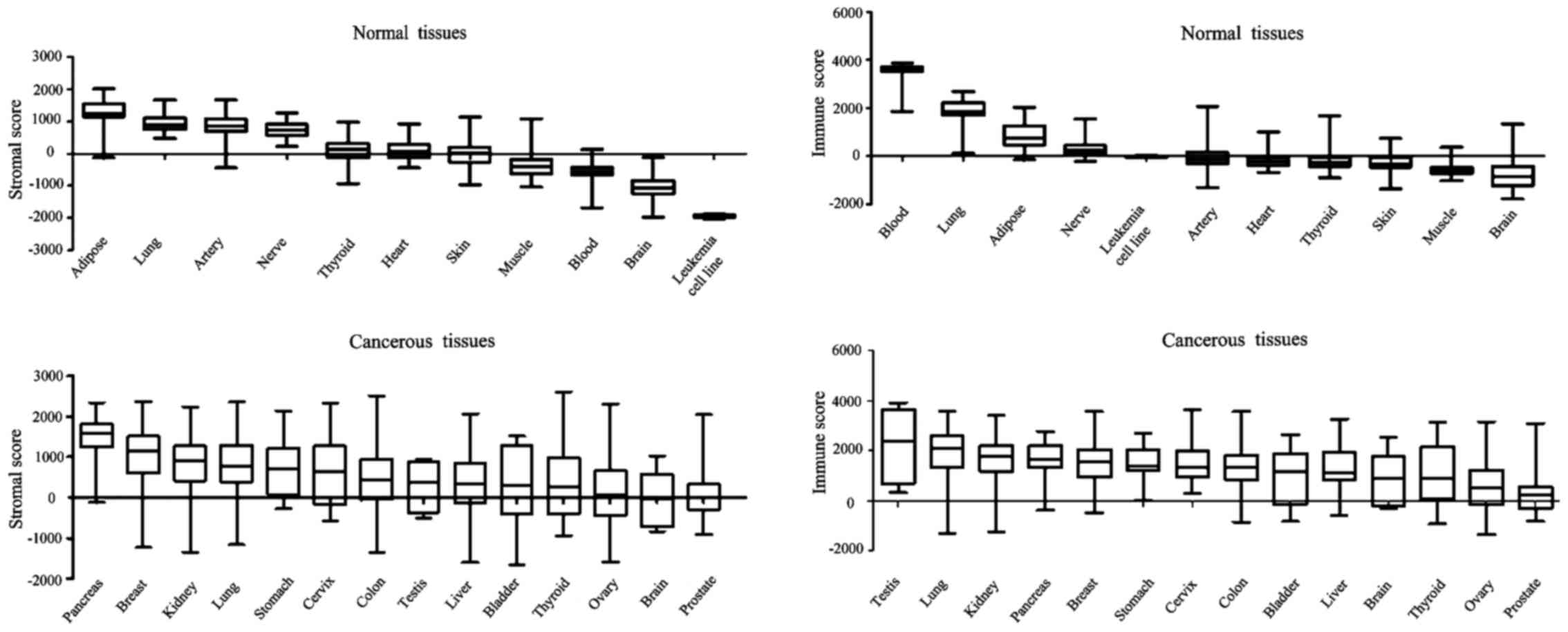
datasets, GSE45878 and GSE2109, were analyzed (Fig. 1). Among normal tissues, adipose had
the highest stromal score, whereas brain had the lowest; lung had
the highest immune score whereas brain had the lowest. The present
findings are consistent with the results from an RNA-sequencing
dataset (E-MTAB-2836). Regarding malignant tissues, almost all of
them presented a positive average ESTIMATE score: Pancreas had the
highest stromal score and prostate the lowest, whilst testis had
the highest immune score and prostate the lowest. Notably, normal
pancreas had a low stromal score and normal testis had a low immune
score (data not shown), whereas cancerous pancreas had a high
stromal score and cancerous testis had a high immune score.
Leukemia presented an extremely narrow range of scores, indicating
the robust performance of the ESTIMATE algorithm.
Stromal and immune scores as a
potential prognostic marker for multiple types of cancer
We subsequently hypothesized that as the two scores
represent common malignancy features, they may be used as potential
markers for cancer prognosis. The prognostic efficiency of stromal
and immune scores in breast cancer, CRC, Ewing's sarcoma, glioma,
hepatocellular carcinoma, leukemia, lymphoma, lung cancer,
melanoma, ovarian cancer, prostate cancer and sarcoma was
investigated. Survival curves were plotted following patient
categorization in groups of high and low ESTIMATE scores. Notably,
the two scores were indicative of patient survival in multiple
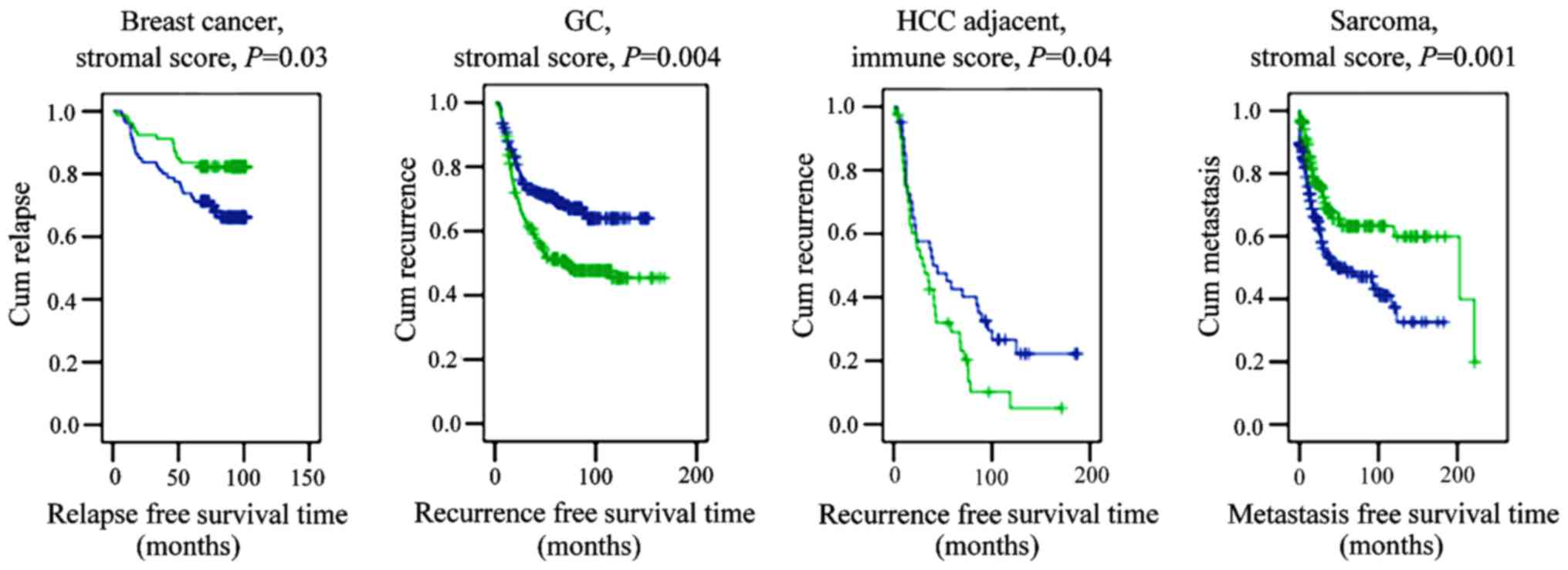
types of cancer (Fig. 2).
Indicatively, the immune score separated patients with long and
short survival in ovarian cancer and melanoma. Additionally, the
stromal score signified the survival time of patients with CRC and
glioma. Furthemore, the scores indicated survival rates within
specific subtypes of cancer. For instance, the stromal score
indicated the survival rate of patients with stage 2 American Joint
Committee on Cancer (AJCC) CRC (22),
moderately differentiated CRC, oligodendroglioma, glioblastoma and
served a prognostic role in patients with rituximab,
cyclophosphamide, doxorubicin, vincristine and prednisone-treated
lymphoma. The immune score indicated the survival time of patients
with basal breast cancer.
 | Figure 2.Stromal and immune scores are
predictive of patient survival in several types of cancer.
Kaplan-Meier curves of patients with breast cancer, CRC, glioma,
lymphoma, melanoma and ovarian cancer. P-values were generated
using the log-rank test. Blue lines indicate low scores; green
lines indicate high scores. AJCC, American Joint Committee on
Cancer; CRC, colorectal cancer; R-CHOP, rituximab,
cyclophosphamide, hydroxydaunorubicin, oncovin and prednisone; Cum,
cumulative. |
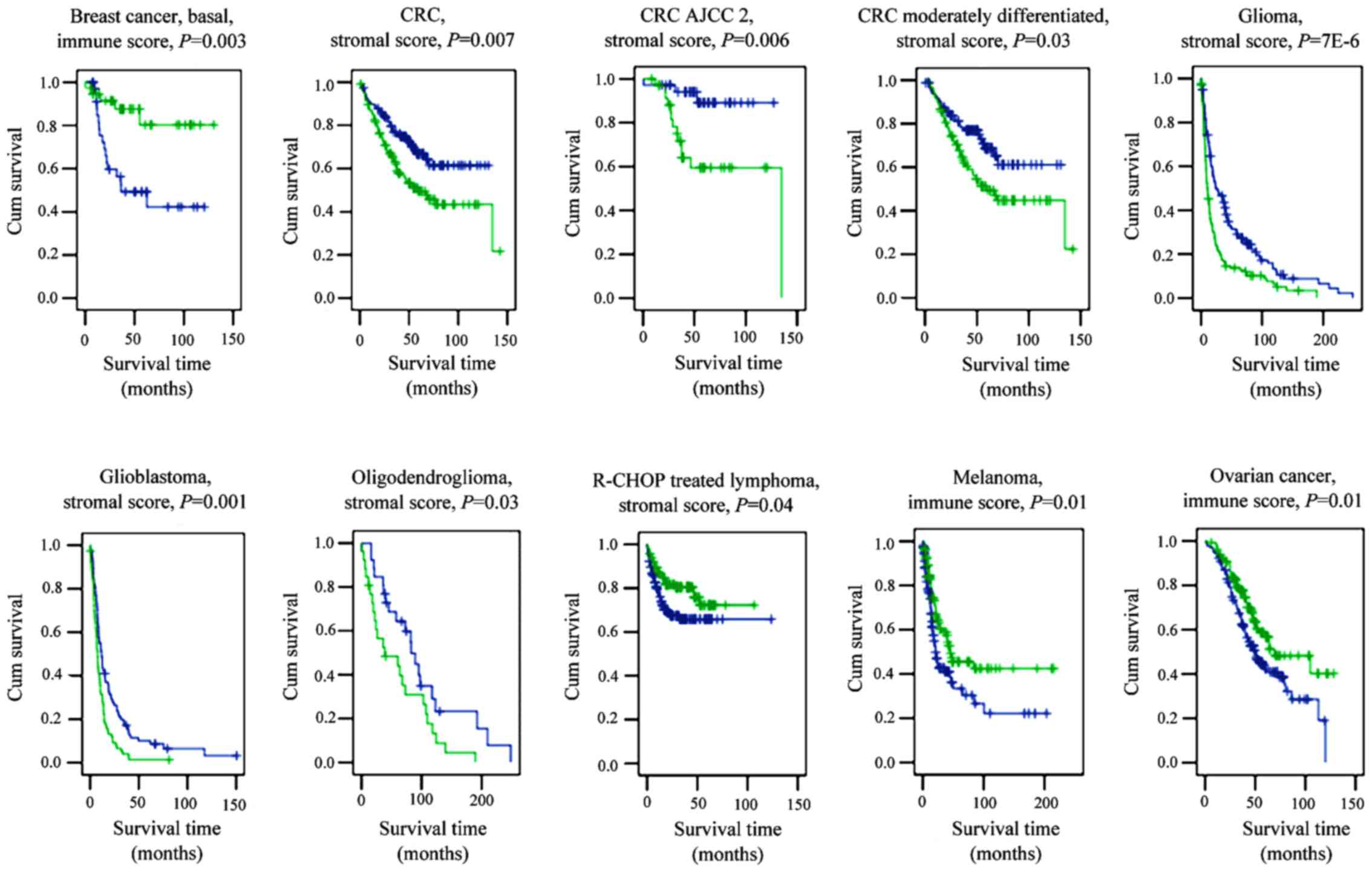
The stromal score separated patients with early
relapse from those with late relapse in basal breast cancer, and
differentiated between patients with early metastasis and late
metastasis in sarcoma (Fig. 3).
Notably, the immune score of hepatocellular carcinoma-adjacent
hepatitis/cirrhotic liver tissue was found to be indicative of
disease recurrence.
Certain GEO and TCGA datasets that contained
additional clinical information were further analyzed by
multivariate analysis. The immune score indicated the survival rate
of patients with breast cancer, melanoma and ovarian cancer
following adjustment for clinical parameters (Table I). Additionally, the stromal score
indicated the survival rate of patients with CRC and glioma, and
predicted cancer recurrence and metastasis in patients with sarcoma
following adjustment for clinical parameters (Table I). Furthermore, the clinical
implication of the immune score in breast cancer and melanoma was
also validated in the TCGA dataset.
 | Table I.Multivariate Cox regression of overall
survival and metastasis data in several types of cancer. |
Table I.
Multivariate Cox regression of overall
survival and metastasis data in several types of cancer.
| Data source | Disease | Variable | P-value | Estimated hazard
ratio |
|---|
| Gene Expression
Omnibus | Colorectal cancer
survival | Stromal score | 0.008 | 1.487 |
|
| Breast cancer
survival | Immune score | 0.001 | 0.544 |
|
| Gastric cancer
survival | Stromal score | 0.002 | 1.314 |
|
| Glioma
survival | Stromal score | 0.009 | 1.437 |
|
| Melanoma
survival | Immune score | 0.000 | 0.541 |
|
| Ovarian cancer
survival | Stromal | 0.033 | 1.327 |
|
|
| Immune score | 0.001 | 0.649 |
|
| Sarcoma
metastasis | Stromal score | 0.002 | 0.702 |
| The Cancer Genome
Atlas | Breast cancer
survival | Stromal score | 0.040 | 1.278 |
|
|
| Immune score | 0.046 | 0.744 |
|
| Cervical squamous
cell carcinoma survival | Stromal score | 0.026 | 1.456 |
|
|
| Immune score | 0.008 | 0.611 |
|
| Skin cutaneous
melanoma survival | Immune score | 0.002 | 0.685 |
Stromal and immune scores predict CRC
progression
The significance of the cancer microenvironment in
tumor progression has been repeatedly indicated. Stromal and immune
cells are major non-tumor components of cancer. Even though the
stromal score is predictive of survival in patients with AJCC stage
2 CRC, the immune score indicated a progression from polyp to CRC.
The average immune scores in normal colon mucosa, polyp, primary
CRC and metastatic CRC were calculated as 1,203, 488, 887 and 500,
respectively. However, no statistically significant difference
between the normal colon mucosa and polyp tissue was observed. The
immune score was significantly lower in primary and metastatic CRC
than in normal colon or polyp tissue. An association between the
downregulation of immune system-associated genes and metastasis in
CRC has been previously reported (23). The immune score is significantly lower
in p53 mutant patients (P=0.02) in the GSE41258 dataset. It has
been demonstrated that p53 regulates immunological activities
(24), indicating that p53 has a
possible regulatory role in CRC progression.
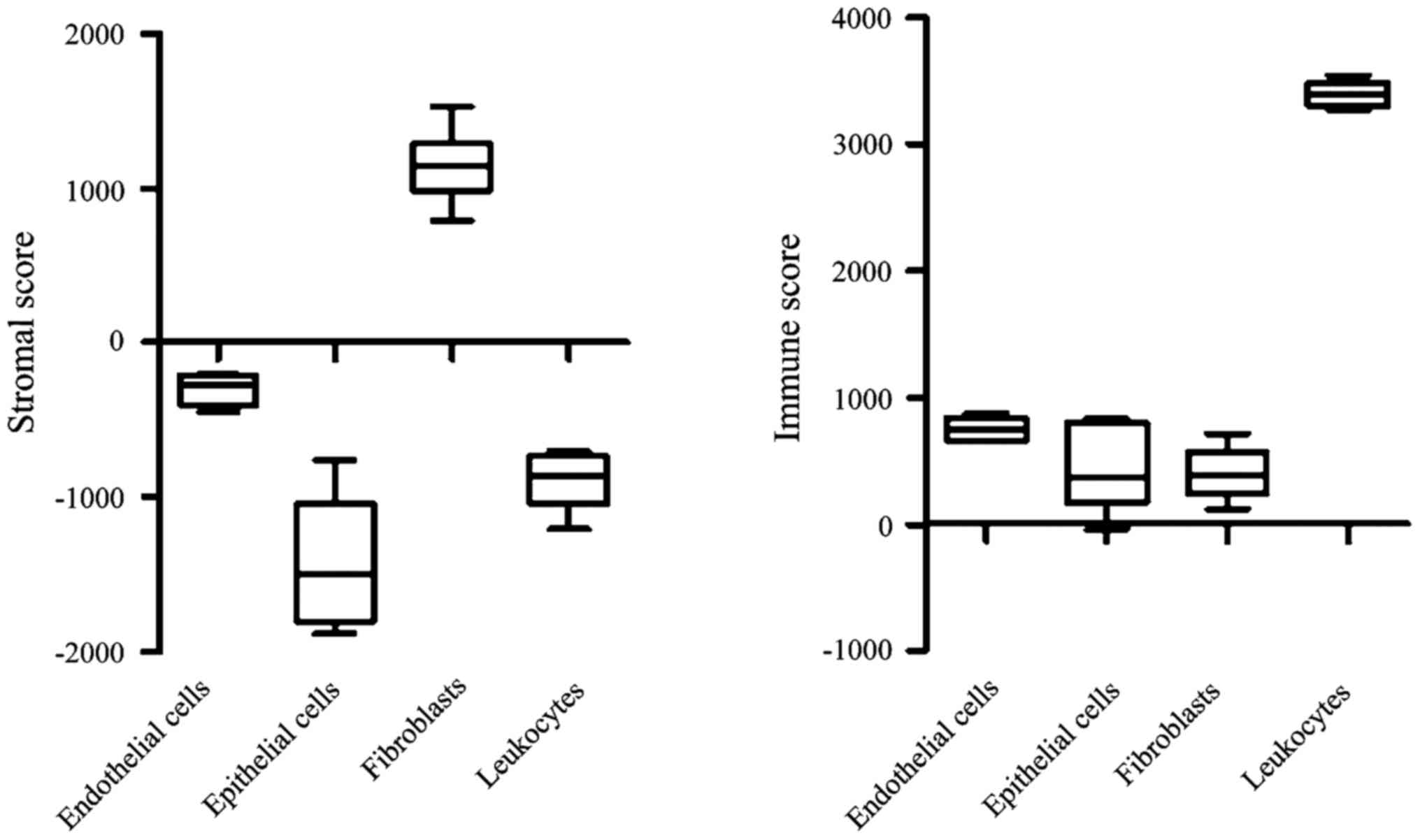
Gene expression analysis of specific cellular
populations isolated from CRC patients revealed that fibroblasts
and leukocytes have the highest stromal and immune scores among
endothelial cells, epithelial cells, fibroblasts and leukocytes
(Fig. 4). These results indicate the
robustness of the ESTIMATE algorithm.
Implication of drug resistance by
immune score
The anticancer activity of fluorouracil (5-FU)
involves the restoration of T-cell immunity (25). The GSE11582 dataset includes the
response of lymphoblastoid cell lines treated with a range of 5-FU
concentrations. Pearson's correlation analysis revealed that the
immune score was positively correlated with 5-FU treatment response
(R=0.2, P=0.008).
Discussion
To the best of our knowledge, this is the first
study in which ESTIMATE scoring was used to differentiate between
tissues. Stromal and immune scores were associated with the
clinical outcome of the patient and chemotherapy drug resistance.
The prognostic value of the two scores was validated using multiple
microarray platforms. Either the stromal or the immune score was
associated with patient survival, relapse and metastasis in
multiple types of cancer. Furthermore, the two scores were
associated with chemotherapeutic drug response. The present study
has indicated a microenvironment view of tissue-based tumor
transcriptomic data and highlighted the contribution of stromal and
immune cells in carcinogenesis. However, the precise molecular
mechanism underlying this phenomenon should be investigated in
future studies.
Tissues are composed of a mixture of cell types.
Currently, tissue-based transcriptomic data do not reflect the
information from multiple cell types. Even though a number of
technological approaches, including flow cytometry and LCM, have
been developed, their application in cancer research is limited
owing to the expensive cost. Therefore, it is crucial to study the
information hidden in available public datasets. Functional modules
in cancer transcriptomic datasets have been previously identified
by gene co-expression network analysis (4). In the present study, two major
components of the cancer microenvironment, stromal and immune
cells, were investigated, and it was demonstrated that the ESTIMATE
scores for these components could predict patient clinical
outcomes. For instance, the stroma score was identified as a
predictor for survival in patients with gastric cancer (a lower
stroma score was indicative of better patient prognosis). Notably,
the ESTIMATE-derived stromal score was found to be more efficient
than the recently described stromal super-module-based method
(12). However, it should be
mentioned that the stroma might serve distinct roles in different
tissues. In the present study it was demonstrated that a higher
stroma score indicated later relapse in breast cancer, which is
consistent with a clustering-derived gene signature method
(8,10). The immune score was also found to
predict survival in basal breast cancer, in agreement with a
previous study that used the Profile Analysis using the Clustering
and Kurtosis method (9). Similarly, a
higher immune score was associated with a longer survival time in
melanoma and ovarian cancer, but earlier recurrence in
hepatocellular carcinoma-adjacent tissue. The degree of
tumour-infiltrating lymphocytes in particular activated CD8+
T-cells, within melanoma positively correlates with better
prognosis (26). The decreased
recruitment of tumor-infiltrating lymphocytes may lead to poor
prognosis in high-risk ovarian cancer patients (27). It has been previously demonstrated
that the poor prognostic signature in hepatocellular
carcinoma-adjacent tissue involved genes associated with
inflammation, including interferon signaling and activation of
nuclear factor-κB and tumor necrosis factor (28). These results indicated that non-tumor
liver tissue could serve a prognostic role in patients with
early-stage disease. These results may suggest the dual
host-protective and tumor-promoting roles of immune cells in
different tumor types (29,30).
The results of the present study indicated that
research into microenvironment-associated cells is warranted in
patients with cancer. Understanding the effect of the
microenvironment on drug sensitivity may improve the efficiency of
targeted therapies (31). For
instance, an association has been demonstrated between the
initiation of metastasis in CRC and the TGF-β stromal program
(7). Even though the ESTIMATE
algorithm is based on cancer tissue data, it was found to be
effective in assessing cellular data as well (Fig. 4). In the present study, the ESTIMATE
algorithm was used on cell line data, identifying a positive
correlation between 5-FU treatment response and the immune score
and indicating the potential mechanism of the drug (25). A lower immune score was observed in
p53-mutant CRC patients, indicating that 5-FU may not be the
optimal treatment choice for p53 mutant patients. Indeed, several
clinical studies have reported that CRC patients with wild-type p53
benefit from 5-FU-based chemotherapy, but those with mutant TP53 do
not (32,33). Thus, robust patient stratification
using microenvironment data may aid the development and application
of cancer therapies (34).
It is reasonable to apply the ESTIMATE scoring to a
specific tissue-based transcriptomic dataset, as the sampling
criteria are identical for every specimen within a study. To the
best of our knowledge, the present study is the first to
demonstrate that ESTIMATE scores are indicative of patient
survival, relapse, metastasis and chemotherapeutic drug resistance.
The two scores may have a prognostic value, indicating that stromal
and immune cells contribute to tumor clinical outcome. It was
further demonstrated that immune cells were associated with CRC
development and that the ESTIMATE scores may become useful
indicators of tissue-based patient prognosis.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81400617 and
81502091), the National Natural Science Foundation of Zhejiang
Provincial China (grant no. LQ14H030001) and the Ningbo Natural
Science Foundation (grant no. 2013A610232).
References
|
1
|
Klausner RD: The fabric of cancer cell
biology-Weaving together the strands. Cancer Cell. 1:3–10. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Navin N, Kendall J, Troge J, Andrews P,
Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et
al: Tumour evolution inferred by single-cell sequencing. Nature.
472:90–94. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bissell MJ and Hines WC: Why don't we get
more cancer? A proposed role of the microenvironment in restraining
cancer progression. Nat Med. 17:320–329. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu W, Li L and Li W: Gene co-expression
analysis identifies common modules related to prognosis and drug
resistance in cancer cell lines. Int J Cancer. 135:2795–2803. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yadav VK and De S: An assessment of
computational methods for estimating purity and clonality using
genomic data derived from heterogeneous tumor tissue samples. Brief
Bioinform. 16:232–241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yoshihara K, Shahmoradgoli M, Martinez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calon A, Espinet E, Palomo-Ponce S,
Tauriello DV, Iglesias M, Céspedes MV, Sevillano M, Nadal C, Jung
P, Zhang XH, et al: Dependency of colorectal cancer on a
TGF-β-driven program in stromal cells for metastasis initiation.
Cancer Cell. 22:571–584. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng Q, Chang JT, Gwin WR, Zhu J, Ambs S,
Geradts J and Lyerly HK: A signature of epithelial-mesenchymal
plasticity and stromal activation in primary tumor modulates late
recurrence in breast cancer independent of disease subtype. Breast
Cancer Res. 16:4072014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Teschendorff AE, Miremadi A, Pinder SE,
Ellis IO and Caldas C: An immune response gene expression module
identifies a good prognosis subtype in estrogen receptor negative
breast cancer. Genome Biol. 8:R1572007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Finak G, Bertos N, Pepin F, Sadekova S,
Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu
A, et al: Stromal gene expression predicts clinical outcome in
breast cancer. Nat Med. 14:518–527. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Isella C, Terrasi A, Bellomo SE, Petti C,
Galatola G, Muratore A, Mellano A, Senetta R, Cassenti A, Sonetto
C, et al: Stromal contribution to the colorectal cancer
transcriptome. Nat Genet. 47:312–319. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu Y, Grabsch H, Ivanova T, Tan IB, Murray
J, Ooi CH, Wright AI, West NP, Hutchins GG, Wu J, et al:
Comprehensive genomic meta-analysis identifies intra-tumoural
stroma as a predictor of survival in patients with gastric cancer.
Gut. 62:1100–1111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bianchini G, Qi Y, Alvarez RH, Iwamoto T,
Coutant C, Ibrahim NK, Valero V, Cristofanilli M, Green MC,
Radvanyi L, et al: Molecular anatomy of breast cancer stroma and
its prognostic value in estrogen receptor-positive and -negative
cancers. J Clin Oncol. 28:4316–4323. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calon A, Lonardo E, Berenguer-Llergo A,
Espinet E, Hernando-Momblona X, Iglesias M, Sevillano M,
Palomo-Ponce S, Tauriello DV, Byrom D, et al: Stromal gene
expression defines poor-prognosis subtypes in colorectal cancer.
Nat Genet. 47:320–329. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meyer PN, Fu K, Greiner T, Smith L,
Delabie J, Gascoyne R, Ott G, Rosenwald A, Braziel R, Campo E, et
al: The stromal cell marker SPARC predicts for survival in patients
with diffuse large B-cell lymphoma treated with rituximab. Am J
Clin Pathol. 135:54–61. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Goossens N, Hoshida Y and Aguirre-Ghiso
JA: Origin and interpretation of cancer transcriptome profiling:
The essential role of the stroma in determining prognosis and drug
resistance. EMBO Mol Med. 7:1385–1387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Quigley DA and Kristensen V: Predicting
prognosis and therapeutic response from interactions between
lymphocytes and tumor cells. Mol Oncol. 9:2054–2062. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rhodes DR and Chinnaiyan AM: Integrative
analysis of the cancer transcriptome. Nat Genet. 37 Suppl:S31–S37.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Team RC: R A language and environment for
statistical computing. Vienna Austria R Foundation for Statistical
Computing. http://www.R-project.orgFebruary 10–2015
|
|
21
|
Harrell FE Jr: rms: Regression Modeling
Strategies. R package version 4.1–3. http://CRAN.R-project.org/package=rms2014
|
|
22
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fehlker M, Huska MR, Jöns T,
Andrade-Navarro MA and Kemmner W: Concerted down-regulation of
immune-system related genes predicts metastasis in colorectal
carcinoma. BMC Cancer. 14:642014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Menendez D, Shatz M and Resnick MA:
Interactions between the tumor suppressor p53 and immune responses.
Curr Opin Oncol. 25:85–92. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bracci L, Schiavoni G, Sistigu A and
Belardelli F: Immune-based mechanisms of cytotoxic chemotherapy:
Implications for the design of novel and rationale-based combined
treatments against cancer. Cell Death Differ. 21:15–25. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tan LY, Martini C, Fridlender ZG, Bonder
CS, Brown MP and Ebert LM: Control of immune cell entry through the
tumour vasculature: A missing link in optimising melanoma
immunotherapy? Clin Trans Immunol. 6:e1342017. View Article : Google Scholar
|
|
27
|
Yoshihara K, Tsunoda T, Shigemizu D,
Fujiwara H, Hatae M, Fujiwara H, Masuzaki H, Katabuchi H, Kawakami
Y, Okamoto A, et al: High-risk ovarian cancer based on 126-gene
expression signature is uniquely characterized by downregulation of
antigen presentation pathway. Clin Cancer Res. 18:1374–1385. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hoshida Y, Villanueva A, Kobayashi M, Peix
J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, et
al: Gene expression in fixed tissues and outcome in hepatocellular
carcinoma. N Engl J Med. 359:1995–2004. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schreiber RD, Old LJ and Smyth MJ: Cancer
immunoediting: Integrating immunity's roles in cancer suppression
and promotion. Science. 331:1565–1570. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Linsley PS, Chaussabel D and Speake C: The
relationship of immune cell signatures to patient survival varies
within and between tumor types. PLoS One. 10:e01387262015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Donner A: Drug resistance: The stroma's
contribution. Nat Chem Biol. 8:7392012. View Article : Google Scholar
|
|
32
|
Russo A, Bazan V, Iacopetta B, Kerr D,
Soussi T and Gebbia N; TP53-CRC Collaborative Study Group, : The
TP53 colorectal cancer international collaborative study on the
prognostic and predictive significance of p53 mutation: Influence
of tumor site, type of mutation, and adjuvant treatment. J Clin
Oncol. 23:7518–7528. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Boyer J, McLean EG, Aroori S, Wilson P,
McCulla A, Carey PD, Longley DB and Johnston PG: Characterization
of p53 wild-type and null isogenic colorectal cancer cell lines
resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin
Cancer Res. 10:2158–2167. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cohen RL and Settleman J: From cancer
genomics to precision oncology-tissue's still an issue. Cell.
157:1509–1514. 2014. View Article : Google Scholar : PubMed/NCBI
|